Abstract
Four LMW-m and one novel chimeric (between LMW-i and LMW-m types) low-molecular-weight glutenin subunit (LMW-GS) genes from Aegilops neglecta (UUMM), Ae. kotschyi (UUSS), and Ae. juvenalis (DDMMUU) were isolated and characterized. Sequence structures showed that the 4 LMW-m-type genes, assigned to the M genome of Ae. neglecta, displayed a high homology with those from hexaploid common wheat. The novel chimeric gene, designed as AjkLMW-i, was isolated from both Ae. kotschyi and Ae. juvenalis and shown to be located on the U genome. Phylogentic analysis demonstrated that it had higher identity to the LMW-m-type than the LMW-i-type genes. A total of 20 single nucleotide polymorphisms (SNPs) were detected among the 4 LMW-m genes, with 13 of these being nonsynonymous SNPs that resulted in amino acid substitutions in the deduced mature proteins. Phylogenetic analysis demonstrated that it had higher identity to the LMW-m-type than the LMW-i-type genes. The divergence time estimation showed that the M and D genomes were closely related and diverged at 5.42 million years ago (MYA) while the differentiation between the U and A genomes was 6.82 MYA. We propose that, in addition to homologous recombination, an illegitimate recombination event on the U genome may have occurred 6.38 MYA and resulted in the generation of the chimeric gene AjkLMW-i, which may be an important genetic mechanism for the origin and evolution of LMW-GS Glu-3 alleles as well as other prolamin genes.
THE wheat storage proteins named glutenin and gliadins are the major storage proteins in wheat endosperm and are the main determinants for bread-making quality (Payne et al. 1987; Ma et al. 2005). These proteins are heterogeneous in composition and exist in at least 50 individual components separable by electrophoretic techniques (Zhang et al. 2006). According to their molecular weight, polymeric glutenins have been subdivided into high-molecular-weight (HMW) glutenins (HMW-GSs, 70,000–90,000 Da) and low-molecular-weight (LMW) glutenins (LMW-GSs, 20,000–45,000 Da) (D'Ovidio and Masci 2004). The LMW-GSs are the main components of storage protein, accounting for ∼60% of the total protein in mature seed. It is known that LMW-GSs are important components of the giant gluten polymers that confer dough elasticity and extensibility (Wrigley 1996).
LMW-GSs have been divided into three groups on the basis of the first amino acid residue of N-terminal sequences, namely LMW-m (methionine), LMW-s (serine), and LMW-i (isoleucine) types (D'Ovidio and Masci 2004). Genetic analysis showed that LMW-GSs were encoded by the complex Glu-3 loci on the short arms of homeologous group 1 chromosomes in hexaploid wheat (Singh and Shepherd 1988; Gupta and Shepherd 1990; Yan et al. 1999). In the past, an increasing number of LMW-GS genes have been isolated and characterized from common wheat (D'Ovidio and Masci 2004) and high allelic variations have been identified at the Glu-3 locus (Gianibelli et al. 2002; Yan et al. 2003a,b,c; An et al. 2005; Li et al. 2006). A recent focus has been to isolate new LMW-GS genes from wheat related species, e.g., Triticum boeoticum (Lee et al. 1999), T. monoccocum (An et al. 2006), Aegilops tauschii (Johal et al. 2004; Pei et al. 2007), and T. dicoccoides (Q. Li et al. 2007; Li et al. 2008). These genes may provide valuable resources for wheat quality improvement and knowledge for understanding the phylogenetics and evolution of the LMW-GS family.
There are four main structural regions in a typical LMW-GS gene (D'Ovidio and Masci 2004): a signal, a short N-terminal, a repetitive domain, and a C-terminal that generally contains three distinctive subregions. The gene sizes vary, depending mainly on the number of repeats in the repetitive domain, normally ranging from 12 to 25 amino acid residues. The extensive allelic variation among both LMW-GS and HMW-GS genes are due mainly to single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) of repetitive units (Johal et al. 2004; Yan et al. 2004; An et al. 2006; Zhang et al. 2006; Li et al. 2008). It was speculated that the genetic mechanisms for these variations may result from unequal crossing over and slippage during replication and dot mutation (Anderson and Greene 1989; D'Ovidio et al. 1996; An et al. 2006).
Aegilops is the closest relative to Triticum among 20 categories of Triticeae species. Among these, Ae. neglecta (2n = 4x = 28, UUMM), Ae. kotschyi (2n = 4x = 28, UUSS), and Ae. juvenalis (2n = 6x = 42, DDMMUU) have been confirmed to possess many useful genes for wheat improvement (Li et al. 2003; Smith et al. 2004). In this study, we isolated and characterized five novel LMW-GS genes from three Aegilops species; in particular, a novel chimeric gene, designed as AjkLMW-i, was recovered from both Ae. kotschyi and Ae. juvenalis. A possible molecular mechanism of the origin and evolution of the Glu-3 gene family is proposed.
MATERIALS AND METHODS
Plant materials:
Ae. neglecta accession PI298897, Ae. kotschyi accession PI226615, and Ae. juvenalis accession PI330485 were kindly provided by the International Maize and Wheat Improvement Centre (CIMMYT), Mexico.
