Abstract
Environmental factors during juvenile growth such as temperature and nutrition have major effects on adult morphology and life-history traits. In Drosophila melanogaster, ovary size, measured as ovariole number, and body size, measured as thorax length, are developmentally plastic traits with respect to larval nutrition. Herein we investigated the genetic basis for plasticity of ovariole number and body size, as well the genetic basis for their allometric relationship using recombinant inbred lines (RILs) derived from a natural population in Winters, California. We reared 196 RILs in four yeast concentrations and measured ovariole number and body size. The genetic correlation between ovariole number and thorax length was positive, but the strength of this correlation decreased with increasing yeast concentration. Genetic variation and genotype-by-environment (G × E) interactions were observed for both traits. We identified quantitative trait loci (QTL), epistatic, QTL-by-environment, and epistatic-by-environment interactions for both traits and their scaling relationships. The results are discussed in the context of multivariate trait evolution.
IN general, life-history traits are very sensitive to the environment. Temperature, competition, predation, and nutrition can alter age and size at maturity, survival, and reproduction (Roff 2002). These life-history traits directly determine demographic fitness and, consequently, their response to the environment is predicted to be subject to natural selection (Via and Lande 1985). To accurately assess the evolutionary history and potential of life histories vis-à-vis the environment, the specific genetic basis of these environmentally sensitive traits must be understood. With an explicit genetic model in hand, functional and molecular genetics can be tied to population genetics and ecology. Such a synthesis will lead to a deeper understanding of evolutionary processes.
Recently, there has been considerable progress in unraveling the molecular genetic basis of life-history plasticity. For example, a genetic basis of environmentally influenced, age-dependent survivorship in a variety of animals (Panowski et al. 2007) and fungi (Bitterman et al. 2003) has been described. Remarkably, some of these genetic pathways are highly conserved (Barbieri et al. 2003). However, much less attention has been paid to the genetic basis of environment-dependent reproduction (Yang et al. 2008) and in particular the role of the preadult environment on adult reproductive capacity.
The quality and quantity of nutrition during embryonic and preadult stages affect adult reproductive capacity in a variety of animals (Rae et al. 2001, 2002; Rhind 2004; Guzmán et al. 2006; Hodin 2008). These effects are often mediated by the morphology and the size of reproductive organs, especially in females. In Drosophila melanogaster, larvae reared on food that varies in yeast concentration differ considerably in total and age-specific fecundity (Tu and Tatar 2003). Adult body size and ovary size, measured as ovariole number, are also modified by larval nutrition such that flies reared on food with less yeast are smaller and have fewer ovarioles than those reared with more yeast (Hodin and Riddiford 2000; Tu and Tatar 2003). The plastic response of body size and ovariole number could functionally underlie reductions in fecundity and thus be subject to natural selection. Variation in adult body size may affect fecundity via effects on adult nutrient acquisition or mating success (Sisodia and Singh 2004). Ovariole number may affect fecundity because ovarioles are the functional units of the ovary. At the tip of each ovariole resides a set of germline stem cells that differentiate into eggs. Eggs can be produced simultaneously in all ovarioles, and thus ovariole number sets an upper limit on fecundity (David 1970).
In addition to the plastic responses of body size and ovariole number, genetic variation in these traits has long been thought to be under natural selection because of their correlation with fecundity (Honek 1993). In D. melanogaster, interpopulation variation in ovariole number is correlated with differences in fecundity (Boulétreau-Merle et al. 1982, but see also Schmidt et al. 2005), and intrapopulation variation is correlated with maximum fecundity (David 1970), but not necessarily total fitness (Wayne et al. 1997). Artificial selection on ovariole number elicits a correlated response in fecundity; among selection lines there is a positive correlation between ovariole number and fecundity (Robertson 1957; Engstrom 1971). Thorax length, ovariole number, and fecundity are also positively correlated among various Drosophilid species (Kambysellis and Heed 1971; R'kha et al. 1997).
Attempts to describe the evolution of either body size or ovariole number in relation to fecundity are complicated by the allometry between these characters across environments, genotypes, or species. One approach to studying the evolution of correlated characters is to measure their variance–covariance matrix (G, Lande and Arnold 1983). This is a powerful approach to predicting short-term evolution, but it does not describe the specific genetic basis of correlated traits, without which we are limited in predicting evolutionary processes. Using G as a predictive tool without knowledge of the causative genetics may lead to a misinterpretation of the underlying developmental and physiological mechanisms that coregulate traits (Houle 1991; Gromko 1995; Pigliucci 2006). A complementary approach is to identify the genetic basis of the traits through quantitative trait locus (QTL) mapping. This approach identifies pleiotropic and plastic loci and may eventually lead to the identification of specific genes underlying quantitative variation (Mackay 2001).
Previous attempts to map pleiotropic and plastic QTL have been successful in many organisms. For example, in Arabidopsis thaliana QTL controlling various aspects of flower morphology under variable photic environments have been identified (Ungerer et al. 2003). In Caenorhabditis elegans, plastic responses of life-history traits across temperatures have been mapped (Gutteling et al. 2007). And in D. melanogaster fitness-related traits have been mapped across multiple larval and adult environments (Fry et al. 1998; Gurganus et al. 1998; Leips and Mackay 2000; Vieira et al. 2000). These studies have all revealed QTL that are both pleiotropic and nonpleiotropic as well as QTL that vary in effect across environments (i.e., plastic) and loci with fixed effects across environments. QTL mapping studies, however, are unable to resolve the classic distinction (Via et al. 1995) between loci that vary in direct response to the environment (the so-called allelic sensitivity model) and loci that modulate the response of other genes in an environment-dependent fashion (the gene regulation model; but see Leips and Mackay 2000). Such distinctions can be made only when the molecular basis of plasticity for a particular trait is understood.
In this study, we investigate the genetic basis of variation of ovariole number and body size plasticity and allometry as a first step in describing the functional genetics and evolution of these traits at a molecular level. While there is considerable information about the molecular and quantitative genetic basis for adult body size (e.g., Wayne et al. 1997, 2001; Leevers and Hafen 2003; Oldham and Hafen 2003; Hafen 2004; Caldwell et al. 2005; Colombani et al. 2005; Mirth et al. 2005) and ovariole number (Coyne et al. 1991; Wayne et al. 1997, 2001; Hodin and Riddiford 1998; Wayne and Mackay 1998; Wayne and McIntyre 2002; Telonis-Scott et al. 2005; Orgogozo et al. 2006) in D. melanogaster and related species, little is known about the genetic basis for nutrient-induced phenotypic plasticity in these traits.
We used QTL mapping to describe the genetic architecture of phenotypic plasticity in ovariole number and thorax length within a population of recombinant inbred lines (RILs). We address the following questions. First, what are the genomic positions and environment-specific effects of QTL and epistatic interactions for ovariole number and thorax length within our mapping population? Second, how many QTL and epistatic interactions are shared between ovariole number and thorax length? And third, what are the genomic positions and environment-specific effects of QTL and epistatic interactions that affect the allometric relationship between ovariole number and thorax length?
To address these questions, we reared a large panel of RILs segregating naturally occurring alleles under controlled density in four larval yeast environments. We document genetic and genotype–environment variation for both traits within our mapping population. We identify QTL and epistatic interactions that underlie these sources of variation for both traits. QTL and epistatic interactions for ovariole number and thorax length are largely independent. Further, we identify QTL and epistatic interactions that affect the allometric relationship between ovariole number and thorax length in an environment-dependent fashion. We discuss these findings in relation to the multivariate evolution of life-history plasticity.
MATERIALS AND METHODS
Fly stocks and genetic map:
We used a panel of 196 advanced intercross RILs randomly drawn from a larger population of 300 RILs (A. Genissel and S. V. Nuzhdin, unpublished data). These RILs were derived from two wild females (lines 89 and 58 whose alleles are hereafter referred to as AA and aa, respectively) caught in an orchard population in Winters, California (38°N, 121°W) during 2001. These 2 lines were isogenized by 40 generations of inbreeding. These parental lines were expanded to a set of 500 isogenic lines and these offspring lines were randomly intermated for 15 generations. Each intermated line was sib-crossed for 15 generations to make the final set of RILs. Prior to the initiation of the present experiment, RILs were kept at 25°, 12 hr light:12 hr dark (12 L:12 D), in low culture density on ∼2% yeast-by-volume fly medium, with live yeast sprinkled on top. These lines were SNP genotyped at 31, 34, and 37 intronic and intergenic markers, respectively, along the X, second, and third chromosomes, using a multiplex oligoligation assay (A. Genissel and S. V. Nuzhdin, unpublished data). Extensive map expansion was observed, relative to the standard Drosophila recombination map (Lindsey and Zimm 1992). The cumulative map length in our population is ∼4000 cM, when using the Kosambi map conversion function, which gives our analysis a high degree of precision in mapping QTL.
