Abstract
In this report, we describe several approaches to improve the scalability and throughput of major genetic crosses in ends-out gene targeting. We generated new sets of targeting vectors and fly stocks and introduced a novel negative selection marker that drastically reduced the frequency of false-positive targeting candidates.
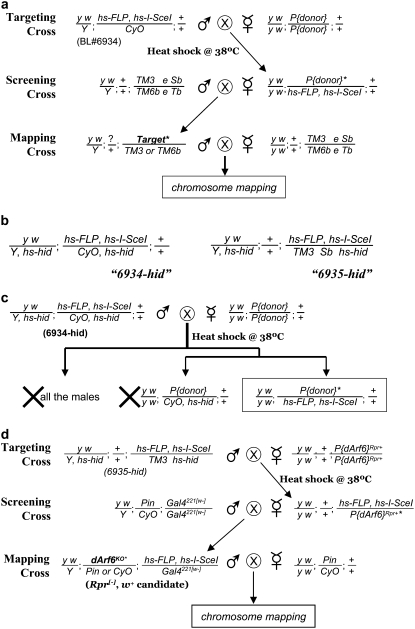
THE development of homologous recombination-based gene targeting is a landmark breakthrough in Drosophila genetics (Rong and Golic 2000; Gong and Golic 2003). In particular, the so-called “ends-out” or replacement-type gene targeting offers a straightforward approach for generating either knockout or knockin alleles. To date, there are already >20 genes that have been modified by ends-out targeting (supplemental Table 1). Nonetheless, the frequency of target-specific homologous recombination in Drosophila varies tremendously, ranging from >1/200 gametes (Manoli et al. 2005) to <1/350,000 (Jones et al. 2007) (also Y. Hong, unpublished data), i.e., a >1800-fold difference. In cases of low targeting efficiency (<1/100,000 gametes), ends-out targeting can be exceedingly time and labor intensive. Here, we optimized the current ends-out targeting scheme by focusing on improving the scalability and throughput of its major genetic crosses. As illustrated in Figure 1a, there are three major genetic crosses in a typical ends-out targeting. In the targeting cross, virgin females of a transgenic line bearing the donor DNA (“P{donor}”) are crossed with hs-FLP, hs-I-SceI males, and their larval progeny are heat-shocked to induce the generation of linear donor DNA fragments by FLPase and I-SceI enzymes. In the screening cross, virgin females from the targeting cross that are of the correct genotype (P{donor}*/hs-FLP, hs-I-SceI) are crossed with proper chromosome balancer males, and preliminary targeting candidates are recovered on the basis of their w+ marker. However, many of these candidates might be false positives due to the failure of excision or nontargeting integration of the donor DNA. In the mapping cross, only preliminary candidates whose w+ marker is mapped to the target gene chromosome are selected for further analysis.
Figure 1.—
Genetic crosses in targeting experiments. (a) Genetic crosses of a typical ends-out targeting experiment. The transgenic donor DNA (“P{donor}”) is on the second chromosome, while the target gene is on the third chromosome. P{donor}*, linearized extrachromosomal donor DNA fragment that only exists transiently—it will either be lost permanently or inserted into a chromosome by targeted or nontargeted integration events. Target*, potential targeting events. Note that the majority of potential targeting events may be nonspecific and not located on the third chromosome (see text). ?, this copy of the second chromosome is inherited from the female in the screening cross. It could be the donor chromosome or hs-FLP, hs-I-SceI, or the recombinant between the two. Nonetheless, this copy of the chromosome is irrelevant in the mapping cross. (b) Genotypes of 6934-hid and 6935-hid. (c) 6934-hid stock eliminates the virgin collection and genotype sorting in the targeting cross. (d) The genetic cross scheme of the dArf6KO targeting experiment. Because dArf6 is on the second chromosome, a transgenic line carrying dArf6KO donor DNA (“P{dArf6}Rpr+”) on the third chromosome was used. w; Pin/CyO; Gal4221[w−] stock was used to set up the screening cross in lieu of a regular Pin/CyO balancer stock. This allowed simultaneous selection against nonspecific targeting candidates while balancing the potential specific targeting candidates from the screening cross. P{dArf6}Rpr+*, linearized extrachromosomal dArf6KO donor DNA fragment. dArf6KO*, potential targeting events.