RT–PCR:
Seed endosperm mRNA was extracted 15 days after flowering from accession PI298897, PI226615, and PI330485. cDNA synthesis was carried out with oligo(dT) from ∼100 ng mRNA using Superscript first-strand synthesis system (Invitrogen). Subsequent PCR amplification was preformed with the synthesized 1-μl cDNA as template and with the primers 1 + 2 and 3 + 4:
5′-ATg AAg ACC TTC CTC gTC TTT g-3′
5′-TCA gTA ggC ACC AAC TCC g-3′
5′-ATC ATC ACA AgC ACA AgC ATC-3′
5′-TTC TTA TCA gTA ggC ACC AAC-3′.
PCR amplifications were performed in a 25-μl reaction volume containing 1.25 units La Taq polymerase (TaKaRa), 50 ng of template DNA, 12.5 μl 2× GC buffer I (MgCl2+ plus), 0.2 mm dNTP, and 0.25 μm of each primer. The reaction was carried out in a PTC-100 (MJ Research): 94° for 5 min to denature the template DNA, 35 cycles at 94° for 1 min, 58° for 1 min, 72° for 90 sec, and a final extension at 72° for 10 min.
PCR products were separated by 1.0% agarose gels and the expected fragments were purified from the gels using Quick DNA extraction kit (TaKaRa). The purified products were ligated into pGEM-T Easy vector (Promega) and transformed into cells of the Escherchia coli DH-5α strain. DNA sequences were obtained from three clones using the primer walking technique (performed by Sangon Biotech, Shanghai).
Sequence comparison and identification of SNPs:
Complete amino acid sequences were used to perform multiple alignment of different LMW-GS proteins by using Bioedit 7.0 software, and the SNPs were identified by means of multiple alignments of different LMW-GS genes from Triticum species.
Expression and detection of cloned LMW-GS genes in E. coli:
Three pair primers were designed to amplify newly cloned LMW subunit DNA sequences:
5′-AAg ggA TCC AAT TTC ACA gC-3′ (XhoI)
5′-AgT CTC gAg TCA gTA ggC ACC-3′ (BamHI)
5′-AAg ggA TCC ATg gAg ACT Ag-3′ (XhoI)
5′-AAT CTC gAg TCA gTA ggC ACC-3′ (BamHI)
5′-Tgg CAT ATg ATg AAg ACC TTC C-3′ (NedI)
5′-TTA CTC gAg gTA ggC ACC AAC-3′ (XhoI).
The template was the cloning vector DNA purified from DH5α, and the PCR amplification was performed as described above. The nucleotide sequence coding for mature proteins of cloned new genes were amplified by these three pairs of primers. The restriction sites (underlined above) were incorporated into the 5′-end of each primer. The purified PCR products were cloned into pMD18-T simple vector (TaKaRa) and sequenced by Sangon. Subsequently, the separated vectors from positive clones were digested by BamHI and XhoI, and purified DNA fragments were then ligated into the expression vector pGEX-4T-2. E. coli BLR (DE3) plysS cells were transformed with the pET-30a and pGEX-4T-2 plasmids containing the cloned LMW subunit genes with and without the signal sequences, respectively. Positive clones confirmed by PCR were selected and then grew on LB medium. After reaching 0.6 OD, the cells were induced by adding 0.5 mm IPTG and incubated for 4 hr at 150 rpm and 37°.
Extraction and detection of expressed proteins by SDS–PAGE and Western blotting were performed according to X. Li et al. (2007).
Phylogenetic analysis:
The homology tree was constructed by using the complete gene sequences and DNAMAN5.2.2 software. The neighbor-joining tree was used to calculate the divergent times of different genomes and genes with MEGA3 (Gaut et al. 1996; Kumar et al. 2004) according to the signal peptide and conservative domain V of LMW-GS genes. The evolutionary rate of 6.5 × 10−9 substitutions/site/year was used according to Allaby et al. (1999).
RESULTS AND DISCUSSION
PCR amplification and cloning of LMW-GS genes:
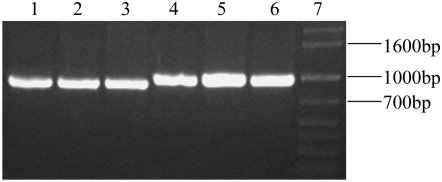
Since there are numerous LMW-GS pseudogenes present in wheat endosperm, cDNA was used as templates for RT–PCR amplification (Figure 1). One clear PCR product of ∼900 bp from Ae. juvenalis, Ae. Kotschyi, and Ae. neglecta was obtained by using the primer 1 + 2 while another PCR product with ∼1000 bp in Ae. neglecta was amplified with the primer 3 + 4. The two fragments, corresponding to the size of the coding region of LMW-GS genes, were sequenced and five novel LMW-GS genes from three Aegilops species were obtained, including four LMW-m (AnLMW-m1-4) type genes from Ae. neglecta and a LMW-i (AjkLMW-i) type gene from both Ae. kotschyi and Ae. Juvenalis, according to the first amino acid residue of the N-terminal sequence. The AjkLMW-i gene of 897 bp was isolated from two Aegilops species containing the common U genome, indicating that it is located on the U genome. The AnLMW-m1 gene amplified by the primer 1 + 2 from Ae. neglecta was 906 bp in length while the AnLMW-m2, AnLMW-m3, and AnLMW-m4 by the primer 3 + 4 was 903, 906, and 903 bp, respectively. The five novel genes were deposited in EMBL with the accession nos. EF536030, EF536031, EF536032, EF536033, and EF536034.