Rearing conditions:
Larvae were reared in four food treatments that contained 0.2, 0.4, 0.6, and 0.8% autoclaved yeast (Lasaffre Yeast, product no. 73050) by volume. Sugar, cornmeal, agar, and tegosept concentrations (11, 8, 5, and 1% by volume, respectively) were kept constant across all treatments. Each rearing vial contained 10 ml of medium. Experimental food was used within 4 days of being made.
Parental lines and RILs were assayed in three replicate blocks, each block representing a successive generation. For each line, ∼50–100 mixed-sex adults were placed into small cages with petri dishes containing apple juice–agar medium as oviposition substrate with ∼0.5 ml yeast paste, made from autoclaved yeast and water, on each petri dish to stimulate oviposition. Adults oviposited for 12–24 hr prior to egg collection. Fifty eggs from each line were transferred to a larval rearing vial of each food treatment; care was taken not to transfer any yeast paste. Rearing vials were maintained at 25°, 12 L:12 D until preservation.
Adults emerging from rearing vials were transferred to vials with 2% autoclaved yeast by volume (11% sugar, 8% cornmeal, 5% agar, and 1% tegosept) plus live yeast and were left in these vials for 3–4 days. This treatment does not affect ovariole number but induces vitellogenesis, making ovariole counts more reliable. Flies were thereafter transferred to cryovials and frozen at −80°.
Phenotyping:
Up to five females (average four) were phenotyped per genotype per treatment per replicate. Mesothorax length (the distance from the tip of the scutellum to the most anterior part of the mesothorax) was measured with an occular micrometer accurate to 0.033 mm. Flies were dissected to score ovariole number for each ovary.
Statistical analyses:
Variance components:
We used two approaches to examine the differences in phenotypic plasticity of ovariole number and thorax length among RILs. The first approach estimates the proportion of genetic variation within each environment and the second approach estimates the extent of genotype–environment variation. By using these two approaches, we are able to assess whether the genotype–environment variation measured by the second approach is due to changes in the magnitude of genetic variation or changes in the rank order of genotypes across environments. In the first approach, we fit the following model for each food treatment and trait separately: y = μ + G + B + G:B + error. In the second approach, we used the following model: y = μ +E + G + B + G:E + E:B + G:B + error. In both approaches, y refers to either ovariole number or thorax length of individual flies, G is the random line effect, B is the random block effect, and G:B is their random interaction. In the second approach, E is the fixed food effect, and G:E and E:B are the random interaction effects. We also assessed the extent of genotype–environment variation among the parental lines using the second approach.
Mixed-effect models were calculated in SAS 9.13 using the PROC MIXED function (SAS Software 2002).
Genetic correlations:
Correlations were calculated between traits within environments and within traits across environments. The correlation between any two pairs was calculated as covij/σiσj, where covij is the covariance of the line means, σi and σj are the square roots of the among-line variances, and i and j represent different traits or environments depending upon the comparison. Ninety-five percent confidence ellipses were calculated using the car package (Fox 2008).
QTL analysis:
Single-marker QTL analysis was performed using multiple imputation (Sen and Churchill 2001) implemented in the R/qtl package (Broman et al. 2003), using R 2.4.1 (R Development Core Team 2006). We used 50 imputations with a step size of 3 cM. QTL models were fit using the within-environment mean phenotype for each RIL. For both phenotypes we used two strategies of QTL analysis. The first approach mapped QTL affecting each phenotype within each environment. The second considered the food treatment and mapped QTL using the null model, y = μ + E + error, the reduced model, y = μ + Mi + E + error, and the full model, y = μ + Mi + E + Mi:E + error, where y is the environment-specific genotype mean for either phenotype, μ is the grand mean, Mi is the effect of the ith QTL, E is the food effect coded as a contrast matrix against the 0.2% yeast treatment, Mi:E is the interaction between the ith QTL and the food effect, and error is the normally distributed residual error. We compared models by taking the differences in the log-likelihood odds (LOD score) at each marker or imputed marker. The difference in LOD scores between the reduced and null models is used to infer QTL that have main effects, averaged across environments. The difference in LOD scores between the full and reduced models represents the contribution of the Mi:E interaction term, and allows us to distinguish QTL that vary their effect across environments from those that have consistent effects across environments.
We tested for QTL with main and environment-specific effects on ovariole number after removing the additive effects of thorax length. To test for main effects, we compared the full model y = μ + Mi + E + T + error to the reduced model, y = μ + E + T + error, where y is ovariole number, and T is environment-corrected thorax length (i.e., the residuals of the relationship between thorax length and environment), thereby removing the colinearity between E and T. To test for environment-specific effects we compared the full model, y = μ + Mi + E + T + Mi:E + error to the reduced model, y = μ + Mi + E + T + error. These models fix the slope of the relationship between ovariole number and thorax length, but allow the intercept of the relationship between ovariole number and thorax length to vary.
To test for QTL affecting the relationship between ovariole number and thorax length within and across environments, we compared the full model, y = μ + Mi + E + T + E:T + Mi:E + Mi:E:T + error, to the reduced model, y = μ + Mi + E + T + E:T + Mi:E + error. These models allow both the slope and the intercept of the relationship between ovariole number and thorax length to vary.
Epistatic QTL analysis:
We used the multiple-imputation method to map epistatic and epistatic-by-environment QTL for ovariole number and thorax length. We also tested for epistatic-by-environment interactions for ovariole number after removing the additive effects of thorax length and epistatic-by-environment interactions affecting the relationship between ovariole number and thorax length. To account for missing genotype data, we used 50 imputations; however, we imputed data only at the empirical markers and not at pseudomarkers. Locations of maximum LOD were later refined to pseudomarkers (see below).
To identify epistatic interactions for ovariole number and thorax length, we calculated the difference in LOD scores between the full model, y = μ + E + Mi + Mj + Mi:Mj + error, and the reduced model, y = μ + E +Mi + Mj + error, where Mi and Mj are the QTL being tested. To identify epistatic-by-environment interactions for ovariole number and thorax length, we calculated the difference in LOD scores between the full model, y = μ + E + Mi + Mj + E:Mi + E:Mj + Mi:Mj + E:Mi:Mj + error, and the reduced model, y = μ + E + Mi + Mj + E:Mi + E:Mj + Mi:Mj + error.
To identify epistatic and epistatic-by-environment interactions for ovariole number after removing the additive effects of thorax length, we calculated the difference in LOD scores between the full and reduced models, as above, except that environment-corrected thorax length (T) was included as an additive covariate. Likewise, we tested for epistatic-by-thorax length and epistatic-by-environment-by-thorax length interactions, by comparing full and reduced models with thorax length as an additive and interactive covariate.
Once an initial set of epistatic QTL was identified, we refined their location using repeated calls to the fitqtl function. We scanned ±30 cM with respect to each identified marker per epistatic pair at a step size of 3 cM with 50 imputations. This procedure allowed us to localize positions of maximum LOD (when they occurred at pseudomarkers between observed markers) as well as to obtain 2-LOD intervals per epistatic QTL.
In several cases, epistatic interactions had high LOD scores due to heteroscedasticity, caused by unequal sample sizes. We discarded any of these epistatic interactions when they had <10 lines representing any one genotype.
Statistical thresholds for QTL and epistatic interaction:
Statistical thresholds for all QTL and epistatic interactions were defined by the GWERk statistic (Chen and Storey 2006). Briefly, the GWERk statistic is a LOD value above which there is a probability (α) of k false positives. We used k = 1 and α = 0.05 for most thresholds. The only exception was main-effect QTL for ovariole number and thorax length where we used k = 0 because k = 1 was too permissive (the LOD value of GWER0 was ∼1.6 for either model). The use of slightly more liberal thresholds (i.e., k = 1) is appropriate because all QTL and epistatic interactions were further subjected to model selection (see below).
To derive the GWERk statistic, we performed 1500 permutations of the phenotypes across environments and RIL genotype for each QTL and epistatic model. For each permutation, we recorded the LOD scores of the highest and second-highest peaks. Because different LOD peaks on the same chromosome might actually reflect the same underlying causative locus, we defined the second-highest peak as the highest peak not on the chromosome (or chromosomes, in the case of epistasis) previously identified for the highest LOD peak (Chen and Storey 2006). The 1 − α quantile of the distribution of highest LOD peaks corresponds to the GWER0 threshold and the 1 − α quantile of the distribution of second-highest LOD peaks corresponds to the GWER1 threshold. The GWERk thresholds are provided in Table 1.
TABLE 1.
