Abstract
Miniature inverted-repeat transposable elements (MITEs) are a particular type of defective class II elements present in genomes as high-copy-number populations of small and highly homogeneous elements. While virtually all class II transposon families contain non-autonomous defective transposon copies, only a subset of them have a related MITE family. At present it is not known in which circumstances MITEs are generated instead of typical class II defective transposons. The ability to produce MITEs could be an exclusive characteristic of particular transposases, could be related to a particular structure of certain defective class II elements, or could be the consequence of particular constraints imposed by certain host genomes on transposon populations. We describe here a new family of pogo-like transposons from Medicago truncatula closely related to the Arabidopsis Lemi1 element that we have named MtLemi1. In contrast to the Arabidopsis Lemi1, present as a single-copy element and associated with hundreds of related Emigrant MITEs, MtLemi1 has attained >30 copies and has not generated MITEs. This shows that a particular transposon can adopt completely different strategies to colonize genomes. The comparison of AtLemi1 and MtLemi1 reveals transposase-specific domains and possible regulatory sequences that could be linked to the ability to produce MITEs.
TRANSPOSABLE elements can be divided in two classes according to their structure and transposition mechanism. Class I elements transpose by a replicative mechanism invoking an RNA molecule that is reverse transcribed prior to integration, while class II elements are mobilized by a cleavage and strand-transfer mechanism usually known as “cut and paste.” Transposition of class II elements is catalyzed by a transposase, which is usually encoded by the transposon itself. However, defective class II elements, conserving the transposon structure but lacking a transposase-coding capacity, also exist. These are usually mutation derivatives of their autonomous counterparts with which they show extensive sequence similarities. Each defective element is usually unique and presents specific mutations. Defective elements can be mobilized in trans by related transposases as long as they contain the sequence motifs recognized by the transposase that are essential for mobilization (e.g., terminal inverted repeats, or TIRs, and, in some cases, subterminal repeated sequences).
Miniature inverted-repeat transposable elements (MITEs) are a particular type of defective transposons. MITEs contain TIRs and can be mobilized by a cut-and-paste mechanism (Kikuchi et al. 2003), and some of them have been shown to be deletion derivatives of autonomous class II transposons (Feschotte and Mouches 2000). It has been shown that transposases of related class II transposons specifically bind to their TIRs (Feschotte et al. 2005; Loot et al. 2006) and mobilize them (Dufresne et al. 2007; Miskey et al. 2007; Yang et al. 2007). Nevertheless, in contrast to typical class II defective elements, in which each defective copy is unique, MITEs exist as populations that are highly homogeneous in size and sequence. Moreover, MITEs are usually present at a high copy number in genomes while typical class II defective elements are usually present at low or moderate copy number. Phylogenetic analyses of MITE populations have shown that these elements are probably generated by a burst of amplifications from a single or very few elements (Santiago et al. 2002), and it has been proposed that they are generated by a still unknown replicative mechanism (Feschotte et al. 2002; Casacuberta and Santiago 2003). At present it is not known in which circumstances MITEs are produced. At least three different hypotheses can be put forward. First, the possibility of amplification could be related to a particular structure of certain defective class II elements. This would imply a two-step process, in which a subset of typical defective class II elements bearing particular characteristics (e.g., small size) would be subsequently amplified (Feschotte et al. 2002; Casacuberta and Santiago 2003). Second, the ability to produce MITEs could be an exclusive characteristic of particular transposases that would generate these elements instead of typical class II defective elements. And third, the production of MITEs instead of typical defective class II elements could be the consequence of particular constraints imposed by certain host genomes on transposon populations, and not be related to particular transposon or transposase structures.
The Emigrant MITE is present in >100 copies in the genome of Arabidopsis (Casacuberta et al. 1998; Santiago et al. 2002). Some Arabidopsis ecotypes also contain a single-copy pogo-like element called Lemi1 (Feschotte and Mouches 2000; Loot et al. 2006). Lemi1 and Emigrant show extensive sequence similarity, suggesting that Emigrant was generated by an internal deletion of Lemi1 (Feschotte and Mouches 2000).We have recently shown that the proteins encoded by Lemi1 specifically bind Emigrant TIRs and subterminal regions, suggesting that Lemi1 provides the enzymatic activities for Emigrant mobilization (Loot et al. 2006).
To obtain new insight into Lemi1/Emigrant transposition and evolution, we searched for the presence of both elements in several plant genomes. We found that the genome of Medicago truncatula contains a putative transposon whose sequence is highly similar to that of the Arabidopsis Lemi1 element, and we have named it MtLemi1. Interestingly, the MtLemi1 element is present in >30 copies in M. truncatula, and some of these copies have deletions of their internal regions and likely represent Lemi1-defective elements. On the contrary, M. truncatula does not contain short Lemi1-related elements that could represent Emigrant MITEs. These results suggest that Lemi1 evolved differently in both genomes, generating MITEs in Arabidopsis and typical class II defective elements in M. truncatula. We compare the structures of MtLemi1 and AtLemi1 to obtain new insight into MITE generation and amplification.