Figure 1.—
PCR amplification products from cDNA of three Aegilops species. Lanes 1–3: PCR product from Ae. kotschyi, Ae. neglecta, and Ae. juvenalis with allele-specific-PCR primer 1 + 2. Lanes 4–6: the same materials with primer 3 + 4. Lane 7: 1 kb plus DNA marker.
Molecular characterization of the cloned genes and expression in E. coli:
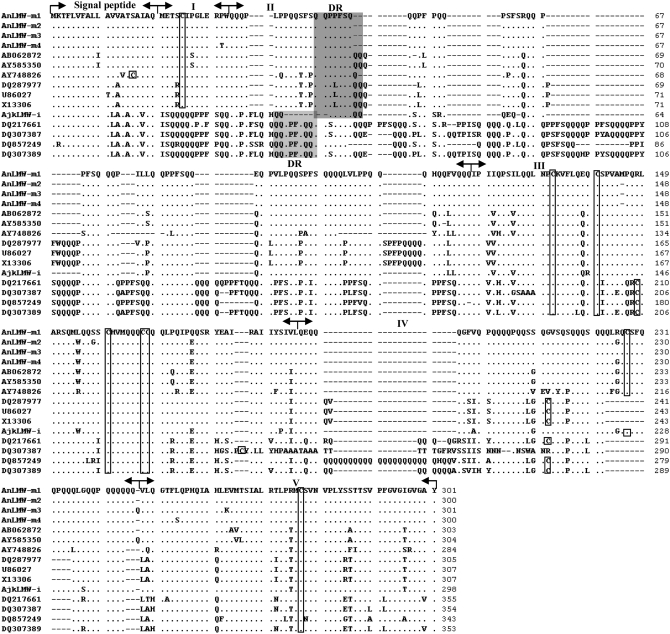
The multiple alignments of 5 cloned genes with other 10 different LMW-GS genes retrieved from EMBL (Figure 2) revealed that, apparently, the AnLMW-m1-4 genes possessed the typical structural characteristics of LMW-m-type genes in the signal peptide, N-terminal domain, repetitive domain, and the three subregions of the C-terminal domain, as well as eight conserved cysteine residues. The signal peptide sequences were highly conserved in the 4 cloned LMW-m genes and no dot mutation or insertion/deletion was presented in this domain. Compared to other LMW-m type genes, their eight cysteine residues and positions showed high similarity, suggesting that the U and M genomes had LMW-GS loci similar to those of the A, B, and C genomes in common wheat.
Figure 2.—
Comparison of deduced amino acid sequences of 15 LMW-GS genes. The mature protein sequence was divided into five domains, namely: I, N-terminal domain; II, repetitive domain; III, cysteine-rich region; IV, glutamine-rich region; and V, C-terminal conservative region. The same sequences and deletions with the AnLMW-m1 subunit are indicated by dots and dashes, respectively. The cysteine residues are represented by a box. The shaded area indicates the DR present in both LMW-m- and LMW-i-type subunits.
Since the previous reported LMW-GS genes were from different Triticum and Aegilops genomes (Johal et al. 2004; An et al. 2006), there contained considerable SNP variations in the repetitive and C-terminal domains. A total of 20 SNPs resulted from point mutation, but indel variations were detected among the four cloned LMW-m-type genes, which scattered in the different domains, except the signal peptide and N-terminal domains (Table 1). Of 20 SNPs, 13 were nonsynonymous SNPs and produced amino acid substitutions.
TABLE 1.
Identification of SNPs in the four LMW-m genes
| LMW-GS gene | 94 | 230 | 249 | 301 | 532 | 583 | 618 | 643 | 720 | 742 | 747 | 795 | 808 | 822 | 859 | 953 | 979 | 993 | 1016 | 1079 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AnLMW-m1 | C | G | A | G | A | G | C | A | C | G | A | G | A | T | A | T | G | T | G | T |
| AnLMW-m2 | C | G | A | G | A | G | T | G | T | G | A | G | A | C | G | T | G | T | G | T |
| AnLMW-m3 | C | G | A | G | A | G | T | A | T | G | A | G | A | C | A | T | A | T | G | T |
| AnLMW-m4 | A | G | A | G | A | G | T | A | T | G | A | G | A | C | G | C | G | T | G | T |
| Nine other LMW-m genes | C | A | G/T | C | G | C | T | A | C | A | G | A | G | C | G | T | G | C | C | C/G |
Italics indicate SNPs in the four LMW-m genes.
As typical LMW-i genes, the deduced amino acid sequences of the AkjLMW-i gene had no N-terminal domain, and began at ISQQQQQ. However, some unique structural characters occurred in the AkjLMW-i gene. It contained only seven cysteine residues, of which the positions were conservative for the LMW-m type, which is not a characteristic of the LMW-i-type genes. The third cysteine residue, a characteristic in the LMW-i-type genes, was substituted with arginine residue in the cysteine-rich domain. In addition, the position of another cysteine residue was similar to that of LMW-m-type genes in the glutamine-rich region as shown in Figure 2. Furthermore, large fragment deletions and substitutions presented in the AkjLMW-i gene were similar to LMW-m-type genes in III, IV, and V domains. Therefore, the cloned AkjLMW-i gene was a novel chimeric gene, which possessed characteristics of both LMW-i (1–43 residues) and LMW-m (44–298 residues) type genes.