Statistical thresholds for specific model terms
| Trait | Model term | k | GWERk |
|---|---|---|---|
| Ovariole no. | Mi | 0 | 3.16 |
| Mi:E | 1 | 2.73 | |
| Mi + T | 1 | 3.10 | |
| Mi:E + T | 1 | 2.78 | |
| Mi:E:T | 1 | 2.87 | |
| Mi:Mj | 1 | 2.43 | |
| Mi:Mj:E | 1 | 4.66 | |
| Mi:Mj:E + T | 1 | 4.17 | |
| Mi:Mj:E:T | 1 | 4.46 | |
| Thorax length | Mi | 0 | 3.11 |
| Mi:E | 1 | 2.76 | |
| Mi:Mj | 1 | 3.28 | |
| Mi:Mj:E | 1 | 4.67 |
QTL model selection:
We used a robust model selection strategy to identify a set of QTL and epistatic interactions that most parsimoniously explain the observed phenotypic distribution of RIL means. We defined the model search space by the QTL and epistatic interactions that exceeded the GWERk at the α = 0.05 threshold. Each QTL or epistatic interaction was defined as an independent term and we fit every possible additive model of these independent terms. For example, if we identified one QTL and one epistatic interaction (e.g., QTLA and the epistatic interaction QTLB × QTLC), there would be four possible models, including the null model (i.e., no QTL or epistatic interaction), that contain the additive effects of QTLA and QTLB × QTLC. Note that interactions between independent terms were not tested (e.g., QTLA × QTLB × QTLC). In general, the number of additive models = 2 raised to the number of independent QTL and epistatic interactions identified. For ovariole number we identified 15 QTL and epistatic interactions and for thorax length we identified 12 QTL and epistatic interactions. Thus, we fit 215 models for ovariole number and 212 models for thorax length. (See Tables 6 and 7 for detailed information on QTL and epistatic terms tested.) For both ovariole number and thorax length, each model contained yeast level, coded as a contrast matrix against the 0.2% treatment. For ovariole number, each model contained environment-corrected thorax length.
TABLE 6.
Positions of putative QTL and epistatic interactions affecting ovariole number
| QTL locationa
|
Conditional LOD
|
Relative importancec
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr. | LOD peak | 2-LOD interval | QTL model termb | Raw LOD | mBIC | BIC | mBIC | BIC | |
| Main-effect QTL | 2R | 13.42 | 13.41–13.43, | Mi | 4.15 | 3.61 | 1.76 | 0.9535 | 0.9629 |
| 55A1 | 55A1–55A1 | ||||||||
| X | 5.98 | 4.14–13.26 | Mi:E + T | 3.27 | 2.37 | 3.59 | 0.8875 | 0.9952 | |
| 5D5 | 4B5–11F6 | ||||||||
| 3L | 17.30 | 15.58–21.85 | Mi + T | 4.32 | — | — | 0.0779 | 0.4157 | |
| 93E2 | 92B1–96F10 | ||||||||
| 3R | 6.56 | 0.00–7.70 | Mi + T | 6.23 | — | 0.82 | 0.2814 | 0.7429 | |
| 65B5 | 61A5–66A11 | ||||||||
| X | 11.90 | 9.26–12.63 | Mi:E:T | 3.39 | 5.88 | 4.45 | 0.9941 | 0.9859 | |
| 11A3 | 8E1–11C2 | ||||||||
| 2L | 7.83 | 6.53–9.09 | Mi:E:T | 3.30 | 7.43 | 5.20 | 0.9992 | 0.9980 | |
| 28D2 | 26E1–30A1 | ||||||||
| 2R | 17.86 | 17.52–19.57 | Mi:E:T | 3.26 | 9.07 | 7.54 | 0.9999 | 0.9999 | |
| 58D4 | 58B1–60B7 | ||||||||
| Epistatic interactions | X | 6.39 | 5.78–7.94 | Mi:Mj | 4.22 | — | 1.98 | 0.0012 | 0.9112 |
| 6B2 | 5C9–7D6 | ||||||||
| 2R | 17.18 | 16.30–18.55 | |||||||
| 57F8 | 57B2–59C2 | ||||||||
| X | 21.74 | 20.77–21.74 | Mi:Mj | 6.13 | — | 3.45 | 0.2717 | 0.9980 | |
| 20C1 | 19E5–20C1 | ||||||||
| 3L | 1.66 | 1.41–1.78 | |||||||
| 62A10 | 62A1–62B3 | ||||||||
| 2L | 2.17 | 0.89–2.63 | Mi:Mj | 3.91 | 8.10 | 6.41 | 0.7691 | 0.9999 | |
| 22D1 | 21E2–23A1 | ||||||||
| 3R | 15.90 | 15.46–16.47 | |||||||
| 92C5 | 92A11–92F4 | ||||||||
| 2L | 2.17 | 0.48–3.09 | Mi:Mj | 3.65 | — | 3.68 | 0.0948 | 0.9981 | |
| 22D1 | 21D1–23D1 | ||||||||
| 3R | 27.61 | 27.43–27.61 | |||||||
| 100D2 | 100C7–100D2 | ||||||||
| 2L | 6.13 | 5.71–6.63 | Mi:Mj | 3.86 | — | 3.54 | 0.0823 | 0.9972 | |
| 26B7 | 25F3–26F2 | ||||||||
| 2R | 15.77 | 13.95–16.30 | |||||||
| 56F11 | 55C9–57B2 | ||||||||
| 2L | 15.23 | 15.10–17.55 | Mi:Mj:E + T | 4.52 | 13.20 | 7.88 | 0.9972 | 0.9999 | |
| 35C4 | 35C2–36D1 | ||||||||
| 3R | 1.23 | 0–8.55 | |||||||
| 83A2 | 81F5–87C7 | ||||||||
| X | 12.84 | 12.69–13.37 | Mi:Mj:E:T | 5.42 | 15.52 | 13.26 | 0.9999 | 0.9999 | |
| 11D6 | 11D1–12A4 | ||||||||
| 2L | 3.57 | 0–3.65 | |||||||
| 24A1 | 21A5–24A4 | ||||||||
| 2L | 15.07 | 14.99–15.10 | Mi:Mj:E:T | 7.54 | 13.38 | 12.82 | 0.9999 | 0.9999 | |
| 35C1 | 35B8–35C2 | ||||||||
| 3L | 12.19 | 11.86–12.20 | |||||||
| 69A3 | 68E3–69A3 | ||||||||
In megabases and cytological band, relative to release 4.3 of the D. melanogaster genome.
See text for description of models.
Italic values represent terms retained in the best mBIC and BIC models.
TABLE 7.
Positions of putative QTL and epistatic interactions affecting thorax length
| QTL locationa
|
Conditional LOD
|
Relative importancec
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr. | LOD peak | 2-LOD interval | QTL model termb | Raw LOD | mBIC | BIC | mBIC | BIC | |
| Main-effect QTL | 3L | 12.17 | 11.86–12.20 | Mi | 5.92 | 2.16 | — | 0.7913 | 0.4453 |
| 69A2 | 68E3–69A3 | ||||||||
| 3L | 16.37 | 14.25–18.01 | Mi | 4.29 | 2.70 | 0.93 | 0.5845 | 0.7665 | |
| 72F1 | 70D4–75B5 | ||||||||
| 3R | 1.23 | 0–6.94 | Mi | 5.62 | — | — | 0.4380 | 0.4385 | |
| 83A2 | 81F5–86D4 | ||||||||
| 2R | 6.95 | 6.73–7.77 | Mi:E | 2.92 | 4.38 | 3.35 | 0.9974 | 0.9908 | |
| 27C6 | 47D8–48F1 | ||||||||
| 3L | 13.38 | 13.35–13.39 | Mi:E | 2.86 | 2.79 | 2.74 | 0.9060 | 0.9664 | |
| 70A8 | 70A7–70A8 | ||||||||
| Epistatic interactions | X | 8.17 | 7.94–9.29 | Mi:Mj | 5.34 | — | 3.15 | 0.3940 | 0.9936 |
| 7E2 | 7D6–8E1 | ||||||||
| X | 17.48 | 16.14–17.81 | |||||||
| 16B10 | 14B9–16F4 | ||||||||
| X | 21.74 | 19.33–21.74 | Mi:Mj | 4.18 | 6.52 | 2.94 | 0.5453 | 0.9905 | |
| 20C1 | 18C8–20C1 | ||||||||
| 2L | 2.17 | 0.48–3.09 | |||||||
| 22D1 | 21D1–23D1 | ||||||||
| X | 21.74 | 19.56–21.74 | Mi:Mj | 3.80 | — | 2.99 | 0.0483 | 0.9987 | |
| 20B3 | 18E3–20C1 | ||||||||
| 3L | 1.66 | 1.05–1.78 | |||||||
| 62A10 | 61E3–62B4 | ||||||||
| 2L | 3.09 | 2.63–5.30 | Mi:Mj | 4.69 | — | 4.78 | 0.0061 | 0.9992 | |
| 23D1 | 23A1–25D4 | ||||||||
| 3L | 5.88 | 5.00–6.02 | |||||||
| 64F5 | 64C4–65A3 | ||||||||
| 2R | 9.66 | 9.29–10.27 | Mi:Mj | 3.97 | — | 3.84 | 0.0031 | 0.9980 | |
| 50E1 | 50C3–51C2 | ||||||||
| 3L | 1.05 | 1.05–1.78 | |||||||
| 61E3 | 61E3–62B4 | ||||||||
| 3L | 14.26 | 14.21–16.37 | Mi:Mj | 3.37 | — | 3.82 | 0.0083 | 0.9971 | |
| 70D5 | 70D4–72E5 | ||||||||
| 3L | 21.47 | 20.34–21.47 | |||||||
| 78D5 | 77C1–78D3 | ||||||||
| X | 5.57 | 4.14–6.19 | Mi:Mj:E | 4.41 | 7.12 | 7.70 | 0.8891 | 0.9994 | |
| 5B2 | 4B5–5F2 | ||||||||
| X | 13.05 | 12.74–13.16 | |||||||
| 11E3 | 11D1–11E11 | ||||||||
In megabases and cytological band, relative to release 4.3 of the D. melanogaster genome.