MATERIALS AND METHODS
DNA extraction, PCR amplification, cloning, and sequencing:
DNA from M. truncatula accessions and Arabidopsis and Brassica species was extracted as described (Dellaporta et al. 1984). Loci containing MtLemi1 insertions were amplified by PCR using primers corresponding to their flanking genomic sequences. For MtLemi1-18, the primers were MtLemi1-18-FW2 (5′-CACTCAAATGCTTTGCTCCA-3′) and MtLemi1-18-rev2 (5′-GTTTAAAACCGGCAAGGTGA-3′); for MtLemi1-4, the primers were MtLemi1-4-FW (5′-GGGTTTGGGCATCAACTTTAGAGT-3′) and MtLemi1-4-rev (5′-CGTGGACCGATAATAACATCTTT-3′); for the MtLemi1-8, the primers were MtLemi1-8-FW (5′-GCCCATAATGCTTGAGGTGT-3′) and MtLemi1-8-rev (5′-TTGTTGCGTTGGTTATTGGA-3′). The transposase-encoding region of BnLemi1 and AaLemi1 elements was amplified using the Lemi1#630 (5′-TCAGCAAAATTTCCAGAGATG-3′) and Lemi1#1629 (5′-TGCATCATTTTCACCAGGATA-3′) primers. Lemi1-defective elements and Emigrant-like elements from Brassica napus (Bn), Arabidopsis arenosa (Aa), Arabidopsis thaliana (At), and M. truncatula (Mt) were amplified using the 5′TIR1 (5′-TACAGTAAAACCTCTATAAATTAATA-3′) primer. PCR fragments were cloned using the TOPO TA cloning kit (Invitrogen) and were sequenced using standard procedures.
Bioinformatic analysis:
The sequences of AtLemi1 (Loot et al. 2006) were used to search MtLemi1-related elements in the NCBI database (http://www.ncbi.nlm.nih.gov/) and in the M. truncatula resources website (http://www.medicago.org/genome/).
Multiple alignments were performed using ClustalW (Thompson et al. 1994) and alignments were manually refined. Synonymous (Ks) and nonsynonymous (Ka) substitutions were calculated using the DnaSP program (Rozas et al. 2003).The analysis of the insertion site specificity was performed using weblogo (http://weblogo.berkeley.edu/ aligments). The analysis of MITEs using the TRANSPO program (Santiago et al. 2002) was done at http://alggen.lsi.upc.es/ using the Mt2.0 release (January 2008) of the M. truncatula genome made accessible by the Medicago Genome Sequencing Consortium at http://www.medicago.org. The program searched for the presence of Emigrant TIR sequences (CAGTAGAAACTCTATAAATTAAT) separated by >200 nt and <600 nt and allowing three mismatches per TIR.
RESULTS
In contrast to Emigrant MITEs, Lemi1 has not transposed in the recent past in Arabidopsis:
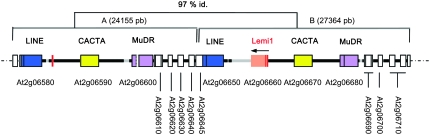
We have recently reported that some Emigrant insertions are polymorphic among different Arabidopsis ecotypes, or even among different individuals of the same ecotype, suggesting that Emigrant MITEs have recently transposed during Arabidopsis evolution (Loot et al. 2006). In contrast, a preliminary analysis did not give any indication of recent mobility of their potentially autonomous counterpart, the Lemi1 element (Loot et al. 2006). Lemi1 is present in only one copy in most Arabidopsis ecotypes, although PCR and Southern analysis suggested that some ecotypes could be devoid of Lemi1 (our unpublished results). To obtain new insight into possible Lemi1 mobility in the recent past, we have analyzed the genomic region of the Arabidopsis Columbia-0 ecotype that contains the Lemi1. In the Columbia-0 ecotype of Arabidopsis, Lemi1 lies in chromosome 2 in a region containing other mobile elements (Figure 1). A detailed analysis revealed that this region (Figure 1, region B) has been duplicated in tandem during Arabidopsis evolution. The duplicated region contains a LINE, a CACTA, and a MuDR transposon. It also includes a 5′ short fragment of the Lemi1 element and its 5′ flanking sequence, but not the rest of the Lemi1 element and its 3′ flanking sequences (Figure 1, region A). The most parsimonious hypothesis for the evolution of this region is that one of the two copies of Lemi1 was severely deleted after the duplication of the genomic region.
Figure 1.—
Structure of AtLemi1 locus in Arabidopsis Columbia-0 ecotype. Schematic of the two copies (A and B) of the duplicated region. The size of each copy is shown in parentheses and the percentage of identity of the shared sequences is also shown at the top. Genes are shown as open boxes, and mobile elements are shown as solid boxes. Dark colors refer to duplicated sequences and lighter colors refer to nonduplicated/deleted sequences. The accession numbers are shown below each gene/mobile element. The orientation of the Lemi1 element is indicated by an arrow.
This traces back the possible mobility of the single Lemi1 copy present in the Arabidopsis Columbia-0 genome to a time preceding the duplication of this genomic region and suggests that, contrary to its associated Emigrant elements, Lemi1 has not transposed during the recent evolution of the Arabidopsis genome. We have previously shown that although Lemi1 and Emigrant elements share the same TIRs, Lemi1 does not maintain all the transposase-binding sites present in Emigrant subterminal regions (Loot et al. 2006). A different binding to the subterminal regions could modify the final protein/DNA structure and explain the different transposition capacity of the Arabidopsis Lemi1 and Emigrant elements.
M. truncatula contains a pogo-like transposon highly similar to Lemi1:
To obtain new insight into the Lemi1/Emigrant evolution, we looked for the presence of similar elements in the databases. Most plant sequenced genomes, such as rice (Feschotte et al. 2003) and grapevine (A. Benjak, A. Fornek and J. M. Casacuberta, unpublished results), are devoid of Lemi1 elements, although rice contains some 34 Mariner-like elements of other families (Feschotte et al. 2003). On the contrary, we have found an important number of Lemi1-related sequences in M. truncatula. We thus decided to analyze these sequences in more detail. Eleven of these sequences contain an almost intact long Orf potentially coding for a pogo-like transposase and are flanked by short TIRs and a TA dinucleotide that could correspond to the target-site duplication generated upon insertion. These sequences could thus correspond to a M. truncatula pogo-like DNA transposon.