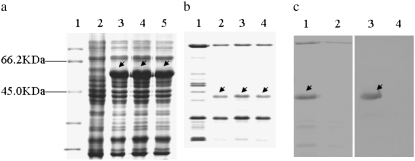
Five cloned LMW-GS genes were expressed in E. coli by two kinds of expression vectors (pGEX-4T-2 and pET-30a) to confirm their authenticity. The nucleotide sequences coding for mature proteins and complete open reading frames with signal sequences were amplified with primers 1 + 2, 3 + 4, and 5 + 6 and then ligated into each expression vector. Three genes (AnLMW-m1, AnLMW-m2, and AnLMW-m3) were successfully expressed with both expression vectors, and the fusion proteins expressed in E. coli were identified by SDS–PAGE (Figure 3, a and b) and Western blotting (Figure 3c). However, the expressed proteins of the other two genes were not detected.
Figure 3.—
SDS–PAGE and West blotting detection of induced fusion proteins in different expression vectors. (a) AnLMW-m1, AnLMW-m2, and AnLMW-m3 genes without signal sequences were expressed in pGEX-4T-2 plasmid. The fusion proteins, including GST-tag with ∼26 kDa, are shown. Lane 1: protein maker. Lane 2: CK (empty vector). Lanes 3–5: the fusion proteins of AnLMW-m1, AnLMW-m2, and AnLMW-m3 subunits are indicated by arrows. (b) AnLMW-m1, AnLMW-m2, and AnLMW-m3 genes with signal sequences amplified by primer 5 + 6 were expressed in pET-30a plasmid. The fusion proteins included an additional sequence coding His-tag with ∼840.86 Da of six amino acid residues in the downstream sequence of the insertion site of the pET-30a vector. Lane 1: CK (empty vector). Lanes 2–4: AnLMW-m1, AnLMW-m2, and AnLMW-m3 subunits indicated by arrows. (c) Western blotting detection of the fusion protein of the AnLMW-m1 gene expressed in E. coli. Lanes 1 and 2 are the induced proteins from bacterial medium, which were transformed with the positive and negative pET-30a plasmids, respectively. Lane 3 is the expressed protein (arrow) strongly hybridizing to the anti-His Tag mouse monoclonal antibody, but without any signal to bacterium.
Phylogenetic analysis among LMW-GS and other storage protein genes:
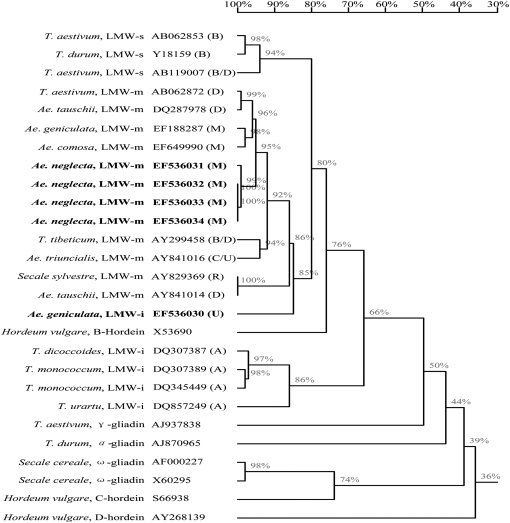
The 27 gene sequences coding for LMW-GS, gliadins, and hordeins from GenBank were used to construct a homology tree, including 3 LMW-s-type, 12 LMW-m-type, and 5 LMW-i-type glutenin genes as well as 4 gliadins and 3 hordein genes from different species and genomes (Figure 4). In general, D- and C-hordein and ω-gliadin genes were clustered in different groups while LMW-GS and B-hordein genes, more closely related to α- and γ-gliadin genes, were clustered in a separate group. With the LMW-GS gene group, three types of LMW-GS genes were divided into different subgroups but LMW-s- and LMW-m-type genes were shown to have higher homology. These results were consistent with their sequence characteristics and recent reports of An et al. (2006) and Pei et al. (2007). Particularly, the AjkLMW-i gene located on the U genome was clustered in the same subgroup with other LMW-m-type genes and shared 86% homology. This was in agreement with its sequence characteristics that most repetitive domains and the C-terminal domain of AjkLMW-i displayed higher similarity with those of LMW-m-type genes. The four LMW-m-type genes isolated from Ae. neglecta were clustered with the other two LMW-m genes from Ae. geniculata (EF188287) and Ae. comosa (EF649990) (Figure 4) and therefore could be assigned to the M genome.
Figure 4.—
Homology tree constructed with complete sequences to show the relationships among LMW-GS and other prolamin genes from different Triticum species and barley (Hordeum vulgare L.). The LMW-GS genes cloned in this study are in boldface type.
On the basis of estimates of divergence of the glutenin gene sequences using an extensive literature (D'Ovidio et al. 1999; Ikeda et al. 2002; Johal et al. 2004; Wicker et al. 2003; Li et al. 2004; An et al. 2006; Zhang et al. 2006; Pei et al. 2007), the M and D genomes were determined to be closely related and to have diverged at 5.42 MYA. Both M and D genomes and the B genome radiated 7.81 MYA. The differentiation between the U and A genomes was dated 6.82 MYA. For the divergence of the three types of LMW-GS genes, the LMW-i-type genes, supposedly the variant form (D'Ovidio and Masci 2004), diverged from the LMW-m and LMW-s genes 9.48 MYA while the LMW-m and LMW-s genes were differentiated at ∼7.81 MYA. The chimeric gene diverging from typical LMW-i-type genes on the U genome occurred 6.38 (±1.62) MYA.