For the targeting cross, the number of P{donor}*/hs-FLP, hs-I-SceI virgin females directly determines the scale of the whole targeting experiment. Genes that are resistant to homologous recombination may require collecting and sorting >15,000 virgins from the targeting cross (Larsson et al. 2004), which is extremely labor intensive due to the time-sensitive nature of virgin collection and the genotyping process. To eliminate this major bottleneck in scaling up the targeting cross, we modified the original hs-FLP, hs-I-SceI stocks by replacing their Y chromosomes and balancer chromosomes with ones that contain hs-hid transgenes (Grether et al. 1995). We named these modified stocks “6934-hid” and “6935-hid” (Figure 1b) after the original stock numbers. Ubiquitous expression of the cell-death gene hid induced by heat shock causes strong lethality. As illustrated in Figure 1c, in a targeting cross using 6934-hid, all male progeny and those female progeny carrying hs-hid balancer chromosome are eliminated. Since P{donor}*/hs-FLP, hs-I-SceI females are the only genotype that survives, 6934-hid and 6935-hid completely eliminate the time-sensitive virgin collection and genotyping process.
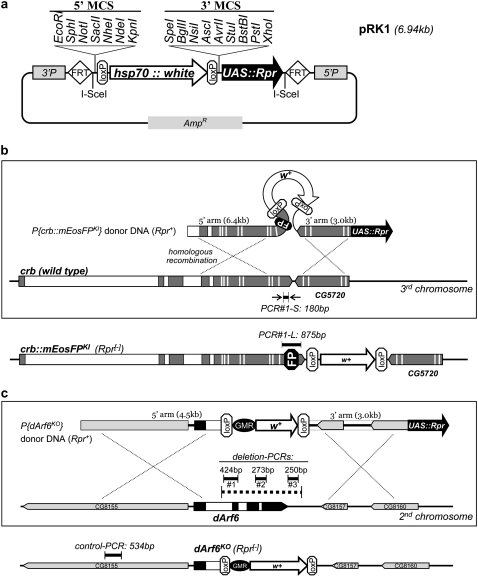
For screening and mapping crosses, we found their throughput was often severely limited by the high background of false positives, which may represent >95–99.9% of preliminary candidates (J. Huang, W. Zhou, and Y. Hong, unpublished results, and see below). Therefore, we introduced a negative selection marker into the current ends-out targeting scheme, so the majority of nontargeted integrations may be directly eliminated before they are subject to any further screening and mapping efforts. Ectopic expression of another cell-death gene reaper (rpr), similar to hid, also causes strong lethality (White et al. 1996). As illustrated in Figure 2b, a UAS-Rpr module can be tagged to the 3′ end of a transgenic donor DNA fragment (e.g., P{crb∷mEosFPKI}). Once the donor DNA fragment is recombined into the target gene locus, UAS-Rpr will be lost due to homologous recombination. In contrast, nontargeted integrations will likely retain the donor DNA fragment with an intact UAS-Rpr module (“Rpr+”). By using proper Gal4 driver stocks to set up the screening cross, Rpr+/Gal4 false-positive candidates will be directly eliminated due to the ectopic expression of Rpr.
Figure 2.—
pRK1 vector and targeting of crb∷mEosFPKI and dArf6KO. (a) Only pRK1 is diagrammed here. The hsp70∷white (w+) transformation marker is flanked by two loxP sites so that w+ can be removed in the final targeted alleles by Cre recombinase. MCS, multiple cloning sites. AmpR, ampicillin-resistant gene. 3′P and 5′P, 3′ and 5′ P-element sequences for transgenic insertion. (b) crb∷mEosFPKI knockin targeting. mEosFP (“FP”) is fused in frame at the position right outside the transmembrane domain of Crb. (c) dArf6KO knockout targeting. Dotted bar indicates the targeted deletion (2.158 kb). In both b and c, shaded or solid boxes are exons and open boxes are introns (introns and exons are not shown for CG8155, CG8157, and CG8160 in c). Fine dotted lines indicate the homologous recombination event. Solid bars indicate the positions and sizes of diagnostic and verification PCRs (for PCR results please see supplemental Figures 1 and 2). Diagrams are not precisely to scale.