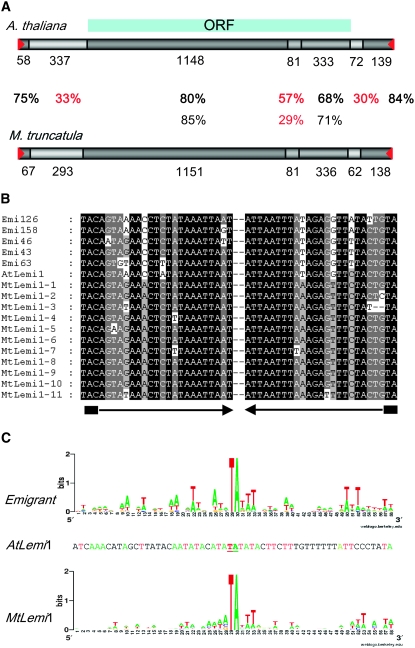
We have derived a consensus sequence from the 11 elements and compared it to the Arabidopsis Lemi1 sequence. Both sequences are highly similar, especially within the TIR and subterminal regions and within most of the coding region (Figure 2A). The putative transposases are 80% identical overall, with long stretches (384 aa) of >85% identity (80% in 1150 bp at the DNA level) (Figure 2A). This level of conservation seems surprisingly high for a transposase from two genomes separated by >90 MY. We thus decided to analyze the conservation of other M. truncatula transposons with respect to their Arabidopsis counterparts. We have not found any Arabidopsis sequence having a significant similarity at the nucleotide level to the recently described M. truncatula PIF elements (Grzebelus et al. 2007) or any M. truncatula sequence having significant similarity to the mudrA-like coding sequence of the Arabidopsis MuDR element (At2g11560). However, we have found M. truncatula sequences having significant similarity to the sequences of the Arabidopsis CACTA elements CACTA1 (AB052792) and CAC5 (AB095515) (not shown), although the level of similarity (65% identity in 471 bp and 66% identity in 533 bp, respectively) is well below of that of the Lemi1-related sequences.
Figure 2.—
Comparison of Lemi1-related element sequences in A. thaliana and M. truncatula genomes. (A) Schematic of the Arabidopsis lemi1 element and a consensus sequence derived from the Lemi1-related sequences obtained from M. truncatula. The degree of sequence identity of the different regions is indicated as a gray scale from light gray (low sequence similarity) to dark gray (high sequence similarity), and the percentage of sequence identity for each region is shown between the two schematics in boldface (at the DNA level) and roman numbers (at the protein level). The sequence length of each region is shown below each box. Terminal inverted repeats are indicated with red triangles. The region containing the transposase open reading frame is represented by the blue box at the top. (B) Alignment of the TIR sequences of five Emigrant elements and the Lemi1 element from Arabidopsis, and the 11 full-length Lemi1-related elements from M. truncatula. TIRs are shown by solid arrows and TSD is indicated with a solid box. (C) Analysis of the preinsertion locus of Emigrant and Lemi1 elements from Arabidopsis and the Lemi1-related elements from M. truncatula. The weblogo representation of the nucleotide frequency (letter size is proportional to its frequency) in each of the 30 nt flanking both sides of the insertions of the 19 elements belonging to the young Emigrant A2 subfamily (Loot et al. 2006) and the 11 potentially full-length Lemi1-related elements from M. truncatula is shown. The flanking sequences of the single Lemi1 element from Arabidopsis are also shown for comparison, using the same code color for the four nucleotides that is used in the weblogo schemes.
The evolutionary rate of coding sequences depends on different factors. The main one is the function, which conditions different constraints. For example, genes coding for RNA-binding proteins (e.g., splicing factors and ribosomal proteins) tend to be particularly well conserved (Alba and Castresana 2005). Thus, to obtain a reference for protein sequence conservation between Arabidopsis and M. truncatula, we have compared the sequences of the putative ribosomal protein L30 and the putative PRP8 splicing factor from these two plants and found that they are 80 and 86% identical, respectively (not shown). This level of conservation is similar to that of the putative transposases encoded by Lemi1 and the M. truncatula Lemi1-related sequences, which show an overall identity of 80% at the protein level.
Transposases bind to the transposase-binding sites located in the TIRs and subterminal regions, and thus closely related transposases should be linked to similar terminal sequences. Indeed, these regions are 75–84% identical in both elements (Figure 2A), and the sequence of the TIRs of the M. truncatula elements are highly similar to those of Arabidopsis Lemi1 and the related Emigrant MITEs (Figure 2B). Moreover, as transposases catalyze the cleavage of the target site, similar transposases should have similar insertion specificity. We thus decided to analyze if the M. truncatula elements target sequences similar to those of the Arabidopsis Lemi1 and the related Emigrant MITEs for integration. We first analyzed the sequence of the preinsertion sites of the Arabidopsis Emigrant elements belonging to the young EmiA2 subfamily, which has been shown to have transposed recently during Arabidopsis evolution (Santiago et al. 2002). Emigrant elements have a strict insertion specificity for a TA dinucleotide, which will be duplicated flanking the element upon integration (Figure 2C). In addition, Emigrant elements show a preference for integration in A/T-rich regions and also tend to integrate in sequences having a T in positions +1 and +3 (Figure 2C). The insertion sequence of the single-copy Lemi1 element follows the consensus for the Emigrant target sequence, in agreement with the hypothesis that both Lemi1 and Emigrant elements are mobilized by the Lemi1-encoded transposase (Loot et al. 2006). This target-site preference is similar to that of other Tc1/mariner elements, such as the Sleeping Beauty transposon that targets the palindromic AT-repeat ATATATAT, although target-site selection seems to be determined primarily on the level of DNA structure and not by specific base-pair interactions (Vigdal et al. 2002).The 11 M. tuncatula elements are also flanked by a TA nucleotide, suggesting that they target this dinucleotide for insertion with high specificity. The analysis of 30 nt upstream and downstream from the insertions suggests that, as it is the case for the Arabidopsis Lemi1 and Emigrant elements, these elements also target regions with a high A/T content and that they have a preference for a T at positions +1 and +3 (Figure 2C). In summary, the M. truncatula pogo-like elements described here are highly similar to the Arabidopsis Lemi1 element. They have similar TIR sequences, encode extremely similar putative transposases, and target similar sequences for integration. All these data suggest that both elements derive from the same Lemi1 ancestral transposon and can be considered as the same transposon evolving in two different genomic contexts. We have thus decided to name them M. truncatula Lemi1 element (MtLemi1) and A. thaliana Lemi1 element (AtLemi1).