Origin of the chimeric gene and molecular mechanisms of allelic variations and evolution at Glu-3 loci:
LMW-GS encoded by Glu-3 loci and HMW-GS by Glu-1 are complex gene families. The molecular characteristics of glutenin genes can reveal primary genetic mechanisms for their allelic variations as well as origin and evolution. First, a number of SNP variations resulted from dot mutation in glutenin genes, as shown in this study and recent reports (An et al. 2006; Zhang et al. 2006), may lead to nonsynonymous SNP and various amino acid substitutions, resulting in extensive allelic variations among LMW subunits. Second, indels, duplications, and inversions of one and more repeats by unequal crossing over or slippage (Anderson and Greene 1989) could result in striking expansion or contraction of glutenin genes. This mechanism could be strongly supported by several unusual HMW and LMW glutenin genes (D'Ovidio et al. 1996; X. Li et al. 2007).
Recently, different chimeric genes among wheat storage protein loci, have been identified, such as a γ-gliadin/LMW-GS chimeric gene from an old Hungarian wheat variety (Nagy et al. 2005) and a HMW-GS y-/x-type chimeric gene from Pseudoroegneria stipifolia (Z. Li et al. 2007). In addition, numerous chimeric genes were separated and characterized from various plant species (Lumbreras et al. 2001; Hedgcoth et al. 2002) and from bacteria, yeast, Drosophila, and mammals (Roth and Wilson 1986; Allgood and Silhavy 1988; Arguello et al. 2006). It was obvious that the chimeric genes were generated by recombination and crossing over among different genes and coding loci as suggested by Nagy et al. (2005).
It is well documented that LMW glutenin subunits are encoded by multigene families at the Glu-3 loci of the A, B, and D chromosomes of common wheat (D'Ovidio and Masci 2004). Some other genomes, e.g., U and M from related species, also contain similar coding loci as shown in this work. The copy numbers of LMW-GS genes in common wheat were estimated to have 10–15 (Harberd et al. 1985) or 35–40 (Sabelli and Shewry 1991; Cassidy et al. 1998). Until now, the precise gene organizations at Glu-3 loci of different genomes are still not clear. A recent report demonstrated that two LMW-i-type genes from the A genome of Triticum monococcum were located separately by >150 kbp (Wicker et al. 2003). Additionally, a single locus could encode different types of LMW-GS genes; for example, both LMW-s and LMW-m subunit genes appeared to be present at the Glu-B3 locus in the common wheat cultivar Norin 61 (Ikeda et al. 2006). The Glu-A3 locus could encode m-type (Lee et al. 1999) and i-type LMW-GS (Cloutier et al. 2001; Zhang et al. 2004; An et al 2006; Li et al. 2008). Consequently, it could be deduced that each Glu-3 locus might encode more than one type of LMW-GS. On the basis of these facts, it is possible that both LMW-i and LMW-m subunit genes locate at the Glu-U3 locus of the U genome and that the crossing over between them results in the generation of the chimeric AjkLMW-i gene.
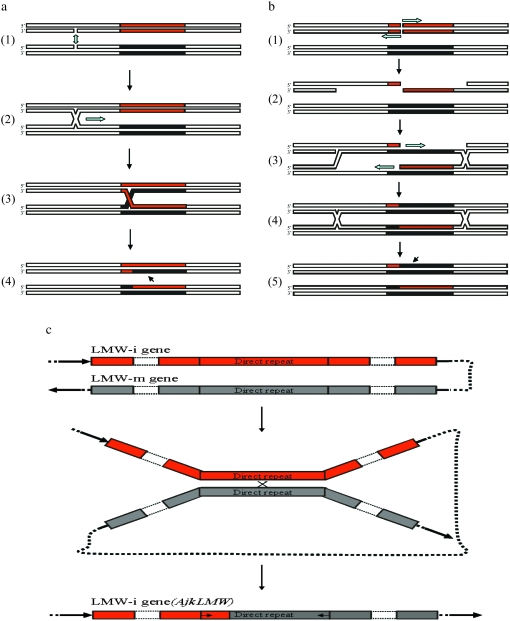
According to our results and previous reports described above, we proposed several molecular mechanisms for the origin and evolution of the AjkLMW-i gene, which may be applicable to other chimeric genes (Figure 5). The Holliday model (single-stranded invasion mode) and double-stranded break-repair pathway theoretically could explain the general genetic mechanisms of homologous recombination and crossing over between LMW-i and LMW-m genes of Glu-3 loci. As shown in Figure 5a, two nicks could occur in both non-sister chromatids in the synaptonemal complex, and then Holliday junction could be produced. As the result of the resolution of the branch in LMW-i- and LMW-m-type genes, two chimeric genes were formed. In the double-stranded break-repair pathway, double strands were broken in the same sites of the LMW-i-type gene and were digested from 5′ to 3′ as shown in Figure 5b. Subsequently, a bare single strand was intruded into the other chromatids and partnered with one of the chromatids. The other chromatid was used as template to synthesize a new sequence. After the resolution of the branch, the new chimeric gene was generated.
