Abstract
Caenorhabditis elegans primarily reproduces as a hermaphrodite. Independent gene conversion events in mutant obligately outcrossing populations of C. elegans [fog-2(lf)] spontaneously repaired the loss-of-function mutation in the fog-2 locus, thereby reestablishing hermaphroditism as the primary means of reproduction for the populations.
SPECIES within the genus Caenorhabditis employ one of two modes of reproduction. Nine of the 11 Caenorhabditis species in culture (Kiontke and Sudhaus 2006) are gonochoristic obligate female/male outcrossers. Gonochorism is thought to be the ancestral state within the genus (Schedl and Kimble 1988; Kiontke et al. 2004). The remaining two species, Caenorhabditis elegans and C. briggsae, have an androdioecious breeding system with populations composed of self-fertile hermaphrodites and males at a low frequency (<0.1%) (Ward and Carrel 1979; Hodgkin and Doniach 1997). The two hermaphroditic Caenorhabditis species are phylogenetically separated by two gonochoristic species, suggesting that hermaphroditism (and an androdioecious breeding system) evolved convergently in C. elegans and C. briggsae (Kiontke et al. 2004). Moreover, the regulation of sperm production in hermaphrodites in these two species differs in important ways. For instance, the fog-2 locus is specifically required for spermatogenesis in C. elegans hermaphrodites (Schedl and Kimble 1988; Nayak et al. 2005). The appearance of fog-2 in the C. elegans genome is thought to be an evolutionarily recent event resulting from a gene duplication that may have ultimately promoted the evolution of hermaphroditism (Clifford et al. 2000; Haag 2005; Nayak et al. 2005). Furthermore, C. elegans also requires fem-2, fem-3, and tra-2 for spermatogenesis in hermaphrodites whereas control of sperm production in C. briggsae hermaphrodites occurs downstream of the fem genes (Hill et al. 2006).
Loss-of-function mutations in fog-2 [fog-2(lf)] in C. elegans result in a change from androdioecy to gonochoristic reproduction (Schedl and Kimble 1988). However, extragenic mutations that suppress, at least to some degree, the fog-2 mutant phenotype, have been found in five different genes: tra-2, fem-3, gld-2, tra-3, and atx-2 (Barton et al. 1987; Schedl and Kimble 1988; Francis et al. 1995a,b; Maine et al. 2004; Nayak et al. 2005). These experiments have used either chemical mutagenesis or RNA interference (RNAi) to discover alleles that restore hermaphroditism in fog-2 mutants. Here we report that spontaneous gene conversion involving the neighboring paralog with an unknown function, ftr-1, can restore the function of fog-2 in experimental populations. These gene conversion events result in a fully functional hermaphrodite that replaces the original fog-2 mutant in experimental populations and may be more frequent than point mutations in restoring the functionality of fog-2(lf) mutants.