MtLemi1 has recently transposed in M. truncatula:
We analyzed the possible mobility of MtLemi1 by two different approaches. First, we looked for evidences of recent transposition during Medicago evolution by looking for possible related empty sites (RESites) in the genome of M. truncatula that could be the result of the transposition of a MtLemi1 element to a previously duplicated genomic region. Second, we looked for possible insertion polymorphisms of a number of MtLemi1 elements among three different M. truncatula genotypes. We found RESites for a number of MtLemi1 elements. In most of the cases, as for MtLemi1-6 and MtLemi1-11, the sequence of the RESite is almost identical to the one of the theoretical empty site, suggesting that the corresponding MtLemi1 element was not present in the recipient sequence prior to duplication and that the RESites are indicative of an insertion polymorphism among the duplicated sequences (supplemental Figure 1).
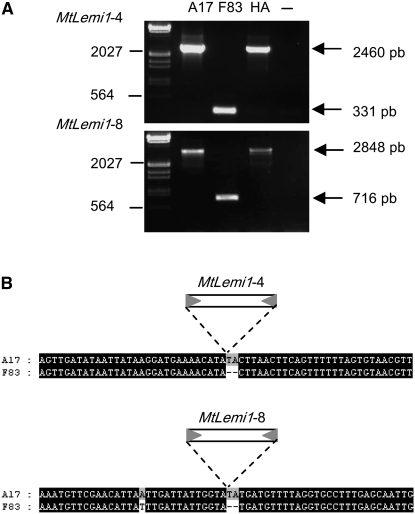
To look for more direct evidences of recent MtLemi1 mobility, we amplified by PCR the 11 loci containing MtLemi1 elements in three different M. truncatula genotypes, the sequenced A17 genotype and two other genotypes (F83 and HA) known to be polymorphic with respect to A17 (M. Crespi, personal communication). In two cases (MtLemi1-4 and MtLemi1-8), the band amplified from F83 was smaller than the band amplified from the other two genotypes, which could be indicative of a MtLemi1 insertion polymorphism (Figure 3A). We have sequenced the DNA fragment amplified from the F83 genotype, which perfectly matches one of the theoretical empty sites (Figure 3B), suggesting that both MtLemi1-4 and MtLemi1-8 inserted in the genome recently, accompanying the differentiation of M. truncatula in different genotypes.
Figure 3.—
MtLemi1 insertion polymorphisms. (A) Amplification of two loci containing an MtLemi1 insertion in M. truncatula genotype A17 in three different accessions. PCR amplification with oligonucleotides flanking the two MtLemi1 elements was performed with DNAs obtained from the indicated M. truncatula genotypes. Control amplifications without DNA are indicated by a dash. (B) Alignment of the 30 bases upstream and downstream from the two MtLemi1-4 and MtLemi1-8 and the corresponding sequences amplified from the F83 M. truncatula accession. The position of the corresponding MtLemi1 element is indicated.
M. truncatula contains defective MtLemi1 elements but not MtLemi1-related MITEs:
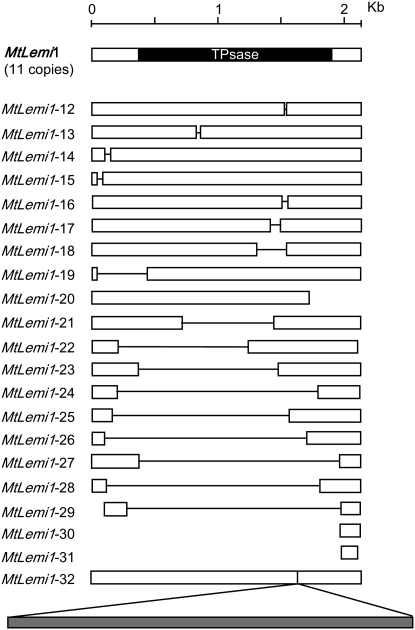
In addition to the 11 full-length MtLemi1 elements, the genome of M. truncatula also contains sequences showing limited sequence homology to AtLemi1. We have analyzed these sequences in detail to determine if they corresponded to defective MtLemi1 elements or Emigrant-like MITEs. Most of these sequences correspond to MtLemi1 elements containing internal deletions of variable length, a few of them consist only of partial 5′ or 3′ MtLemi1 sequences, and one of them consists of a complete MtLemi1 element containing an insertion of 2907 nt previously annotated as a solo-LTR, which probably corresponds to a gypsy-like retrotransposon (Figure 4). A complete list of MtLemi1 elements and their coordinates is given in supplemental Table S1. The structure of these 21 incomplete MtLemi1 elements is unique, the size and the position of the deletions/insertions being specific for each element. This is a characteristic of typical defective class II elements that allows discriminating them from MITEs. In addition, each of these elements is present as a single copy in M. truncatula and all together form a moderate-copy-number family of repetitive elements. These MtLemi1-related elements thus have all the characteristics of typical class II defective elements, not those of MITEs. Additional blast searches with Emigrant sequences or with the noncoding fraction of the MtLemi1 element failed to detect any other MtLemi1-related sequence that could correspond to a M. truncatula MITE family related to MtLemi1. As an alternative approach to searching for Lemi1-related MITEs, we used the program TRANSPO that searches a defined sequence (e.g., the available M. truncatula genome sequence) for the presence of particular TIRs (e.g., Lemi1/Emigrant TIRs) separated by a defined sequence length irrespective of its sequence, allowing the detection of any MITE subfamily that would have conserved the TIRs but not the internal sequence (Santiago et al. 2002). This approach identified only three short MtLemi1-related elements containing different deletions that were already included in our analysis and failed to detect any Lemi1-related MITE family.