Figure 5.—
Several hypotheses on the genetic mechanisms that may generate the chimeric gene. (a) Holliday model (single-stranded invasion mode). Red and blank frame represented LMW-i- and LMW-m-type genes, respectively. Two nicks took place in two non-sister chromatids in synaptonemal complex arrowed in 1. Subsequently, Holliday junction and the branch migration were produced and are indicated by single arrow in 2. As the result of the resolution of the branch in the LMW-i- and LMW-m-type genes in 3, the chimeric gene was formed as indicated with a black arrow in 4. (b) Double-stranded break-repair pathway. Double strands were broken in the same sites of the LMW-i-type gene and digested from 5′ to 3′ as arrowed in 1. Subsequently, a bare single strand intruded into the other chromatids and partnered with one of the chromatids, and the other chromatid was used as template to synthesize a new sequence as shown in 2–4. After the resolution of the branch, the new chimeric gene was produced as arrowed in 5. (c) Illegitimate recombination mechanism. Through the DR coding for nonapeptide (QQPPFSQQQ), the intrastrand recombination occurred in the U genome, and then the novel chemeric gene was generated by crossing over between LMW-i- and LMW-m-type genes. This illegitimate recombination event was estimated to have occurred ∼6.38 (±1.62) MYA.
In addition to a basic homologous recombination mechanism, illegitimate (nonhomologous) recombination may be an important molecular mechanism involved in the generation of the chimeric AjkLMW-i gene, which requires only a few base pairs of sequence identity (Wicker et al. 2003) or even no sequence homology (Arguello et al. 2006). Our study revealed that a direct repeat (DR) was presented in both typical LMW-i and LMW-m-type genes, as shown in Figure 2, which could facilitate the illegitimate recombination events. Furthermore, intrastrand recombination between direct repeats could result in a DR deletion and generation of the novel chimeric AjkLMW-i gene (Figure 5c). According to the constructed neighbor-joining tree, this novel gene could originate from a primitive LMW-i-type gene of the U genome and the illegitimate recombination event probably occurred ∼6.38 MYA.
It is believed that both homologous recombination and illegitimate recombination will result in formation of novel genes. However, recent studies have shown that illegitimate recombination may be an important genetic mechanism driving genome expansion and contraction and modular protein evolution throughout the tree of life (Patthy 1999; Devos et al. 2002; Katju and Lynch 2003). More recently, an X-linked testes chimeric gene by illegitimate recombination in Drosophila has been identified (Arguello et al. 2006), and illegitimate recombination can result in a large fragment deletion in Glu-D-1-1 genes and generation of new alleles (Zhang et al. 2008). In wheat, eight chimeric genes resulting from γ-gliadins and LMW-GS genes were found, which supposedly resulted from crossing over between the Gli-1 and Glu-3 loci (Nagy et al. 2005). However, since Gli-1 and Glu-3 are closely linked and their mapping distance is ∼100 kb (Spielmeyer et al. 2000), homologous recombination and exchange between them are rare although interlocus recombination was observed in different Triticum species (Dubcovsky et al. 1997). Furthermore, it is very difficult to produce crossing over between genes at the same locus via homologous recombination. Thus, nonhomologous illegitimate recombination is most likely to be responsible for forming these chimeric genes. The recent findings of a chimeric HMW-GS gene from the St genome of P. stipifolia (Z. Li et al. 2007) and a large fragment deletion in a HMW glutenin subunit gene resulting from illegitimate recombination through bacterial expression (Zhang et al. 2008) strongly support this hypothesis. Considering that different direct repeats are commonly present in storage protein genes, and that the i- and m-types of LMW-GS genes may locate separately at a single Glu-3 locus, these could facilitate occurrence of illegitimate recombination events. It could be concluded, therefore, that illegitimate recombination is a more important genetic mechanism for the origin and evolution of LMW-GS and other storage protein genes, although multiple possible genetic hypotheses exist.
Acknowledgments
This research was financially supported by grants from National Natural Science Foundation of China (30571154 and 30771334) and the Ministry of Science and Technology of China (2002CB111300 and 2006AA10Z186).
References
- Allaby, R. G., M. Banerjee and T. A. Brown, 1999. Evolution of the high molecular weight glutenin loci of the A, B, D, and R genomes of wheat. Genome 42 296–307. [PubMed] [Google Scholar]
- Allgood, N. D., and T. J. Silhavy, 1988. Illegitimate recombination in bacteria, pp. 309–330 in Genetic Recombination, edited by R. Kucherlapati and G. R. Simth. American Society for Microbiology, Washington, DC.