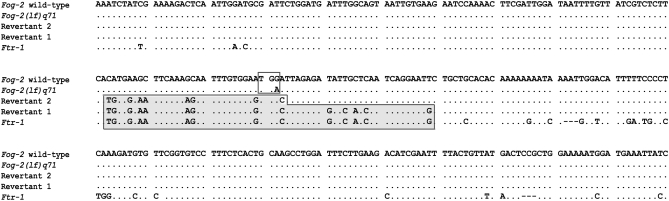
During an experimental evolution study comprising 74 fog-2(lf) lines derived from the same ancestral pair, we identified two independent instances in which fog-2 mutants (normally obligate outcrossers) had reverted spontaneously to hermaphroditism. Revertant 1 appeared in an experimental phase that involved repeated population bottlenecks of two individuals per generation in conjunction with knockdown of the mismatch repair gene msh-2 by a standard RNAi feeding protocol (Kamath et al. 2000). Revertant 2 appeared during the second phase of the experiment involving population expansion in the absence of msh-2 RNAi. In each instance, a putative case of reversion to hermaphroditism was detected by observing extremely biased sex ratios in the offspring generation, namely, the near complete absence of males (male–female crosses yield 50:50 offspring sex ratios whereas males are rare or absent in selfing hermaphroditic populations of C. elegans). Reversion to functional hermaphroditism was confirmed by the production of self progeny by individually plating L4 larvae. To determine the genetic basis of reversion to hermaphroditism, the fog-2 gene was PCR amplified and sequenced in (i) the wild-type C. elegans laboratory strain, N2, (ii) the fog-2 mutant strain, and (iii) the two experimental fog-2 mutant strains that reverted to hermaphroditism. Each of the two sex reversal events resulted from a gene conversion whereby a short segment of a paralogous gene ftr-1 recombined with the fog-2(lf) mutant allele, replacing the premature stop codon with a tryptophan codon (Figure 1). Both gene conversion events are relatively short, replacing at minimum 56 and 32 nucleotides of fog-2 sequence with ftr-1 sequence, respectively (maximum possible lengths of the gene conversion tracts are 145 and 121 bp, respectively). The length of these gene conversion tracts are well within the average range of converted lengths found between paralogs in the C. elegans genome (Semple and Wolfe 1999) although considerably shorter than the >200-bp conversion tracts detected in an assay of DNA double-strand break repair employing an extrachromosomal DNA template (Plasterk and Groenen 1992).
Figure 1.—
Nucleotide sequence alignments representing two independent gene conversion events at the fog-2 locus by ftr-1 resulting in a switch from obligate outcrossing to hermaphroditism in two fog-2(lf) mutant lines. In-frame nucleotide positions 200–499 of exon 3 (total length 640 bp) are displayed. The small, clear box displays the nonsense mutation G → A in the fog-2(lf)q71 allele resulting in a nonfunctional gene relative to the wild type. The larger shaded boxed area represents the minimum gene conversion tracts by the upstream ftr-1 locus in sex-revertants 1 and 2. Indels are indicated by dashed lines and dots represent identical nucleotides to the fog-2 wild-type sequence.
A comparison of fog-2 and ftr-1 found signatures of past gene conversion in their evolution. Although the overall sequence divergence between fog-2 and ftr-1 over their homologous coding regions is 16%, a few large segments are completely identical between the two genes. Using Geneconv, a software that employs statistical tests to detect gene conversion, we found three statistically significant regions (P-values = 0.0000, 0.0021, and 0.0415) ranging from 39 to 75 nucleotides in length that are identical between fog-2 and ftr-1 (Sawyer 1999). However, the directionality of these past gene conversion events is unknown, with the possibility that either ftr-1 or fog-2 sequence tracts have served as the donor sequence.
Our sample size is clearly too small to draw any definitive conclusions about the relative rates of point mutations and gene conversion in the C. elegans genome. One of the gene conversion events occurred during msh-2 knockdown by RNAi, which might be expected to increase the rates of gene conversion. Conversely, it is also expected to increase the nucleotide substitution rate and hence the rate at which fog-2 reverts to wild type by point mutation. However, the fact that we found gene conversion events and no direct reversion to wild type by point mutation suggests that gene conversion is at least as common in the C. elegans genome as point mutations, if not more frequent. The genomic proximity of the ftr-1 and fog-2 loci (Figure 2) may also facilitate a high frequency of gene conversion between them. Indeed, studies of gene conversion events in both C. elegans (Semple and Wolfe 1999) and yeast (Drouin 2002) have found a negative correlation between the frequency of gene conversion events and the distance between gene pairs (unlinked vs. linked genes). Finally, the chromosomal location of fog-2 and ftr-1 may further enhance the rate of gene conversion. Both genes reside close to the right end of chromosome V. Chromosomal arms in C. elegans are known to have higher recombination rates relative to the center (Barnes et al. 1995; Hillier et al. 2007) and crossing over increases the probability of gene conversion (Jeffreys and May 2004).