Figure 4.—
Structure of the MtLemi1 elements. A scheme representing the structure of the 11 full-length MtLemi1 copies, with a solid box indicating the position of the region potentially encoding the transposase, shown at the top together with a reference scale. Schemes representing each of the 21 Mtlemi1-defective elements described here are shown below. Open boxes indicate conserved sequences while deletions are indicated by horizontal lines. The insertion of a retroelement-related sequence in MtLemi1-32 is shown as a shaded box.
We thus conclude that, in contrast to what is found in Arabidopsis, the M. truncatula Lemi1 element has produced MtLemi1-defective elements but not MITEs.
MtLemi1 contains specific domains:
The alternative production of typical defective elements or MITEs could be the consequence of particular characteristics of the AtLemi1 and MtLemi1 elements. We thus looked for possible differences between AtLemi1 and MtLemi1. In spite of their overall very high level of identity, MtLemi1 and AtLemi1 have three short regions that are less conserved (percentage of similarities shown in red in Figure 2A). Two of these regions flank the coding region, upstream and downstream, and the third is located within the coding region (Figure 2A). The high variability of the region downstream of the coding region could be explained by the absence within this region of important functional elements. Indeed, this region precedes the TIRs and subterminal regions known to contain transposase-binding sites (Loot et al. 2006). The variable sequence preceding the coding region does not contain transposase-binding sites either and could also be devoid of functionality. Nevertheless, most plant promoters are located immediately upstream from the transcribed regions and thus it is reasonable to think that this region could contain the transposase promoter. In this case, the high sequence variability of this region may suggest a different pattern of expression for the transposases of AtLemi1 and MtLemi1.
The third variable region is a 81-nt-long sequence located within the coding region that coincides with a short sequence that is deleted in some Arabidopsis ecotypes (Loot et al. 2006). This region is only 57% identical between AtLemi1 and MtLemi1, while the rest of the coding region is 80% (in the first 1148 nt) and 68% (in the last 333 nt) identical. Interestingly, this difference in sequence identity is even more evident at the protein level. While the two putative transposases are 80% identical overall, their N-terminal and C-terminal domains being 85 and 71% identical, the level of identity drops to 29% in this short region. This lower conservation at the protein level could be an indication of low selective constraints or, alternatively, of positive selection. We have thus decided to analyze the nonsynonymous-to-synonymous rate ratio along the coding region of the AtLemi1 of the Arabidopsis Coimbra1 ecotype, one of the Arabidopsis ecotypes that contains a complete Lemi1 element (Loot et al. 2006), and the MtLemi1-7 element, one of the MtLemi1 elements with transposase-coding capacity. While the first 1152 nt and the last 339 nt of the coding region show a low Ka/Ks ratio (0.13 and 0.27, respectively) indicative of negative purifying selection, the short 81-nt variable region has a Ka/Ks ratio well over 1 (2.32) that could indicate positive selection for this region. Similar values were found when comparing other MtLemi1 complete elements. Nevertheless, we could not confirm that this region has been subjected to positive selection using maximum-likelihood methods such as PAML (Yang 2007; data not shown).
Analysis of the Lemi1/Emigrant system in other plant species:
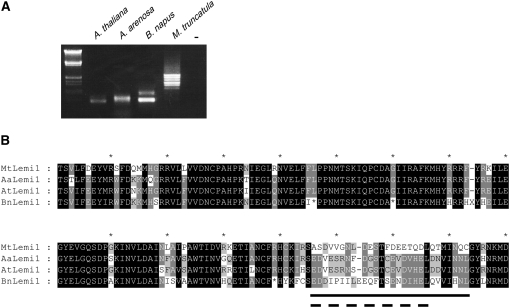
To investigate whether the production of MITEs correlates with the presence in the Lemi1 transposase of particular sequence motifs, we analyzed the Lemi1/Emigrant system in other species. Previous results suggested that different Brassica species also contain Emigrant-related MITEs (Casacuberta et al. 1998), which could indicate that the ability of the Arabidopsis Lemi1 to produce Emigrant MITEs is shared by its related Lemi1 elements in other genomes of the Brassicaceae group. To confirm this hypothesis, we have investigated the possible presence of Emigrant-like elements or, alternatively, Lemi1-defective elements in B. napus and A. arenosa, by PCR amplification with oligonucleotides corresponding to the Lemi1/Emigrant TIRs using Arabidopsis and M. truncatula as controls. Interestingly, while the amplification from M. truncatula reveals several bands of different mobility corresponding to different MtLemi1-defective elements, the amplification from B. napus and A. arenosa produces only small-size bands, similar to the one obtained from Arabidopsis (Figure 5A). Sequencing of these bands indeed confirmed that they correspond to Emigrant-like elements (not shown). This confirms that the ability to produce MITEs is shared by the Lemi1 elements of different Brassica species and that these genomes do not contain typical Lemi1-defective elements similar to those present in M. truncatula.