To implement the UAS-Rpr selection, we made a new set of ends-out targeting vectors, pRK1 and pRK2 (Figure 2a) that were based on the integration and modification of pEndsOut2 and pBS70W (available from http://dgrc.cgb.indiana.edu/). Both pRK1 and pRK2 contain a UAS-Rpr module, while pRK2 also has a GMR (Hay et al. 1994)-enhanced w+ marker to further facilitate the recovery of targeting candidates. We made two targeting constructs, a crb∷mEosFPKI knockin construct (Figure 2b) and a dArf6KO knockout construct (Figure 2c), on the basis of pRK1 and pRK2, respectively. Crb is a transmembrane protein essential for developing cell polarity (Tepass et al. 1990). We plan to study the trafficking and dynamics of Crb by tagging it with a photo-convertible fluorescent protein mEosFP (Wiedenmann et al. 2004). dArf6 (Arf51F) is a small GTPase that may play key roles in Drosophila muscle and nervous system development, although no dArf6 mutants are currently available. dArf6KO targeting aims to delete 2.158 kb of the dArf6 locus that includes all the coding exons plus the 3′-UTR (Figure 2c). We obtained multiple transgenic lines from both targeting constructs at normal frequency (supplemental Table 2), indicating that UAS-Rpr in pRK1 and pRK2 was sufficiently silent in the absence of the Gal4 driver and did not adversely affect the routine P-element-based transgenic process. All the transgenic donor lines were larval or pupal lethal when crossed with neuronal-specific drivers Gal4477 and Gal4221 (supplemental Table 2) (Grueber et al. 2003). Gal4221/UAS-Rpr also consistently produced very few adult escapers of a fully penetrated wing inflation phenotype (supplemental Figure 1d).
To evaluate the effectiveness of UAS-Rpr without any bias, we first carried out crb∷mEosFPKI knockin experiments without UAS-Rpr selection (similar to Figure 1a). From ∼5 × 104 screening cross progenies, we recovered 270 male candidates, of which 14 were mapped to the third chromosome where crb is located. We then screened 125 non-third chromosome candidates and all 14 third chromosome candidates for the presence of UAS-Rpr by crossing them into Gal4221. As summarized in Table 1, only 3/125 of the non-third chromosome candidates (i.e., false positives) are Rpr[−],while 11/14 of the third chromosome candidates are Rpr[−] of which 7 were confirmed by PCR to have the correct targeting events (supplemental Figure 1a). Extrapolating from these data, selecting against UAS-Rpr in the screening cross of crb∷mEosFPKI would eliminate >96% (253/263) of false positives (Table 1). In addition, UAS-Rpr selection also eliminates tandem-insertion mutants (Gong and Golic 2003), which can be difficult to distinguish from true targeting candidates by simple PCR assays (supplemental Figure 1, b and c). The homologous recombination frequency of crb∷mEosFPKI is ∼1/7000 if only considering the male candidates.
TABLE 1.