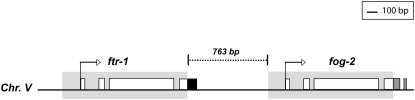
Figure 2.—
Schematic depicting the regions of homology between paralogs fog-2 and ftr-1. Shaded narrow rectangles denote exons; horizontal lines represent introns and duplicated flanking regions where applicable. The duplicated region is shaded. Duplicated segments as determined by shared sequence homology between the two paralogs are also depicted by the correspondence of regions with identical color and pattern. The figure is drawn to scale. ftr-1 comprises four exons encoding 314 amino acids. The exon–intron structure of fog-2, comprising five exons (encoding for 327 aa) exhibits both similarities and dissimilarities relative to ftr-1. Homology between fog-2 and ftr-1 commences ∼170 bp upstream of the start codon, encompassing the first three exons and introns and terminating at nucleotide position 91 of the terminal exon (total length 186 bp). The last 95 bp of the terminal exon of ftr-1 as well as its 3′ downstream region bear no homology to the corresponding C-terminal region of fog-2. The KS value between ftr-1 and fog-2 over the region of homology comprising the duplication span (1248 bp) is 0.22 with the Nei–Gojobori method (Nei and Gojobori 1986) and 0.26 if corrected for multiple hits under the Jukes–Cantor model (Jukes and Cantor 1969). Regarding fog-2, the latter 66 bp of exon 4 (total 157 bp), and intron 4 (45 bp) and exon 5 (23 bp) in their entirety comprise unique sequence bearing no obvious homology to ftr-1. To determine an alternative genomic source for this nonhomologous sequence tract in the C-terminal end of fog-2, this 134 bp of unique ORF (comprising both exonic and intronic regions) in isolation as well as in conjunction with 500 bp of the fog-2 3′ downstream region was queried against the C. elegans genome sequence in WormBase using a BlastN search. In addition, we queried a 37-aa-long sequence coded by the unique exonic regions of fog-2 against WormBase using a tBlastN search. All three queries failed to yield any alternate hits, suggesting that this stretch of sequence unique to fog-2 may have been assimilated into its reading frame via recruitment of novel neighborhood sequence from its new genomic location. The loci of both paralogs, ftr-1 and fog-2, reside on chromosome V as tandem genes with positive strand orientation and are separated by a 763-bp stretch of unique sequence. The genomic proximity of fog-2 and ftr-1 suggests unequal exchange or slippage as the mechanism of duplication and conforms to the general pattern of genomic location observed for evolutionarily young gene duplicates in C. elegans (Katju and Lynch 2003, 2006).
These gene conversion events during experimental evolution in the laboratory raise the question whether similar events (i.e., gene conversion between fog-2 and members of the ftr family) are important in nature. Most Caenorhabditis species are obligate outcrossers and it is tempting to speculate that in some environments where outcrossing is favored, a loss-of-function mutation in fog-2 could be advantageous in C. elegans. A high rate of gene conversion would make such loss-of-function mutations more reversible than by point mutations alone. However, this is unlikely to have been important in the recent evolutionary history of C. elegans. Despite the fact that male sperm readily overwhelms hermaphroditic sperm in the event of a male-hermaphrodite mating, fog-2 mutants are at a severe disadvantage in mixed populations of the two (Chasnov and Chow 2002; Stewart and Phillips 2002), even under experimental conditions imposing a high mutational load when outcrossing may be more beneficial (Cutter 2005; Manoel et al. 2007). Moreover, mating behavior in C. elegans appears to have degenerated relative to other obligate outcrossers in the genus, such as C. remanei and Caenorhabditis spp. 4 (Rene Garcia et al. 2007). Nonetheless, gene conversion between ftr-1 and fog-2 has the potential to shape genetic variation at these loci in natural populations, thereby modifying the number of sperm produced by hermaphrodites with important implications for the degree of inbreeding vs. outcrossing in nature.