Figure 5.—
The Lemi1/Emigrant system in different species. (A) PCR amplification of Lemi1-defective elements from Arabidopsis Columbia-0 ecotype (A. thaliana), A. arenosa, B. napus, and M. truncatula. A control amplification without DNA (−) is also included. (B) Comparison of different Lemi1 transposase sequences. Alignment of a C-terminal region of the deduced Lemi1 transposase sequence from M. truncatula (MtLemi1), A. arenosa (AaLemi1), A. thaliana (AtLemi1), and B. napus (BnLemi1) sequence. Gaps are shown and STOP codons are indicated by a star. The most divergent domain is shown by a solid line and the region deleted in some Arabidopsis ecotypes is shown by a dashed line.
We then asked if the transposase encoded by the Lemi1 elements present in B. napus and A. arenosa resembles that of AtLemi1 or that of MtLemi1, especially in the short domain of 27 amino acids characteristic of these two elements. We have amplified by PCR most of the Lemi1 transposase-coding region from B. napus and A. arenosa and compared the deduced protein sequences with those of AtLemi1 and MtLemi1 elements. An alignment of the four protein sequences shows that the transposases encoded by all these four elements are highly similar except for the short region of 27 amino acids already found to be different between AtLemi1 and MtLemi1 (underlined in Figure 5B). Within this region, the two Arabidopsis Lemi1 sequences are almost identical (93% identity), while both sequences are completely different from the one of M. truncatula (29% identity). The sequence obtained from B. napus shows an intermediate degree of conservation (47%). This sequence overall is less conserved and contains several STOP codons, suggesting that it represents an older and inactivated Lemi1 element. Nevertheless, BnLemi1 maintains almost invariably the final part of the sequence (Figure 5). While B. napus, A. arenosa, and A. thaliana are closely related species and their similarity within the variable region could be the consequence of their phylogenetic relationship, the sequence of this short motif seems also to accompany the ability of Lemi1 to produce MITEs and could imply a role for this transposase domain in the production of MITEs.
DISCUSSION
An important fraction of the genome of M. truncatula has been sequenced and is currently available. However, although a handful of M. truncatula transposons have been described (Charrier et al. 1999; Jurka 2000; Holligan et al. 2006; Vitte and Bennetzen 2006; Grzebelus et al. 2007; Macas and Neumann 2007) to date, most of them have not been fully characterized. Here we present a global analysis of a pogo-like family of transposons, closely related to the AtLemi1 element that we have named as MtLemi1. The MtLemi1 family comprises at least 11 potentially full-length elements and 21 defective elements. Some of these elements have probably retained the capacity to transpose, as we have found examples of insertion polymorphisms among M. truncatula genotypes. The Tc1/mariner superfamily of transposons is widespread in eukaryotes and, in particular, in plants (Feschotte et al. 2003). However, the Lemi1 family, which belongs to the pogo subgroup of the Tc1/mariner superfamily, seems more restricted. It is present in only one copy in most A. thaliana ecotypes (Feschotte and Mouches 2000; Loot et al. 2006), and here we show that it is also present in other Brassicaceae species, but it is not present in other sequenced genomes such as rice (Feschotte et al. 2003). We have found sequences related to Lemi1 in several Solanaceae as well as in cotton and grapevine (not shown) but they probably represent very old and mutated elements. The only plant genome in which we have been able to find potentially complete Lemi1 elements is M. truncatula. The level of sequence similarity between MtLemi1 and AtLemi1 is extremely high and unexpected for a transposase from two genomes separated by >90 MY. This could suggest a strong selective pressure on Lemi1 transposase indicative of an important role for the host maintenance. Alternatively, this result could also indicate a horizontal transfer of the Lemi1 transposon during evolution. Although most DNA transposon superfamilies are shared between different eukaryotic supergroups, and some are closely related to prokaryote insertion sequences, which suggests that they are old components of genomes and have been transmitted vertically, examples of horizontal transfer have been reported (Feschotte and Pritham 2007). The recent report of the horizontal transfer of a Mutator-like element between two grass lineages shows that plant transposons can also be transferred between different plant species (Diao et al. 2006). A more in-depth analysis of the presence of the Lemi1 transposon in different plants will be needed to discriminate between these two hypotheses. In any case, the very high similarity of the transposase and the structure of the Lemi1 element in these two plants allows comparing the evolutionary dynamics of these two elements as if it were a single element within two different genomic contexts.
The work presented here shows that Lemi1 has attained a moderate copy number in M. truncatula and, remarkably, that it is not associated with Emigrant MITEs. Moreover, both the potentially autonomous and the defective MtLemi1 elements have moved recently and may have retained the capacity to transpose. On the contrary, the AtLemi1 element is present as a single copy in most Arabidopsis ecotypes, it has not transposed in recent times, and it is associated with >100 Emigrant MITEs that have transposed recently during Arabidopsis evolution (Casacuberta et al. 1998; Santiago et al. 2002). Moreover, our results show that these characteristics are shared by the Lemi1 element present in other Arabidopsis or Brassica species. These results demonstrate that a particular transposon, the Lemi1 pogo-like element in this case, may adopt completely different strategies to colonize different genomes. More precisely, Lemi1 itself does not proliferate in Arabidopsis but produces hundreds of Lemi1-related Emigrant elements, while it produces typical defective elements and attains >30 copies in M. truncatula.