- An, X., Y. Yan, Y. Xiao, Q. Li, S. L.K. Hsam et al., 2005. Genetic diversity of European spelt wheat (Triticum spelta L.) revealed by glutenin subunit variations at Glu-1 and Glu-3 loci. Euphytica 146 193–201. [Google Scholar]
- An, X., Q. Zhang, Y. Yan, Q. Li, Y. Zhang et al., 2006. Cloning and molecular characterization of three novel LMW-i glutenin subunit genes from cultivatied einkorn (Triticum monococcum L.). Theor. Appl. Genet. 113 383–395. [DOI] [PubMed] [Google Scholar]
- Anderson, O. D., and F. C. Greene, 1989. The characterization and comparative analysis of high-molecular-weight glutenin genes from genomes A and B of a hexaploid bread wheat. Theor. Appl. Genet. 77 689–700. [DOI] [PubMed] [Google Scholar]
- Arguello, J. R., Y. Chen, S. Yang, W. Wang and M. Long, 2006. Origination of an X–Linked testes chimeric gene by illegitimate recombination in Drosophila. PLoS Genet. 2 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy, B. G., J. Dvorak and O. D. Anderson, 1998. The wheat low-molecular-weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theor. Appl. Genet. 96 743–750. [Google Scholar]
- Cloutier, S., C. Rampitsch, G. A. Penner and O. M. Lukow, 2001. Cloning and expression of a LMW-i glutenin Gene. J. Cereal Sci. 33 143–154. [Google Scholar]
- D'Ovidio, R., and S. Masci, 2004. The low-molecular-weight glutenin subunits of wheat gluten. J. Cereal Sci. 39 321–339. [Google Scholar]
- D'Ovidio, R., D. Lafiandra and E. Porceddu, 1996. Identification and molecular characterization of a large insertion within the repetitive domain of a high-molecular-weight glutenin subunit gene from hexaploid wheat. Theor. Appl. Genet. 93 1048–1053. [DOI] [PubMed] [Google Scholar]
- D'Ovidio, R., C. Marchitelli, C. L. Ercoli and E. Porceddu, 1999. Sequence similarity between allelic Glu-B3 genes related to quality properties of durum wheat. Theor. Appl. Genet. 98 455–461. [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. F. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., M. Echaide, S. Giancola, M. Rousset, M. C. Luo et al., 1997. Seed-storage-loci in RFLP maps of diploid, tetraploid, and hexaploid wheat. Theor. Appl. Genet. 95 1169–1180. [Google Scholar]
- Gaut, B. S., B. R. Morton, B. C. McCaig and M. T. Clegg, 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianibelli, M. C., C. W. Wrigley and F. Macritchie, 2002. Polymorphism of low Mr glutenin subunits in Triticum tauschii. J. Cereal Sci. 35 277–286. [Google Scholar]
- Gupta, R. B., and K. W. Shepherd, 1990. Two-step one-dimensional SDS-PAGE analysis of LMW subunits of glutenin. 1. Variation and genetic control of the subunits in hexaploid wheats. Theor. Appl. Genet. 80 65–74. [DOI] [PubMed] [Google Scholar]
- Harberd, N. P., D. Bartels and R. D. Thompson, 1985. Analysis of the gliadin moltigene loci in bread wheat using nullisomic-tetrasomic lines. Mol. Gen. Genet. 198 234–242. [Google Scholar]
- Hedgcoth, C., A. M. El-Shehawi, P. Wei, M. Clarkson and D. Tamalis, 2002. A chimeric open reading frame associated with cytoplasmic male sterility in alloplasmic wheat with Triticum timopheevi mitochondria is present in several Triticum and Aegilops species, barley, and rye. Curr. Genet. 41 357–365. [DOI] [PubMed] [Google Scholar]
- Ikeda, T. M., T. Nagamine, H. Fukupka and H. Yano, 2002. Identification of new low-molecular-weight glutenin subunit genes in wheat. Theor. Appl. Genet. 104 680–687. [DOI] [PubMed] [Google Scholar]
- Ikeda, T..M., E. Araki, Y. Fujita and H. Yano, 2006. Characterization of low-molecular- weight glutenin subunit genes and their protein products in common wheats. Theor. Appl. Genet. 112 327–334. [DOI] [PubMed] [Google Scholar]
- Johal, J., M. C. Gianibelli, S. Rahman, M. K. Morell and K. R. Gale, 2004. Characterization of low-molecular-weight glutenin genes in Aegilops tauschii. Theor. Appl. Genet. 109 1028–1040. [DOI] [PubMed] [Google Scholar]
- Katju, V., and M. Lynch, 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lee, Y. K., M. Ciaffi, R. Appels and M. K. Morel, 1999. The low-molecular-weight glutenin subunit proteins of primitive wheats. II. The genes from A-genome species. Theor. Appl. Genet. 98 126–134. [Google Scholar]
- Li, Q., H. Ye, Q. Yang and S. Jiang, 2003. Relationship between several biochemical indexes and resistance of Aegilops species to oat-bird cherry aphids (Homoptera: Aphididae). Sci. Agric. Sin. 2 994–999. [Google Scholar]
- Li, Q., Y. Yan, A. Wang, X. An, Y. Zhang et al., 2006. Detection of HMW glutenin subunit variations among 205 cultivated emmer accessions (Triticum turgidum ssp. dicoccum). Plant Breed. 125 120–124. [Google Scholar]
- Li, Q., X. An, Y. Xiao, Q. Zang, Y. Zang et al., 2007. Cloning and molecular characterization of LMW glutenin subunit genes in Triticum dicoccoides. Sci. Agric. Sin. 40 457–463. [Google Scholar]
- Li, W., Y. Wan, Z. Liu, K. Liu, X. Liu et al., 2004. Molecular characterization of HMW glutenin subunit allele 1Bx14: further insights into the evolution of Glu-B1–1 alleles in wheat and related species. Theor. Appl. Genet. 109 1093–1104. [DOI] [PubMed] [Google Scholar]