Acknowledgments
We are grateful to A. Villeneuve and two anonymous reviewers for valuable comments and suggestions. We thank the Bruce Bowerman laboratory in the Department of Biology at the University of Oregon for kindly providing the fog-2 mutant line and LeAnne Lovato for technical assistance. The wild-type N2 nematode strain used in this work was provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources (NCRR). V.K. was supported by a National Science Foundation Doctoral Dissertation Improvement grant DEB-0308782. Further support was provided by a NIH IMSD grant (GM060201-07) to E.M.L. and a Centers of Biomedical Research Excellence grant P20-RR18754 from the NIH NCRR to U.B.
References
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M. K., T. Schedl and J. Kimble, 1987. Gain-of-function mutations of fem-3, a sex determination gene in Caenorhabditis elegans. Genetics 115 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov, J. R., and K. L. Chow, 2002. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics 160 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, R., M. Lee, S. Nayak, M. Ohmachi, F. Giorgini et al., 2000. FOG-2, a novel F- box-containing protein, associates with the GLD-1 RNA-binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127 5265–5276. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., 2005. Mutation and the experimental evolution of outcrossing in Caenorhabditis elegans. J. Evol. Biol. 18 27–34. [DOI] [PubMed] [Google Scholar]
- Drouin, G., 2002. Characterization of the gene conversions between the multigene family members of the yeast genome. J. Mol. Evol. 55 14–23. [DOI] [PubMed] [Google Scholar]
- Francis, R., M. K. Barton, J. Kimble and T. Schedl, 1995. a gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, R., E. Maine and T. Schedl, 1995. b Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, E. S., 2005. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology, in Wormbook, edited by The C. elegans Research Community. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Hill, R. C., C. E. de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10 531–538. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., and T. Doniach, 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A. C., and C. A. May, 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36 151–156. [DOI] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969. Evolution of protein molecules, pp. 21–123 in Mammalian Protein Metabolism, edited by H. N. Munro. Academic Press, New York.
- Kamath, R. S., M. Martinez-Campos, P. Zipperlein, A. G. Frazer and J. Ahringer, 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2006. On the formation of novel genes by duplication in the Caenorhabditis elegans genome. Mol. Biol. Evol. 23 1056–1067. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke, K., and W. Sudhaus, 2006. Ecology of Caenorhabditis species, in Wormbook, edited by The C. elegans Research Community. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Maine, E., D. Hansen, D. Springer and V. E. Vought, 2004. Caenorhabditis elegans atx-2 promotes germline proliferation and oocyte fate. Genetics 168 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel, D., S. Carvalho, P. C. Phillips and H. Teotónio, 2007. Selection against males in Caenorhabditis elegans under two mutational treatments. Proc. R. Soc. Lond. Ser. B 274 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 418–426. [DOI] [PubMed] [Google Scholar]
- Plasterk, R. H. A., and J. T. M. Groenen, 1992. Targeted alterations of the Caenorhabditis elegans genome by transgene instructed DNA double strand break repair following Tc1 excision. EMBO J. 11 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rene Garcia, L., B. LeBoeuf and P. Koo, 2007. Diversity in mating behavior of hermaphroditic and male-female Caenorhabditis nematodes. Genetics 175 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S. A, 1999. GENECONV: a computer package for the statistical detection of gene conversion. Washington University, St. Louis. http://www.math.wustl.edu/∼sawyer.
- Schedl, T., and J. Kimble, 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, C., and K. H. Wolfe, 1999. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J. Mol. Evol. 48 555–564. [DOI] [PubMed] [Google Scholar]
- Stewart, A. D., and P. C. Phillips, 2002. Selection and maintenance of androdioecy in Caenorhabditis elegans. Genetics 160 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S., and J. S. Carrel, 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73 304–321. [DOI] [PubMed] [Google Scholar]