Most class II transposons generate defective elements that frequently are deletion derivatives of their autonomous counterparts. This seems to be a relatively frequent phenomenon, which is supposed to be the consequence of abortive gap repair (Rubin and Levy 1997) or slip mispairing during double-strand break repair (Yan et al. 1999; Conrad et al. 2007) upon excision. On the contrary, the generation of MITEs seems to be a relatively rare phenomenon, and the underlying mechanism is not known. It has been proposed that MITEs are generated by a two-step mechanism in which small defective elements are subsequently subjected to a burst of amplification, giving rise to a high number of highly similar elements (Feschotte et al. 2002; Casacuberta and Santiago 2003). However, nothing is known about the conditions that have to be met for such an amplification to occur. The two-step mechanism predicts that MITEs arise from the amplification of typical class II defective transposons, and thus both types of elements are expected to coexist in the same genome. This is what it is found, for example, for the impala element of Fusarium oxisporium, where defective impala elements coexist with the related mimp1 MITEs (Dufresne et al. 2007). On the contrary, this does not seem to be the case for the Lemi1 element, as only typical defective elements are found in M. truncatula whereas only MITEs are found in Arabidopsis and other Brassicae, indicating that the existence of typical defective elements is not a prerequisite for the production of MITEs. This would suggest that particular transposases, particular genomes, or particular transposase/genome combinations are prone to produce MITEs while most transposases/genomes produce typical defective class II elements.
MtLemi1 and AtLemi1 are highly similar except in three short divergent regions. The divergence of one of them could be explained by a lack of functional elements within it, as this region is located downstream of the coding region and does not contain the transposase-binding sites that are within the TIRs and subterminal regions (Loot et al. 2006). On the contrary, the other two variable regions probably include functional elements and their divergence could have consequences on Lemi1 activity. DNA polymerase II promoters are usually located upstream of the coding sequences and it is thus reasonable to think that the region preceding the Lemi1-coding sequence would contain the transposase promoter. The 337-nt DNA region immediately upstream from the transposase-coding sequence is only 33% identical between AtLemi1 and MtLemi1. Although promoters are not highly conserved among different plant species, this low conservation could also be indicative of a different transcriptional regulation of the transposases of both Lemi1 elements, which could have important implications in Lemi1 functionality. For example, replicative transposition of class II elements depends on the repair of the double-strand break generated upon excision by homology-dependent repair instead of nonhomologous end-joining (NHEJ) mechanisms. Homologous recombination and NHEJ act at different stages of the cell cycle (Lee et al. 1997; Takata et al. 1998), and there is probably a correlation between the pathway used to repair a transposon-induced double-strand break and the cell cycle stage at which the repair takes place (Walisko et al. 2006). Thus, a particular pattern of expression during the cell cycle could modify the ability of a transposase to replicatively amplify short sequences and generate MITEs.
The third variable region between MtLemi1 and AtLemi1 is located within the transposase-coding sequence. Both transposases are 85% identical in most of their sequence, but this identity drops to close to 29% in a short sequence of 27 amino acids. This low sequence conservation could be an indication of a lower selective pressure for this region. However, the sharp difference in sequence conservation between this region and the rest of the sequence could also be an indication of a particular and differentiated function for this region in the two genomes. As an example, it could mediate the interaction of MtLemi1 and AtLemi1 transposases with different proteins.
The Ka/Ks ratio, which is very low for most of the coding sequence but increases to 2.32 for this short region, suggests a high rate of amino acid replacements that could be due to positive selection. This would indicate a functional importance for this region. The analysis of the transposase sequence and the type of associated Lemi1-defective elements in different Brassicae species showed that there is a correlation between particular protein motives within this variable region and the existence of MITEs. Thus, this variable protein region could be involved through its interaction with unknown cellular partners in the production of MITEs.
Transposable elements have a high mutagenic potential and the genomes have developed mechanisms to control them and minimize their impact. This puts transposons under a selective pressure to escape to this control and closes the circle of what has been named a long-term “genetic conflict.” The outcome of this arms race could be the rapid evolution of both transposons and cellular proteins or domains of them, which would be positively selected to allow or, alternatively, to avoid, their mutual interaction. The evidence for positive selection of domains of yeast nonhomologous end-joining proteins have been interpreted in this way, and it has been proposed that these host proteins play a role in the host defense against the proliferation of transposable elements (Sawyer and Malik 2006). The production of a high-copy-number population of MITEs, mobilized by a single-copy element, could be a strategy developed by some transposons to efficiently colonize a genome, avoiding the inactivation by gene silencing mechanisms of the transposase source (Casacuberta and Santiago 2003; Feschotte and Pritham 2007). Under this scenario, host defenses would be under a selective pressure to recognize and inactivate transposases capable of producing MITEs, forcing their interaction domains to rapid evolution. This putative interaction domain has been deleted in the AtLemi1 element of some Arabidopsis ecotypes (Loot et al. 2006), and it is tempting to suggest that this could represent the definitive inactivation of Lemi1 transposase and the end of the genetic conflict between Arabidopsis and the Lemi1/Emigrant invader.
Different genomes can show differences in their permissivity to transposon movement and amplification, and constraints imposed by A. thaliana and M.truncatula to Lemi1 evolution could have forced this element to persist through a different strategy. Thus, in addition to the possible presence of host proteins able to directly interact with the Lemi1 transposase, both genomes may differ in many aspects influencing the regulation of Lemi1. The fact that both A. thaliana and M. truncatula are model plant species that can be easily transformed, and that many different genetic and genomic resources exist for both, should allow the testing of the different hypotheses pointed out here for the generation of MITEs by transforming both plants with chimerical Lemi1 transposons containing specific regions of AtLemi1 and MtLemi1.