See text for description of models.
Italic values represent terms retained in the best mBIC and BIC models.
Models were fit using the fitqtl function in R/qtl. Likelihood-ratio statistics were extracted and Bayesian information criterion (BIC) (Schwartz 1978) and modified (m)BIC (Bogdan et al. 2004) statistics were computed for each model. These model selection criterions penalize the log-likelihood statistic for the number of parameters, thereby correcting for model over fitting. In simulation studies, model selection based on BIC tends to overestimate the number of QTL (Broman and Speed 2002), and mBIC is designed to correct this especially when there is epistasis (Bogdan et al. 2004). We choose to calculate both BIC and mBIC because it is unclear how either method performs when there are QTL-by-environment, epistatic-by-environment, or higher-order interactions present. BIC statistics were calculated as
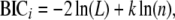 |
where BICi is the BIC statistic for the ith model, L is log-likelihood of the ith model, k is the number of parameters in the ith model, and n is the sample size (in all cases, 745). mBIC statistics were calculated as
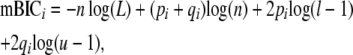 |
where mBICi is the mBIC statistic for the ith model, pi is the number of main-effect QTL terms in the ith model, qi is the number of epistatic terms in the ith model, l is the number of possible main-effect QTL (102), and u is the number of possible epistatic terms (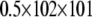 ). The model with the lowest penalized log-likelihood ratio, for each penalization method, was considered the best model for that method. The relative probability of a model was calculated as
). The model with the lowest penalized log-likelihood ratio, for each penalization method, was considered the best model for that method. The relative probability of a model was calculated as
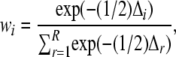 |
where Δi is the difference in penalized log-likelihood between the ith model and the best model, and R is the full set of models (Burnham and Anderson 2002). When w for the best model is small, we interpret that there are several nearly equivalent models. Conversely, when w for the best model is close to unity, we interpret that the best model is unambiguously the best model. We calculated the relative importance of each QTL or interaction as the sum of w for each model that a term appeared in. A relative importance close to unity is interpreted as highly supported, and one close to zero is interpreted as unsupported (Burnham and Anderson 2002).
We calculated the LOD scores of the model terms in the best mBIC and BIC models, conditional on that model. The conditional LOD scores were calculated as the difference between the best model and that model without the particular term.
Effects and genetic means:
We calculated the effects of the terms in the best mBIC and BIC models, conditional on that model, using the fitqtl function. These estimated effects are assumed to be approximately orthogonal to each other (Sen and Churchill 2001). Genetic means across all terms in the model were calculated by multiplying the estimated effects by the model matrix (Neter et al. 1985). Residuals were calculated by subtracting observed from fitted values. For each term, errors were subsequently calculated as the fitted values per effect plus the residuals.
RESULTS
Variance components:
A significant G × E interaction was found between the parental strains for ovariole number (F = 21.463,87, P < 0.001) but not for thorax length (F = 2.333,87, P = 0.08). The extent of G × E between the parental lines for ovariole number and thorax length is immediately apparent when comparing their reaction norms (Figure 1, A and C).
Figure 1.—
Reaction norms of ovariole number and thorax length to yeast concentration. (A and C) Squares and diamonds represent parental lines 89 and 58, respectively, ±95% C.I., and circles represent the mean of the RILs for ovariole number (A) and thorax length (C). (B and D) Each line represents the RIL reaction norm for ovariole number (B) and thorax length (D).
Within each environment, there was significant genetic variation for ovariole number and thorax length among the RILs (Tables 2 and 3). G × E was statistically significant for both ovariole number and thorax length (Table 4), indicating genetic variation for the plastic responses of both traits (Figure 1, B and D).
TABLE 2.
Mixed-effect models for ovariole number within each environment
| 0.2% YBVa
|
0.4% YBVa
|
0.6% YBVa
|
0.8% YBVa
|
|||||
|---|---|---|---|---|---|---|---|---|
| Termb | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d |
| G | 83.67 | 4.48 | 118 | 3.13 | 148.31 | 4.21 | 127.56 | 4.10 |
| F = 1.61190, 220 | (2.25–10.25) | F = 1.31193, 252 | (1.46–10.69) | F = 1.50190, 220 | (2.12–12.06) | F = 1.43193, 254 | (2.17–10.45) | |
| B | 254 | 0.25 | 22.45 | 0 | 1165.55 | 2.55 | 1056.01 | 2.01 |
| F = 5.622, 241 | (0.045–928) | F = 0.262, 268 | (0–0) | F = 1.612, 220 | (0.65–161.4) | F = 13.222, 220 | (0.51–132.95) | |
| G:B | 56.49 | 13.50 | 93.47 | 17.37 | 102.78 | 20.41 | 91.77 | 16.27 |
| F = 4.68210, 1126 | (10.66–17.67) | F = 5.78248, 1714 | (14.14–21.86) | F = 7.01247, 1660 | (16.2–25.46) | F = 4.40250, 1706 | (13.08–20.80) | |
| Error | 12.07 | 12.19 | 16.18 | 16.33 | 14.66 | 14.75 | 20.87 | 21.06 |
| (11.24–13.27) | (15.25–17.49) | (13.79–15.81) | (19.71–22.57) | |||||
Percentage of yeast by volume (YBV) in the larval rearing medium.
G, genotype; B, block; G:B, genotype-by-block.
Values represent type III and F values.
Values represent variance components and confidence intervals.
TABLE 3.
Mixed models for thorax length within each environment
| 0.2% YBVa
|
0.4% YBVa
|
0.6% YBVa
|
0.8% YBVa
|
|||||
|---|---|---|---|---|---|---|---|---|
| Termb | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d | Mean squarec | Var. comp.d |
| G | 261.6 | 20.62 | 198.13 | 6.69 | 163.85 | 5.63 | 188.73 | 8.75 |
| F = 2.05190, 222 | (13.30–36.22) | F = 1.45193, 252 | (3.68–15.72) | F = 1.45190, 251 | (3.09–13.36) | F = 1.76193, 253 | (5.71–15.09) | |
| B | 491.8 | 0.75 | 1221 | 3.13 | 3610 | 3.19 | 570.66 | 1.04 |
| F = 4.382, 247 | (0.14–2433) | F = 10.142, 272 | (0.80–20.42) | F = 18.142, 270 | (0.83–18.00) | F = 5.942, 272 | (0.25–12.41) | |
| G:B | 138.11 | 33.12 | 142.24 | 24.56 | 116.41 | 20.10 | 110.50 | 18.38 |
| F = 3.87210, 1126 | (25.54–44.66) | F = 4.95248, 1714 | (19.84–31.20) | F = 4.78247, 1660 | (16.20–25.60) | F = 4.59250, 1706 | (14.86–23.31) | |
| Error | 35.69 | 36.38 | 28.74 | 28.92 | 24.33 | 24.58 | 24.08 | 24.11 |
| (33.51–36.64) | (27.07–30.96) | (22.99–26.36) | (22.57–25.81) | |||||
Percentage of yeast by volume (YBV) in the larval rearing medium.
G, genotype; B, block; G:B, genotype-by-block.
Values represent type III mean squares ×104 and F values.
Values represent variance components and confidence intervals ×104.
TABLE 4.
Mixed-model results across environments for RILs
| Ovariole no.
|
Thorax length
|
|||
|---|---|---|---|---|
| Terma | Mean squareb | Variance componentc | Mean squareb | Variance componentcd |
| E | 18,221 | — | 6.51 | — |
| F = 42.483, 7.39 | F = 64.333, 6.88 | |||
| G | 217.8 | 2.32 | 0.0396 | 5.05 |
| F = 1.47195, 562 | (1.31–5.17) | F = 1.61195, 553 | (3.02–10.11) | |
| B | 707.2 | 0.32 | 0.1189 | 0.47 |
| F = 1.832, 8.46 | (0.05–55381) | F = 1.322, 7.68 | (0.05–36369) | |
| G:E | 82.2 | 6.21 | 0.0130 | 10.57 |
| F = 3.7557, 6783 | (5.24–7.47) | F = 3.75557, 6783 | (8.95–12.68) | |
| E:B | 462 | 0.88 | 0.1130 | 2.12 |
| F = 20.816, 6783 | (0.35–4.82) | F = 32.636, 6783 | (0.86–10.99) | |
| G:B | 109 | 6.22 | 0.0185 | 11.84 |
| F = 4.91300, 6783 | (5.07–7.81) | F = 5.33300, 6783 | (9.72–14.74) | |
| Error | — | 22.56 | — | 35.06 |
| (21.82–23.35) | (33.91–36.28) | |||
E, environment; G, genotype; B, block; G:E, genotype-by-environment; E:B, environment-by-block; G:B, genotype-by-block.