Genetic and PCR analyses of targeting candidates of crb∷mEosFPKI and dArf6KO
| Targeted allele | Targeting candidates | Rpr test | FRT+ | loxP+ | X chr | PCR verified |
|---|---|---|---|---|---|---|
| crb∷mEosFPKI | Non-third chr candidates | Rpr+ 122 | 17/122 | 122/122 | 0/122 | ND |
| Rpr[−] 3 | 0/3 | 3/3 | 0/3 | ND | ||
| Third chr candidates | Rpr+ 3 | 0/3 | 3/3 | — | 2a/3 | |
| Rpr[−] 11 | 0/11 | 11/11 | — | 7/11 | ||
| dArf6KOb | Non-second chr candidates | Rpr+ 120 | 54/120 | ND | 0/120 | ND |
| Rpr[−] 2 | 0/2 | ND | 0/2 | ND | ||
| Second chr candidates | Rpr+ 2 | 0/2 | ND | — | ND | |
| Rpr[−] 0 | — | — | — | — |
Rpr+, scored by lethality or strong wing phenotypes in the presence of Gal4221[w−] or Gal4477[w−]. The total number of Rpr[−] false positives of crb∷mEosFPKI can be estimated as 10 [6 from non-third chromosome candidates (3 × (256/125)], plus 4 from third chromosome candidates). FRT+ or loxP+, scored by eye color variegation in the presence of constitutively expressed FLPase or Cre recombinase. Approximately 87% [(263 − (17 × 2))/263] of crb∷mEosFPKI false positives and ∼57% [(124 − 54)/124] of dArf6KO false positives showed damaged FRT sites. X chr, candidates that were mapped to the X chromosome. Here, none of the nontarget chromosome false positives were mapped to the X chromosome. Since the 4th chromosome is extremely small, therefore unlikely to harbor any nonspecific targeting events, “X chr” data indicate that virtually all of the nontarget chromosome false positives retained their donor DNA on the original chromosome, due to either damaged FRT sites or insufficient excision of donor DNA. ND, not done; —, not applicable.
Tandem insertion mutants.
Only dArf6KO candidates recovered from screening crosses with regular Pin/CyO balancer stock are listed here (see Table 2).
We then decided to carry out a large-scale dArf6KO targeting experiment by taking full advantage of the new reagents and methods described here, as we failed at dArf6KO targeting on the basis of the original pEndsOut2 vector by screening ∼1.6 × 105 screening cross progenies (W. Zhou and Y. Hong, unpublished results). We recloned the same 5′ and 3′ homologous arms into the pRK2-based vector. By using 6935-hid to set up the targeting cross, we easily collected >2 × 104 virgin females. Twelve thousand of them were mated with w/Y; Pin/CyO; Gal4221[w−] males to set up the screening cross (Figure 1d). From >7 × 105 screening cross progenies (Table 2) we recovered 315 w+ males, of which 5 were verified as specific targeting candidates by PCR (Table 2, supplemental Figure 2, a and b). As a control, 200 virgin females from the same targeting cross were crossed with regular w/Y, Pin/CyO males. Of 124 w+ male candidates recovered, 1.6% (2/124) were Rpr[−], but none harbored the true targeting event (Tables 1 and 2). Thus, UAS-Rpr selection achieved an impressive ∼60-fold reduction of false positives. Effectively, dArf6KO targeting was accomplished at a scale equivalent to screening/mapping >18,000 (315 × 60) preliminary candidates in the absence of UAS-Rpr selection. In addition, the dramatically reduced number of preliminary candidates, combined with their dark-red eye color due to GMR-enhanced w+ expression in pRK2, made the screening process much easier and faster. The homologous recombination frequency of dArf6KO can be estimated as ∼1/140,000 if only considering the male candidates. Homozygotes of dArf6KO are viable but are male and female sterile, so it is possible that dArf6 only plays a specific and indispensable role in germline development. When we were preparing this article, a P-element-induced deletion allele of dArf6 was published and Dyer et al. (2007) observed the same male and female sterile phenotypes in their homozygous dArf6 mutant flies.
TABLE 2.
dArf6KO targeting with and without using UAS-Rpr as a negative selection marker
| Screening cross set up | Progenies screened | Male candidates | Second chr candidates | Rpr+ | FRT+ | PCR verified |
|---|---|---|---|---|---|---|
| (×) w/Y; Pin/CyO; Gal4221[w−] | >7 × 105 | 315 | 30/315 | — | ND | 5/30 |
| (×) w/Y; Pin/CyO | ∼7300 | 124 | 2a/124 | 122/124 | 54/124 | ND |
chr, chromosome; ND, not done; —, not applicable.