In summary, the results presented here show that the Lemi1 transposon has followed different strategies for maintainance in different genomes and open the possibility of analyzing the factors influencing the generation and amplification of MITEs by introducing modified Lemi1/Emigrant elements in the two model plants A. thaliana and M. truncatula.
Acknowledgments
We are grateful to Martin Crespi, Francisco Madueño, and Fabien Nogue for providing us with M. truncatula and A. arenosa material. We are also grateful to Andrej Benjak for his help in bioinformation analysis. We also thank José Castresana for helpful discussions and Elena Casacuberta, Lluïsa Espinás, and Paloma Más for their critical reading of the manuscript. This work was funded by the Spanish Ministerio de Ciencia y Tecnología (grant BIO2003-01211) and the Ministerio de Educación y Ciencia (grant BFU2006-04005) and the Xarxa de Referència en Biotecnologia from the Generalitat de Catalunya.
References
- Alba, M. M., and J. Castresana, 2005. Inverse relationship between evolutionary rate and age of mammalian genes. Mol. Biol. Evol. 22 598–606. [DOI] [PubMed] [Google Scholar]
- Casacuberta, J. M., and N. Santiago, 2003. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311 1–11. [DOI] [PubMed] [Google Scholar]
- Casacuberta, E., J. M. Casacuberta, P. Puigdomenech and A. Monfort, 1998. Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterisation of the Emigrant family of elements. Plant J. 16 79–85. [DOI] [PubMed] [Google Scholar]
- Charrier, B., F. Foucher, E. Kondorosi, Y. d'Aubenton-Carafa, C. Thermes et al., 1999. Bigfoot: a new family of MITE elements characterized from theMedicagogenus. Plant J. 18 431–441. [DOI] [PubMed] [Google Scholar]
- Conrad, L. J., L. Bai, K. Ahern, K. Dusinberre, D. P. Kane et al., 2007. State II dissociation element formation following activator excision in maize. Genetics 177 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S., J. Wood and J. Hicks, 1984. Maize DNA miniprep, pp. 36–37 in Molecular Biology of Plants: A Laboratory Course Manual, edited by R. Malmberg, J. Messing and I. Sussex. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Diao, X., M. Freeling and D. Lisch, 2006. Horizontal transfer of a plant transposon. PLoS Biol. 4 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, M., A. Hua-Van, H. Abd el Wahab, S. B. M'Barek, C. Vasnier et al., 2007. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics 175 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., and C. Mouches, 2000. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol. Biol. Evol. 17 730–737. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., and E. J. Pritham, 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., N. Jiang and S. R. Wessler, 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3 329–341. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., L. Swamy and S. R. Wessler, 2003. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., M. T. Osterlund, R. Peeler and S. R. Wessler, 2005. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 33 2153–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzebelus, D., S. Lasota, T. Gambin, G. Kucherov and A. Gambin, 2007. Diversity and structure of PIF/Harbinger-like elements in the genome of Medicago truncatula. BMC Genomics 8 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holligan, D., X. Zhang, N. Jiang, E. J. Pritham and S. R. Wessler, 2006. The transposable element Landscape of the model legume Lotus japonicus. Genetics 174 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J., 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16 418–420. [DOI] [PubMed] [Google Scholar]
- Kikuchi, K., K. Terauchi, M. Wada and H.-Y. Hirano, 2003. The plant MITE mPing is mobilized in anther culture. Nature 421 167–170. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., R. A. Mitchell, A. Cheng and E. A. Hendrickson, 1997. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 17 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loot, C., N. Santiago, A. Sanz and J. M. Casacuberta, 2006. The proteins encoded by the pogo-like Lemi1 element bind the TIRs and subterminal repeated motifs of the Arabidopsis Emigrant MITE: consequences for the transposition mechanism of MITEs. Nucleic Acids Res. 34 5238–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas, J., and P. Neumann, 2007. Ogre elements: a distinct group of plant Ty3/gypsy-like retrotransposons. Gene 390 108–116. [DOI] [PubMed] [Google Scholar]
- Miskey, C., B. Papp, L. Mates, L. Sinzelle, H. Keller et al., 2007. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 27 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Rubin, E., and A. A. Levy, 1997. Abortive gap repair: underlying mechanism for Ds element formation. Mol. Cell. Biol. 17 6294–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, N., C. Herraiz, J. R. Goni, X. Messeguer and J. M. Casacuberta, 2002. Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol. Biol. Evol. 19 2285–2293. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. L., and H. S. Malik, 2006. Eukaryotic transposable elements and genome evolution special feature: positive selection of yeast nonhomologous end-joining genes and a retrotransposon conflict hypothesis. Proc. Natl. Acad. Sci. USA 103 17614–17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata, M., M. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto et al., 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal, T. J., C. D. Kaufman, Z. Izsvák, D. F. Voytas and Z. Ivics, 2002. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323 441–452. [DOI] [PubMed] [Google Scholar]
- Vitte, C., and J. L. Bennetzen, 2006. Eukaryotic transposable elements and genome evolution special feature: analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisko, O., Z. Izsvak, K. Szabo, C. D. Kaufman, S. Herold et al., 2006. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl. Acad. Sci. USA 103 4062–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., I. M. Martinez-Ferez, S. Kavchok and H. K. Dooner, 1999. Origination of Ds elements from Ac elements in maize: evidence for rare repair synthesis at the site of Ac excision. Genetics 152 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G., F. Zhang, C. N. Hancock and S. R. Wessler, 2007. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 10962–10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]