- Li, X., Y. Zhang, L. Gao, A. Wang, K. Ji et al., 2007. Molecular cloning, heterologous expression and phylogenetic analysis of a novel y-type HMW glutenin subunit gene from G genome of Triticum timopheevi. Genome 50 1130–1140. [DOI] [PubMed] [Google Scholar]
- Li, X., A. Wang, Y. Xiao, Y. Yan, Z. He et al., 2008. Cloning and characterization of a novel low molecular weight glutenin subunit gene at the Glu-A3 locus from wild emmer wheat (Triticum turgidum L. var. dicoccoides). Euphytica 159 181–190. [Google Scholar]
- Li, Z., X. Zhang, H. Zhang, S. Cao, D. Wang et al., 2007. Isolation and characterization of a novel variant of HMW glutenin subunit gene from the St genome of Pseudoroegneria stipifolia. J. Cereal Sci. 47 429–437. [Google Scholar]
- Lumbreras, V., M. M. Albà, T. Kleinow, C. Koncz and M. Pagès, 2001. Domain fusion between SNF1-related kinase subunits during plant evolution. EMBO Rep. 2 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W., R. Appels, F. Bekes, O. Larroque, M. K. Morell et al., 2005. Genetic characterisation of dough rheological properties in a wheat doubled haploid population: additive genetic effects and epistatic interactions. Theor. Appl. Genet. 111 410–422. [DOI] [PubMed] [Google Scholar]
- Nagy, I. J., I. Takàcs, A. Juhász, L. Tamás and Z. Bedö, 2005. Identification of a new class of recombinant prolamin genes in wheat. Genome 48 840–847. [DOI] [PubMed] [Google Scholar]
- Patthy, L., 1999. Genome evolution and the evolution of exon-shuffling: a review. Gene 238 103–114. [DOI] [PubMed] [Google Scholar]
- Payne, P. I., M. A. Nightingale, A. F. Krattiger and L. M. Holt, 1987. The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J. Sci. Food Agric. 40 51–65. [Google Scholar]
- Pei, Y., A. Wang, X. An, X. Li, Y. Zhang et al., 2007. Characterization and comparative analysis of three low molecular weight glutenin C-subunit genes isolated from Aegilops tauschii. Can. J. Plant Sci. 87 273–280. [Google Scholar]
- Roth, D. B., and J. H. Wilson, 1986. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli, P. A., and P. R. Shewry, 1991. Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor. Appl. Genet. 83 209–216. [DOI] [PubMed] [Google Scholar]
- Singh, N. K., and K. W. Shepherd, 1988. Linkage mapping of the genes controlling endosperm proteins in wheat. 1. Genes on the short arms of group 1 chromosomes. Theor. Appl. Genet. 75 628–641. [Google Scholar]
- Smith, C. M., S. Starkey, B. S. Gill, H. Havlícková and V. Holubec, 2004. Identification of Aegilops germplasm with multiple aphid resistance. Euphytica 135 265–273. [Google Scholar]
- Spielmeyer, W., O. Moullet, A. Laroche and E. S. Lagudah, 2000. Highly recombinogenic regions at seed storage protein loci on chromosome 1DS of Aegilops tauschii, the D-genome donor of wheat. Genetics 155 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrigley, C. W., 1996. Giant proteins with flour power. Nature 381 738–739. [DOI] [PubMed] [Google Scholar]
- Yan, Y., S. Prodanovic, N. Mladenov and M. Milovanovic, 1999. Genetic control of low molecular weight glutenin subunits in wheat by A-PAGE analysis. Cereal Res. Commun. 27 251–257. [Google Scholar]
- Yan, Y., S. L. K. Hsam, J. Yu, Y. Jiang, I. Ohtsuka et al., 2003. a HMW and LMW glutenin alleles among putative tetraploid and hexaploid T. spelta progenitors. Theor. Appl. Genet. 107 1321–1330. [DOI] [PubMed] [Google Scholar]
- Yan, Y., S. L. K. Hsam, J. Z. Yu, Y. Jiang and F. J. Zeller, 2003. b Allelic variation of the HMW glutenin subunits in Aegilops tauschii accessions detected by sodium dodecyl sulphate (SDS-PAGE), acid polyacrylamide gel (A-PAGE) and capillary electrophoresis. Euphytica 130 377–385. [Google Scholar]
- Yan, Y., S. L. K. Hsam, J. Z. Yu, Y. Jiang and F. J. Zeller, 2003. c Genetic polymorphisms at Gli-Dt gliadin loci in Aegilops tauschii as revealed by acid polyacrylamide gel and capillary electrophoresis. Plant Breed. 122 120–124. [Google Scholar]
- Yan, Y., J. Zheng, Y. Xiao, J. Yu, Y. Hu et al., 2004. Identification and molecular characterization of a novel y-type Glu-Dt1 glutenin gene of Aegilops tauschii. Theor. Appl. Genet. 108 1349–1358. [DOI] [PubMed] [Google Scholar]
- Zhang, W., M. C. Gianibelli, L. R. Rampling and K. R. Gale, 2004. Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 108 1409–1419. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Q. Li, Y. Yan, J. Zheng, X. An et al., 2006. Molecular characterization and phylogenetic analysis of a novel glutenin gene (Dy10.1t) from Aegilops tauschii. Genome 49 735–745. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. Z., X. H. Li, A. L. Wang, X. L. An, Q. Zhang et al., 2008. Novel x-type HMW glutenin genes from Aegilops tauschii and their implications on the wheat origin and evolution mechanism of Glu-D1–1 proteins. Genetics 178 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]