These two candidates harbored nontargeted integration of donor DNA on the second chromosome and were Rpr+.
Compared with the “rapid scheme” in which preliminary candidates were screened for the loss of FRT sites (Rong et al. 2002; Gong and Golic 2003), UAS-Rpr selection is more efficient since it directly eliminates false positives. In addition, we found that the majority of the false positives (57–87%) had damaged FRT sites (Table 1); therefore, they could only be eliminated by UAS-Rpr selection but not by the FRT test. Since the I-SceI sites are positioned rather close to the FRT sites in ends-out targeting constructs, we speculate that the frequent FRT damage seen here was most likely due to the double-strand DNA repair process triggered by the premature cut of I-SceI sites (Bellaiche et al. 1999; Gong and Golic 2003). Separating FRT and I-SceI sites further away in future pRK-based targeting vectors should further reduce the frequency of false positives. Consistently, the Golic lab reported that the frequency of false positives was low using the pW25 targeting vector in which FRT and I-SceI were separated by 100–150 bp (Gong and Golic 2004). Since pRK-based vectors may not be suitable for making Gal4 knockin alleles, pW25 series vectors should be excellent alternatives.
In summary, for a targeting experiment with an expected homologous recombination frequency of ∼1/100,000 gametes, we estimate that our 6934-hid/6935-hid stocks and UAS-Rpr selection reduced the work load of genetic crosses to a level comparable to a routine P-excision experiment. In addition, our new targeting vectors, such as pRK2, should significantly facilitate the molecular cloning and transgenesis of targeting constructs due to the enhanced multiple cloning sites and w+ expression. Overall, these new reagents and methods should significantly increase the success rate of targeting experiments on genes that are resistant to homologous recombination.
Acknowledgments
We are grateful to Jeff Sekelsky for pBS70W and pEndsOut2 plasmids, Leon Perniciaro for help in screening in dArf6KO targeting candidates, Ulrich Nienhaus for EosFP constructs, Fabrice Rogers for hs-hid stocks, and Fen-Biao Gao, Sige Zou, Peizhang Xu, and Koen Venken for comments on the manuscript. Y.N.J is an investigator of the Howard Hughes Medical Institute. pRK1 and pRK2 will be donated to the Drosophila Genomic Resource Center (DGRC) and 6934-hid and 6935-hid stocks will be donated to the Bloomington Stock Center. This work is supported by start-up funds from the University of Pittsburgh School of Medicine (Y.H.).
References
- Bellaiche, Y., V. Mogila and N. Perrimon, 1999. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics 152 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, N., E. Rebollo, P. Dominguez, N. Elkhatib, P. Chavrier et al., 2007. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development 134 4437–4447. [DOI] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2004. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics 168 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether, M. E., J. M. Abrams, J. Agapite, K. White and H. Steller, 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9 1694–1708. [DOI] [PubMed] [Google Scholar]
- Grueber, W. B., L. Y. Jan and Y. N. Jan, 2003. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112 805–818. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120 2121–2129. [DOI] [PubMed] [Google Scholar]
- Jones, W. D., P. Cayirlioglu, I. G. Kadow and L. B. Vosshall, 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445 86–90. [DOI] [PubMed] [Google Scholar]
- Larsson, M. C., A. I. Domingos, W. D. Jones, M. E. Chiappe, H. Amrein et al., 2004. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43 703–714. [DOI] [PubMed] [Google Scholar]
- Manoli, D. S., M. Foss, A. Villella, B. J. Taylor, J. C. Hall et al., 2005. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436 395–400. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass, U., C. Theres and E. Knust, 1990. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 61 787–799. [DOI] [PubMed] [Google Scholar]
- White, K., E. Tahaoglu and H. Steller, 1996. Cell killing by the Drosophila gene reaper. Science 271 805–807. [DOI] [PubMed] [Google Scholar]
- Wiedenmann, J., S. Ivanchenko, F. Oswald, F. Schmitt, C. Rocker et al., 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 101 15905–15910. [DOI] [PMC free article] [PubMed] [Google Scholar]