Values represent type III mean squares and F values.
Values represent variance components, and values in parentheses represent 95% confidence intervals.
Variance components and confidence intervals ×104.
Genetic correlations:
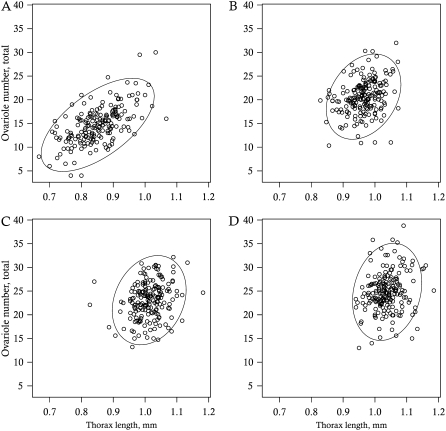
Correlations within traits, between environments, decreased with increasing differences in yeast level (Table 5). These ranges of correlations indicate a moderate amount of crossing reaction norms (Roff 1997). The correlations within environments, between traits, decreased monotonically with increasing yeast level (Table 5, Figure 2) and were significantly different from zero in all environments (nominal P < 2e−16, P = 2.5e−5, P = 0.0013, and P = 0.0089 for 0.2, 0.4, 0.6, and 0.8% yeast by volume, respectively). A significant genetic correlation between ovariole number and thorax length within populations has not been previously reported (Wayne et al. 1997; Telonis-Scott et al. 2005), where measurements were made on flies reared under high yeast concentrations.
TABLE 5.
Genetic correlation matrix
| 0.2% YBVa | 0.4% YBV | 0.6% YBV | 0.8% YBV | |
|---|---|---|---|---|
| 0.2% YBV | 0.59b | 0.44 | 0.45 | 0.13 |
| 0.4% YBV | 0.45 | 0.28 | 0.32 | 0.29 |
| 0.6% YBV | 0.33 | 0.43 | 0.27 | 0.26 |
| 0.8% YBV | 0.25 | 0.34 | 0.38 | 0.19 |
Percentage of yeast by volume (YBV) in the larval rearing medium.
Values above and below the diagonal represent the cross-environmental genetic correlation for ovariole number and thorax length, respectively. Values along the diagonal represent the genetic correlation between ovariole number and thorax length within environments.
Figure 2.—
Correlation between ovariole number and thorax length at 0.2% (A), 0.4% (B), 0.6% (C), and 0.8% (D) yeast by volume. Circles represent RIL means, ovals represent 95% confidence intervals.
QTL analysis:
QTL analysis for ovariole number and thorax length identified the same QTL within any given environment and when phenotypes were concatenated across environments with environment used as an additive covariate. However, LOD scores varied for particular QTL across environments (supplemental Figures 1 and 2) and were reduced when environment was used as an additive covariate. These affects on LOD scores may be attributed to limits in statistical power (e.g., for ovariole number the power to detect a QTL with effect size of 1, at a nominal α = 0.01, within any given environment is 0.21 compared to 0.83 when pooled across environments, given an empirical standard deviation of ∼3.8 and assuming equal allele frequencies). Given the low power of QTL detection with our mapping population within each environment and low LOD scores within any given environment, variation in LOD scores for particular QTL across environments cannot resolve QTL × E interactions.
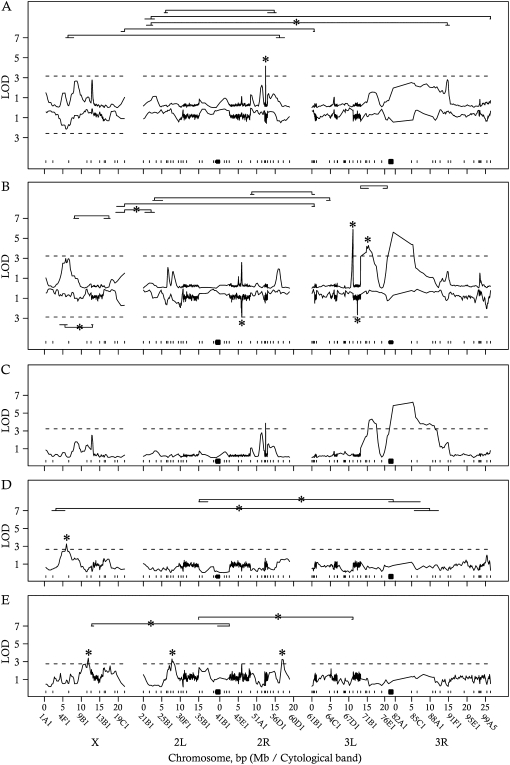
We identified a single QTL for ovariole number (Figure 3A, Table 6) and three QTL for thorax length (Figure 3B, Table 7). We found no QTL × E interactions for ovariole number and two QTL × E interactions for thorax length (Figure 3B, Table 7).
Figure 3.—
QTL scan results for ovariole number (A), thorax length (B), ovariole number conditioned on the additive effects of thorax length (C), ovariole number by environment conditioned on the additive effects of thorax length (D), and ovariole-thorax length-by-environment interaction (E). The x-axis represents genomic position, in megabases (Mb) along the three major Drosophila chromosomes. The y-axis represents LOD scores. (A and B) The top half of the graph represents LOD scores for QTL and epistatic interactions with additive effects across environments, and the bottom half of the graph represents LOD scores for QTL and epistatic interactions with variable effects across environments. See text for details of the QTL model. Brackets represent epistatic interactions. The tip of each bracket represents the 2-LOD support interval for that locus, conditional on the highest LOD score of its interacting partner. Asterisks (*) represent loci retained in the best mBIC model. Horizontal, dashed lines represent permutation thresholds (see text for details). Ticks and squares represent marker location and approximate centromere location, respectively.
We identified two marginal QTL when we tested for ovariole number QTL after conditioning on the additive effects of thorax length (Figure 3C, Table 6). We also identified one QTL × E interaction (Figure 3D, Table 6). When we tested for QTL affecting the relationship between ovariole number and thorax length, we identified no marginal QTL (results not shown) and three QTL × E interactions (Figure 3E, Table 6).
Epistatic QTL analysis:
Five epistatic interactions were identified for ovariole number (Figure 3A, Table 6); however, no epistatic-by-environment interactions were identified for ovariole number. Six epistatic interactions and two epistatic-by-environment interactions were identified for thorax length (Figure 3, Table 7). We identified no additional epistatic interactions for ovariole number when removing the additive effects of thorax length. We identified two epistatic-by-environment interactions for ovariole number when the additive effects of thorax length were removed (Figure 3, Table 6). Additionally, we identified two epistatic-by-environment interactions that affect the relationship between ovariole number and thorax length (Figure 3, Table 6).
We refined the location of these epistatic interactions and calculated their 2-LOD support intervals (Figure 3, Tables 6 and 7). In general, the positions of maximal LOD were close (typically ±3 cM) to the physical marker originally identified.
Final model building:
We identified different best models for both ovariole number and thorax length, depending on the penalization method. For ovariole number, the best mBIC model had a relative probability (w) of 0.19 and the best BIC model had a relative probability of 0.38. For thorax length, the best models had relative probabilities of 0.28 and 0.23 for mBIC and BIC, respectively. These values indicate that the best model is nearly equivalent to other models. Nonetheless, for simplicity, we restrict the remainder of our analysis to the best mBIC model, for either ovariole number or thorax length, which included fewer epistatic interactions than the best BIC and tended to include fewer main-effect QTL. These models represent the minimum number of QTL segregating in our population.
The relative importance of QTL and epistatic interactions generally reflects their inclusion in the best models. However, the unconditional LOD scores of particular QTL or interactions do not necessarily reflect their relative importance or inclusion into the best models (Tables 6 and 7). QTL with low unconditional LOD scores but high relative importance and conditional LOD scores are QTL that are sensitive to genetic background effects. We also observe some QTL with high raw LOD scores and low relative importance. Generally, these are QTL with main and epistatic effects (e.g., thorax length QTL, 3R at 1.23 Mb).
Genetic means and effects:
The estimated genetic means of model terms retained in the best mBIC models are presented for ovariole number (Figure 4), thorax length (Figure 5), and their relationship (Figures 6 and 7). Effects of each term and their variance–covariance matrix are presented in the supplemental data. The effects and variance–covariance matrix for the best BIC models are in the supplemental data.
Figure 4.—
QTL, QTL-by-environment, and epistatic-by-environment effects from the best mBIC model for ovariole number. Points represent genotype means, ±95% C.I.'s. Unless otherwise noted, estimates are relative to the 0.2% yeast-by-volume treatment. (A) 2R at 13.42 Mb. (B) X at 5.98 Mb. (C) 3R at 15.9 Mb and 2L at 2.172 Mb. (D) 3R at 1.2 Mb and X at 5.98 Mb.
Figure 5.—
Effects from the best mBIC model for thorax length. Points represent genotype means, ±95% C.I.'s. Unless otherwise noted, estimates are relative to the 0.2% yeast-by-volume treatment. (A) 3L at 12.17 Mb. (B) 3L at 16.7 Mb. (C) 2R at 6.9 Mb. (D) 3R at 13.4 Mb. (E) 2L at 2.2 Mb and X at 21.7 Mb. (F) X at 13.1 Mb and X at 5.6 Mb.
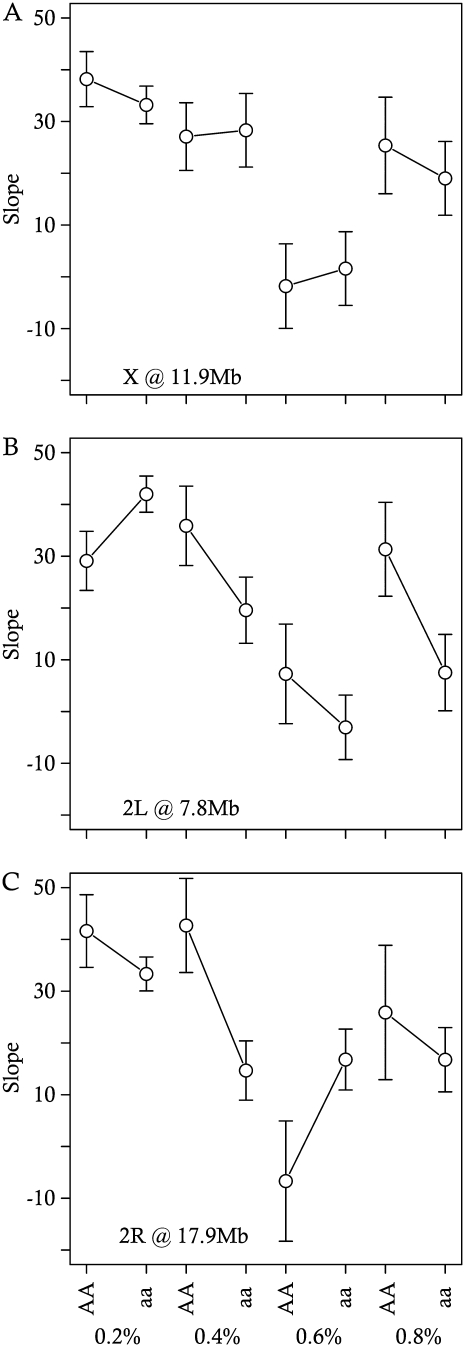
Figure 6.—
Estimated slopes, ±95% C.I.'s, of the relationship between ovariole number and thorax length within environments and alleles at each QTL. (A) X at 11.9 Mb. (B) 2L at 7.8 Mb. (C) 2R at 17.9 Mb.
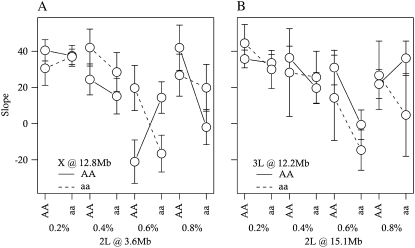
Figure 7.—
Estimated slopes, ±95% C.I.'s, of the relationship between ovariole number and thorax length within environments and alleles for each epistatic interaction. (A) X at 12.8 Mb and 2L at 3.6 Mb. (B) 3L at 12.2 Mb and 2L at 15.1 Mb.
DISCUSSION
We performed a genetic mapping experiment to identify QTL affecting phenotypic plasticity of ovariole number and thorax length in response to yeast concentration in the larval medium. We identified QTL and epistatic interactions for both traits with consistent effects across environments (Figures 3–5). Some of these QTL and epistatic interactions overlap with previously reported QTL affecting ovariole number and other body size correlates (Zimmerman et al. 2000; Wayne et al. 2001; Wayne and McIntyre 2002). QTL- and epistatic-by-environment interactions were observed for thorax length (Figures 3 and 5, Table 7). Interactions of these types were present for ovariole number when we conditioned on thorax length (Figures 3 and 4, Table 6). Additionally, we identified QTL-by-environment and epistatic-by-environment interactions that affect the relationship between ovariole number and thorax length (Figures 3, 6, and 7; Table 6). The allelic effect of these interactions increases with increasing yeast concentration, a pattern that is congruent with the monotonic decrease in the genetic correlation between these traits. These results suggest that the correlated evolution of ovariole number and thorax length may be highly sensitive to environmental factors.
Genotype–environment variation and QTL–environment effects:
Phenotypic plasticity is a ubiquitous phenomenon in nature (Schlichting and Pigliucci 1998; West-Eberhard 2003) and variation in plasticity is widely documented (Scheiner 1993). Genetic variation in plasticity (G × E) must be accounted for by alleles that vary in effect across environments (Via et al. 1995) regardless if G × E is caused by changes in rank order of genotypes across environments or by changes in the magnitude of genetic variation across environments. In our mapping population, we see changes in the rank order of genotypes and the magnitude of variance among environments for ovariole number and thorax length (Tables 2, 3, and 5) and directly tested for QTL that vary in effect across environments.
We identified two QTL- and two epistatic-by-environment interactions for thorax length (Figures 3 and 5, Table 7). All of these interactions show monotonic changes in magnitude across environments (Figure 5). Moreover, all interactions but one (Figure 5D) show stronger effects in low-yeast environments than in high-yeast environments. These results are consistent with a higher amount of among-line variation for thorax length in low- compared to high-yeast environments (Table 3).
The dose-dependent effects of these QTL and epistatic interactions suggest that these loci are physiologically responding directly to yeast concentration. In D. melanogaster, larval nutrient manipulation primarily affects body size through changes in cell number rather than cell size (De Moed et al. 1997). Thus, the candidate genes in these QTL are likely to affect cell-cycle progression or cell-autonomous, nutrient-dependent growth. Within the QTL-by-environment interaction 2R at 6.95 Mb is dare, a protein involved in ecdysone synthesis (Freeman et al. 1999). Recently ecdysone has been implicated in larval growth control and adult body size (Caldwell et al. 2005; Colombani et al. 2005; Mirth et al. 2005) and these effects are mediated by nutrient-dependent insulin signaling.
For ovariole number, no QTL- or epistatic-by-environment interactions were identified in our initial genome scans (Figure 3A) despite the high amount of G × E variation (Table 4). There are three possible reasons for this discrepancy. First, G × E variation for ovariole number may be controlled by many loci of very small effect or by small environment-specific effects of the observed main-effect QTL and epistatic interactions. Although we cannot directly rule out this hypothesis, we note that small QTL-by-environment effects were observed for thorax length (Figure 5, C and D), which suggests that we have adequate statistical power in our experiment. Second, G × E variation for ovariole number may be controlled by high-order epistasis (i.e., greater than two loci). While this is a testable hypothesis, in our design we do not have sufficient power to make such inferences. Third, G × E variation for ovariole number may be affected by allometric relationships. Here we examined this hypothesis by testing for QTL- and epistatic-by-environment interactions when thorax length was included as a covariate.
For this analysis, we first discuss the use of thorax length as an additive covariate; in the following section (see The basis of genetic correlations) we discuss the use of thorax length as an interactive covariate. The use of thorax length as an additive covariate assumes a fixed relationship between ovariole number and thorax length across environments and loci. Loci identified using this mapping strategy can, but do not necessarily, affect the joint means of ovariole number and thorax length (Tables 6 and 7, Figure 3). Regardless if the loci identified affect the joint means of ovariole number and thorax length or not, the causative genes within these QTL may mediate the development of one trait through another or through a mutual pathway (Li et al. 2006). While we have conditioned our ovariole number scans on thorax length, the results and interpretation remain the same if we conditioned thorax length scans on ovariole number. These QTL do not imply a directionality of effects from one trait to another, but rather a codependency of both traits on environmental and genetic effects.
We identified one QTL- and two epistatic-by-environment interactions for ovariole number when we conditioned on the additive effects of thorax length (Figure 3, Table 6). Within these QTL are plausible candidate genes that are known to, or likely to, affect nutrient-dependent growth of adult body parts. For instance, within the epistatic-by-environment interaction 2L at 15.23 Mb–3R at 1.23 Mb are Idgf1, Idgf2, Idgf3, and Ras85D, respectively. The IDGFs are known to affect imaginal disc growth and are secreted by the fat body (Kawamura et al. 1999). RAS signaling (Caldwell et al. 2005) mediated by PI3K activity in the prothoracic gland is known to affect insulin signaling in the larval fat body (Colombani et al. 2005). It is plausible that IDGF expression is mediated by nutrient-dependent insulin signaling.
Because we used environment-corrected thorax length as a covariate, QTL identified for ovariole number conditional on the use of thorax length as an additive covariate affect ovariole number relative to the within-environment average thorax length. Such QTL may play an important role in maintaining genetic variation within populations. For instance, stabilizing selection on thorax length (Roff 1981) may act to maintain allelic variation for ovariole number, and vice versa. To see this, note that these loci do not significantly affect ovariole number prior to conditioning on thorax length. However, different alleles at these loci have an effect on ovariole number relative to average thorax length. Thus, these alleles shift the intercept term of the relationship between ovariole number and thorax length. Because thorax length is centered, the value of ovariole number at the intercept represents the value at the average thorax length. Thus, if there is stabilizing selection on thorax length, segregating allelic variation for ovariole number could be maintained.
The basis of genetic correlations:
Genetic correlations are generated by pleiotropy and by linkage disequilibrium between loci. These mechanisms are generally indistinguishable in QTL mapping and we refer to both mechanisms as pleiotropic QTL. Pleiotropic QTL can affect a set of traits in two ways. First, they may affect the sign and the magnitude of the relationship between traits. Second, they may affect the joint means of a set of traits. We limit our discussion of pleiotropic QTL to the former case because, in our best mBIC models, there are no QTL or epistatic interactions that appear to affect the joint means of traits.
We identified pleiotropic QTL and epistatic interactions that affect the relationship between ovariole number and thorax length in an environment-dependent manner (Figures 3, D and E, 6, and 7; Table 6). These QTL- and epistatic-by-environment interactions were identified by including thorax length as an interactive covariate in our ovariole number scans, thereby allowing us to model trait relationships that vary as a function of environmental and genetic effects. In this sense, we have identified loci that affect the environment-specific, allometric relationship between these traits. Pleiotropic QTL that affect the sign and magnitude of trait relationships have been rarely reported (e.g., Cheverud 2001; Cheverud et al. 2004; Pavlicev et al. 2008; and see Flatt and Kawecki 2004 for the case of a single gene), but provide a context to understand the nature of genetic correlations.
In our mapping population, the net correlation between ovariole number and thorax length monotonically decreases with increasing yeast concentration in the larval medium (Figure 2, Table 5). The net correlation, however, is always positive as would occur if these traits were mutually constrained by resource acquisition or development time (Houle 1991). The strong pattern of change in this correlation suggests that this constraint is relaxed with increasing resources. Furthermore, the relative degree of this relaxation may be under genetic control. The magnitude of allelic effects at these loci is consistently stronger in high-yeast environments (Figures 6 and 7). We may further interpret the decrease in genetic correlation across environments as a product of segregating, environmentally sensitive allelic variation. A small genetic correlation between traits may reflect a high amount of genetic variance in trait relationships and does not necessarily imply that traits are independent of each other.
Allelic variation at loci that affect allometric relationships between traits can be directly acted upon by selection or drift. The action of these evolutionary processes on allometric relationships and genetic correlations have been documented (e.g., Weber 1990; Phillips et al. 2001), and the genetic control of allometry has important implications for interspecific variation (Langlade et al. 2005). The presence of loci that affect allometric relationships and genetic correlations may, however, alter the topology of the evolutionary landscape, thereby producing inaccessible regions of phenotypic space (Blows and Walsh 2008).
Evolution of plastic life-history correlates:
We used QTL mapping to identify the genetic architecture of plasticity in two morphological traits related to fitness, ovariole number and thorax length. Epistasis, pleiotropy, and G × E were pervasive and this architecture represents standing genetic variation within a natural population. Theoretical models (e.g., Gillespie and Turelli 1989; Turelli and Barton 2004) predict that these factors act to maintain variation in populations, and our results confirm these predictions.
Our results also suggest that the evolution of ovariole number and thorax length is highly integrated and environmentally sensitive; the evolution of one character will affect the evolution of the other and will do so in an environment-specific fashion. Assessing the evolutionary potential and history of these characters will ultimately depend on understanding their developmental bases, their relationship to fitness, and the extent of environmental heterogeneity in larval resources.
Acknowledgments
We thank the members of the Tatar and Rand labs for logistic support and T. Flatt, K. Montooth, C. Meiklejohn, D. Rand, A. Schmitt, and D. Weinriech for insightful discussion and comments on the early versions of the manuscript. This manuscript was greatly improved by the comments of two anonymous reviewers and by D. Houle and J. B. Walsh. This work was supported by National Institutes of Health grants GM065513 and GM61773 to S.V.N. and AG021953 to M.T. and by an Ellison Medical Foundation grant to M.T.
References
- Barbieri, M., M. Bonafé, C. Franceschi and G. Paolisso, 2003. Insulin/IGF-1- signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metabol. 285 E1064–E1071. [DOI] [PubMed] [Google Scholar]
- Bitterman, K. J., O. Medvedik and D. A. Sinclair, 2003. Longevity regulation in Saccharomyces cerevisiae: liking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 67 376–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows, M., and B. Walsh, 2008. Spherical cows grazing in Flatland: constraints to selection and adaptation, in Adaptation and Fitness in Animal Populations Evolutionary and Breeding Perspectives on Genetic Resource Management, edited by J. van der Werf, H.-U. Graser, R. Frankham and C. Gondro (in press).
- Bogdan, M., J. K. Gosh and R. W. Doerge, 2004. Modifying the Schwartz Bayesian information criterion to locate multiple interacting quantitative trait loci. Genetics 167 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulétreau-Merle, J., R. Allemand, Y. Cohet and J. R. David, 1982. Reproductive strategy in Drosophila melanogaster: significance of a genetic divergence between temperate and tropical populations. Oecologia 53 323–329. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., and T. P. Speed, 2002. A model selection approach for the identification of quantitative trait loci in experimental crosses. J. R. Stat. Soc. B 64 641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K., H. Wu, S. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 889–890. [DOI] [PubMed] [Google Scholar]
- Burnham, K. P., and D. R. Anderson, 2002. Model Selection and Multimodel Inference. Springer-Verlag, New York.
- Caldwell, P. E., M. Walkiewicz and M. Stern, 2005. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr. Biol. 15 1785–1795. [DOI] [PubMed] [Google Scholar]
- Chen, L., and J. D. Storey, 2006. Relaxed significance criteria for linkage analysis. Genetics 173 2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud, J. M., 2001. The genetic architecture of pleiotropic relations and differential epistasis, pp. 411–433 in The Character Concept in Evolutionary Biology, edited by G. P. Wagner. Academic Press, San Diego.
- Cheverud, J. M., T. H Ehrich, T. T. Vaughn, S. F. Koreishi, R. B. Linsey et al., 2004. Pleiotropic effects on mandibular morphology II: differential epistasis and genetic variation in morphological integration. J. Exp. Zool. Mol. Dev. Evol. 302B 424–435. [DOI] [PubMed] [Google Scholar]
- Colombani, J., L. Bianchini, S. Layalle, E. Pondeville, C. Dauphin-Villemant et al., 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310 667–670. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., J. Rux and J. R. David, 1991. Genetics of morphological differences and hybrid sterility between Drosophila sechellia and its relatives. Genet. Res. 57 113–122. [DOI] [PubMed] [Google Scholar]
- David, J. R., 1970. Le nombre d'ovarioles chez la drosophile en relation avec la fécondité et la valeur adaptive. Arch. Zool. Exp. Gen. 111 357–370. [Google Scholar]
- De Moed, G. H., G. De Jong and W. Scharloo, 1997. The phenotypic plasticity of wing size in Drosophila melanogaster: the cellular basis of its genetic variation. Heredity 79 260–267. [DOI] [PubMed] [Google Scholar]
- Engstrom, L. E., 1971. Studies of the effects of two-way selection for ovariole number in Drosophila melanogaster. Ph.D. Dissertation, University of Illinois, Urbana-Champaign, IL.
- Flatt, T., and T. J. Kawecki, 2004. Pleiotropic effects of Methoprene-tolerant (Met), a gene involved in juvenile hormone metabolism, on life history traits in Drosophila melanogaster. Genetica 122 141–160. [DOI] [PubMed] [Google Scholar]
- Fox, J., 2008. car: companion to applied regression. R package version 1.2–8. http://www.r-project.org.
- Freeman, M. R., A. Dobritsa, P. Gaines, W. A. Segraves and J. R. Carlson, 1999. The dare gene: steroid hormone production, olfactory behavior, and neural degeneration in Drosophila. Development 126 4591–4602. [DOI] [PubMed] [Google Scholar]
- Fry, J. D., S. V. Nuzhdin, E. G. Pasyukova and T. F. Mackay, 1998. QTL mapping of genotype-environment interaction for fitness in Drosophila melanogaster. Genet. Res. 71 133–141. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., and M. Turelli, 1989. Genotype-environment interactions and the maintenance of polygenic variation. Genetics 121 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromko, M. H., 1995. Unpredictability of correlated response to selection: pleiotropy and sampling interact. Evolution 49 685–693. [DOI] [PubMed] [Google Scholar]
- Gurganus, M. C., J. D. Fry, S. V. Nuzhdin, E. G. Pasyukova, R. F. Lyman et al., 1998. Genotype-by-environment interaction at quantitative trait loci affecting sensory bristle number in Drosophila melanogaster. Genetics 149 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling, E. W., J. A. Riksen, J. Bakker and J. E. Kammenga, 2007. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98 28–37. [DOI] [PubMed] [Google Scholar]
- Guzmán, C., R. Cabrera, M. Cárdenas, F. Larrea, P. W. Nathanielsz et al., 2007. Protein restriction during fetal and neonatal development in the rat alters reproductive function and accelerates reproductive ageing in female progeny. J. Physiol. 572 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen, E., 2004. Interplay between growth factor and nutrient signaling: lessons from Drosophila TOR. Curr. Top. Microbiol. Immunol. 279 153–167. [DOI] [PubMed] [Google Scholar]
- Hodin, J., 2008. She shapes events as they come: plasticity in insect reproduction, in Insects and Phenotypic Plasticity, Vol. II. Science Publishers, Enfield, NH(in press).
- Hodin, J., and L. M. Riddiford, 1998. The ecdysone receptor and ultraspiricle regulate the timing and progression of ovarian morphogenesis during Drosophila metamorphosis. Dev. Genes Evol. 208 304–317. [DOI] [PubMed] [Google Scholar]
- Hodin, J., and L. M. Riddiford, 2000. Different mechanisms underlie phenotypic plasticity and interspecific variation for a reproductive character in drosophilids (Insecta: Diptera). Evolution 54 1638–1653. [DOI] [PubMed] [Google Scholar]
- Honek, A., 1993. Intraspecific variation in body size and fecundity in insects: a general relationship. OIKOS 66L 483–492. [Google Scholar]
- Houle, D., 1991. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution 45 630–648. [DOI] [PubMed] [Google Scholar]
- Kambysellis, M. P., and W. B. Heed, 1971. Studies of oogenesis in natural populations of Drosophilidae. I. Relation of ovarian development and ecological habitats of the Hawaiian species. Am. Nat. 105 31–49. [Google Scholar]
- Kawamura, K., T. Shibata, O. Saget, D. Peel and P. J. Bryant, 1999. A new family of growth factors produced by the fat body and active on Drosophila imaginal discs. Development 126 211–219. [DOI] [PubMed] [Google Scholar]
- Lande, R., and S. J. Arnold, 1983. The measurement of selection on correlated characters. Evolution 37 1210–1226. [DOI] [PubMed] [Google Scholar]
- Langlade, N. B., X. Feng, T. Dransfield, L. Copsey, A. I. Hanna et al., 2005. Evolution through genetically controlled allometry space. Proc. Natl. Acad. Sci. USA 102 10221–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leevers, S. J., and E. Hafen, 2003. Growth regulation by insulin and TOR signaling in Drosophila, pp. 167–192 in Cell Growth: Control of Cell Size, edited by M. Hall, M. Raff and G. Thomas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Leips, J., and T. F. Mackay, 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., S. W. Tsaih, K. Shockley, I. M. Stylianou, J. Wergedal et al., 2006. Structural model analysis of multiple quantitative traits. PLoS Genet. 2 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, New York.
- Mackay, T. F. C., 2001. Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2 11–20. [DOI] [PubMed] [Google Scholar]
- Mirth, C., J. W. Truman and L. M. Riddiford, 2005. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr. Biol. 15 1796–1807. [DOI] [PubMed] [Google Scholar]
- Neter, J., W. Wasserman and M. H. Kutner, 1985. Applied Linear Statistical Models, Ed. 2. Richard D. Irwin, Homewood, IL.
- Oldham, S., and E. Hafen, 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13 79–85. [DOI] [PubMed] [Google Scholar]
- Orgogozo, V., K. W. Broman and D. L. Stern, 2006. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 173 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski, S. H., S. Wolff, H. Aguilaniu, J. Durieux and A. Dillin, 2007. PHA- 4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 477 550–555. [DOI] [PubMed] [Google Scholar]
- Pavlicev, M., J. P. Kenney-Hunt, E. A. Norgard, C. C. Roseman, J. B. Wolf et al., 2008. Genetic variation in pleiotropy: differential epistasis as a source of variation in the allometric relationship between long bone lengths and body weight. Evolution 62 199–213. [DOI] [PubMed] [Google Scholar]
- Phillips, P. C., M. C. Whitlock and K. Fowler, 2001. Inbreeding changes the shape of the genetic covariance matrix in Drosophila melanogaster. Genetics 158 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci, M., 2006. Genetic variance-covariance matrices: a critique of the evolutionary quantitative genetic research program. Biol. Philos. 21 1–23. [Google Scholar]
- R Development Core Team, 2006. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Rae, M. T., S. Palassio, C. E. Kyle, A. N. Brooks, R. G. Lea et al., 2001. Effect of maternal undernutrition during pregnancy on early ovarian development and subsequent follicular development in sheep fetuses. Reproduction 122 915–922. [PubMed] [Google Scholar]
- Rae, M. T., C. E. Kyle, D. W. Miller, A. J. Hammond, A. N. Brooks et al., 2002. The effects of undernutrition, in utero, on reproductive function in adult male and female sheep. Anim. Reprod. Sci. 72 63–71. [DOI] [PubMed] [Google Scholar]
- Rhind, S. M., 2004. Effects of maternal nutrition on fetal and neonatal reproductive development and function. Anim. Reprod. Sci. 82–83 169–181. [DOI] [PubMed] [Google Scholar]
- R'kha, S., B. Moreteau, J. A. Coyne and J. R. David, 1997. Evolution of a lesser fitness trait: egg production in the specialist Drosophila sechelia. Genet. Res. Camb. 69 17–23. [DOI] [PubMed] [Google Scholar]
- Robertson, F. W., 1957. Studies in quantitative inheritance X. Genetic variation of ovary size in Drosophila. J. Genet. 55 410–427. [Google Scholar]
- Roff, D., 1981. On being the right size. Am. Nat. 108 405–422. [Google Scholar]
- Roff, D., 1997. Evolutionary Quantitative Genetics. Chapman & Hall, New York.
- Roff, D., 2002. Life History Evolution. Sinauer Associates, Sunderland, MA.
- SAS Software, 2002. Version 9.13 of the SAS System for Windows. SAS Institute, Cary, NC.
- Scheiner, S. M., 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24 35–68. [Google Scholar]
- Schlichting, C. D., and M. Pigliucci, 1998. Phenotypic Evolution: A Reaction Norm Perspective. Sinauer Associates, Sunderland, MA.
- Schmidt, P. S., L. Matzkin, M. Ippolito and W. F. Eanes, 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59 1721–1732. [PubMed] [Google Scholar]
- Schwartz, G., 1978. Estimating the dimension of a model. Ann. Stat. 6 461–464. [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia, S., and B. N. Singh, 2004. Size dependent sexual selection in Drosophila ananasae. Genetica 121 207–217. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott, M., L. M. McIntyre and M. L. Wayne, 2005. Genetic architecture of two fitness-related traits in Drosophila melanogaster: ovariole number and thorax length. Genetica 125 211–222. [DOI] [PubMed] [Google Scholar]
- Tu, M., and M. Tatar, 2003. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 125 327–333. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and N. H. Barton, 2004. Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G × E interactions. Genetics 166 1053–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, M. D. Purugganan and T. F. Mackay, 2003. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via, S., and R. Lande, 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39 505–522. [DOI] [PubMed] [Google Scholar]
- Via, S., R. Gomulkiewicz, G. De Jong, S. M. Scheiner, C. D. Schlichting et al., 1995. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol. Evol. 10 212–217. [DOI] [PubMed] [Google Scholar]
- Vieira, C., E. G. Pasyukova, Z. B. Zeng, J. B. Hackett, R. F. Lyman et al., 2000. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., and T. F. C. Mackay, 1998. Quantitative genetics of ovariole number in Drosophila melanogaster. II. Mutational variation and genotype-environment interaction. Genetics 148 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., and L. McIntyre, 2002. Combining mapping and arraying: a novel approach to candidate gene identification. Proc. Natl. Acad. Sci. USA 99 14903–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., J. B. Hackett and T. F. C. Mackay, 1997. Quantitative genetics of ovariole number in Drosophila melanogaster: I. Segregating variation and fitness. Evolution 51 1156–1163. [DOI] [PubMed] [Google Scholar]
- Wayne, M. L., J. B. Hackett, C. L. Dilda, S. V. Nuzhdin, E. G. Pasyukova et al., 2001. Quantitative trait locus mapping of fitness-related traits in Drosophila melanogaster. Genet. Res. Camb. 77 107–116. [DOI] [PubMed] [Google Scholar]
- Weber, K. E., 1990. Selection on wing allometry in Drosophila melanogaster. Genetics 126 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard, M. J., 2003. Developmental Plasticity and Evolution. Oxford University Press, Oxford.
- Yang, C. H., P. Belawat, E. Hafen, L. Y. Jan and Y. N. Jan, 2008. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science 319 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, E., A. Palsson and G. Gibson, 2000. Quantitative trait loci affecting components of wing shape in Drosophila melanogaster. Genetics 155 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]