Abstract
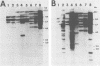
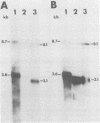
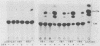
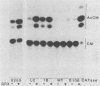
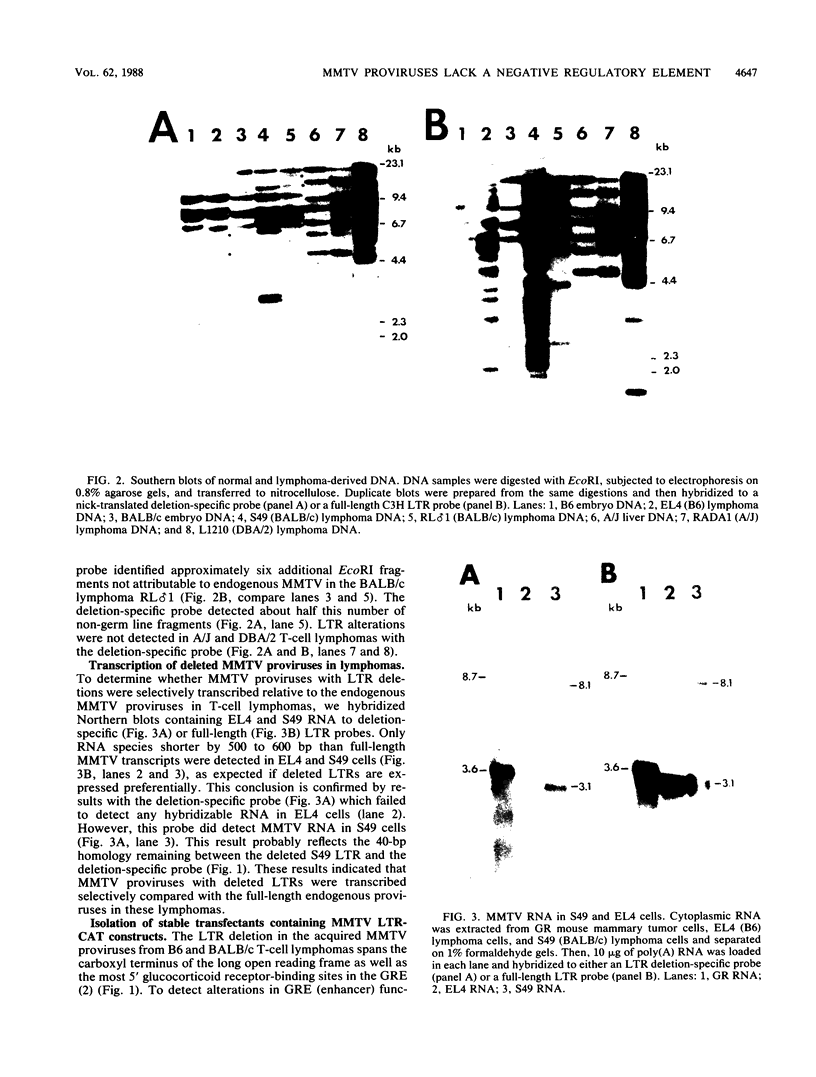
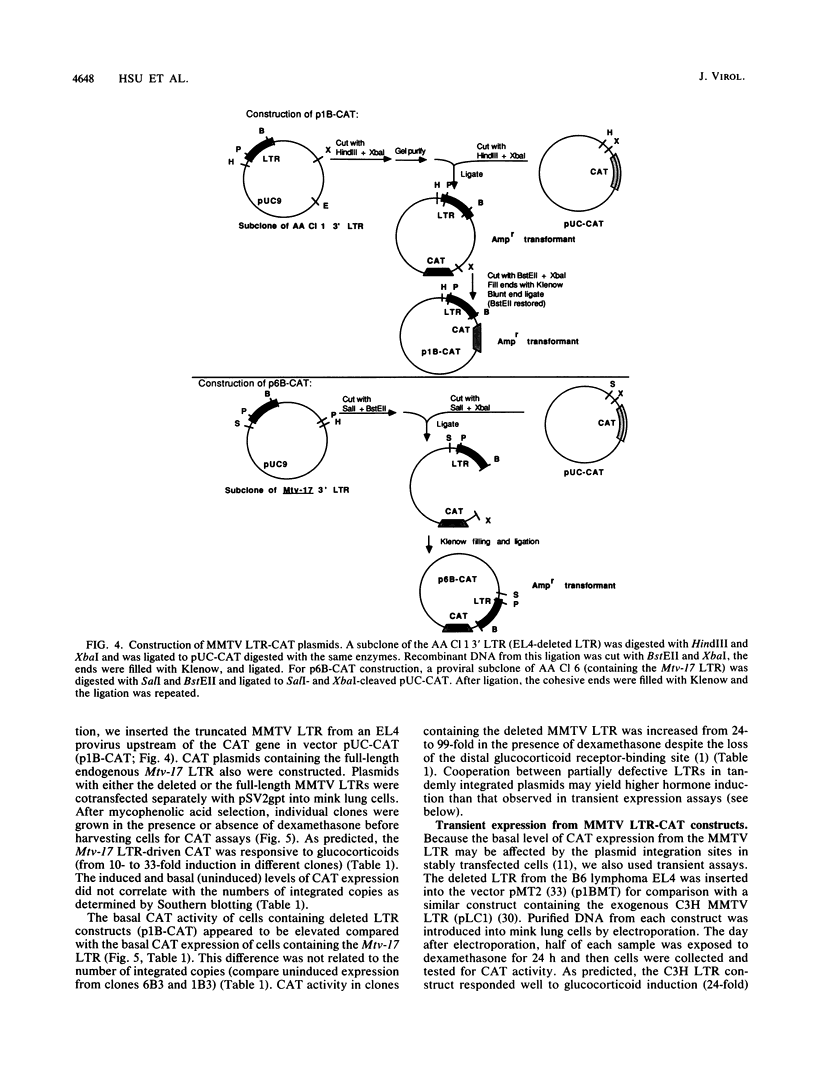
The nucleotide sequences of long terminal repeats (LTRs) from several mouse mammary tumor virus (MMTV) proviruses acquired in mouse T-cell lymphomas were determined. All MMTV proviruses cloned from a C57BL/6 lymphoma contained an identical LTR deletion of 491 base pairs (approximately -655 to -165), whereas an MMTV provirus from a BALB/c T-cell lymphoma had a 430-base-pair deletion in the same U3 region. MMTV proviruses with LTR deletions were acquired in these tumors 10 times more frequently than proviruses with intact LTRs. Because the deletions removed a portion of the glucocorticoid response element or "regulated" enhancer, the transcriptional activity of the deleted MMTV LTRs was assessed in both transient expression and stable transfection experiments. Plasmids were constructed in which the deleted or full-length MMTV LTRs were placed upstream of the chloramphenicol acetyltransferase gene. Results from transfection experiments with these constructs showed that the basal expression of the deleted MMTV LTR in the absence of glucocorticoids was higher than that of the full-length Mtv-17 or C3H MMTV LTRs under the same conditions. Moreover, the C3H LTR with a similar deletion (-637 to -255) also promoted high basal levels of chloramphenicol acetyltransferase activity. These results, coupled with the observation in lymphomas of high basal levels of transcription from MMTV proviruses with deleted LTRs, suggested that these proviruses lack negative regulatory elements in their LTRs. Loss of the negative regulatory element may contribute to the selective propagation of proviruses with deleted LTRs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buetti E., Kühnel B. Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J Mol Biol. 1986 Aug 5;190(3):379–389. doi: 10.1016/0022-2836(86)90009-4. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Maler B. A., Yamamoto K. R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983 Jun;33(2):489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Choi Y. W., Henrard D., Lee I., Ross S. R. The mouse mammary tumor virus long terminal repeat directs expression in epithelial and lymphoid cells of different tissues in transgenic mice. J Virol. 1987 Oct;61(10):3013–3019. doi: 10.1128/jvi.61.10.3013-3019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Dickson C., Smith R., Brookes S., Peters G. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell. 1984 Jun;37(2):529–536. doi: 10.1016/0092-8674(84)90383-0. [DOI] [PubMed] [Google Scholar]
- Dudley J. P., Arfsten A., Hsu C. L., Kozak C., Risser R. Molecular cloning and characterization of mouse mammary tumor proviruses from a T-cell lymphoma. J Virol. 1986 Jan;57(1):385–388. doi: 10.1128/jvi.57.1.385-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. P. Mouse mammary tumor proviruses from a T-cell lymphoma are associated with the retroposon L1Md. J Virol. 1988 Feb;62(2):472–478. doi: 10.1128/jvi.62.2.472-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J., Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984 Jan;49(1):92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Pearson K., Buetti E., Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1(1):3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein S. C., Ross S. R., Yamamoto K. R. Chromosomal position effects determine transcriptional potential of integrated mammary tumor virus DNA. J Mol Biol. 1982 Apr 15;156(3):549–565. doi: 10.1016/0022-2836(82)90266-2. [DOI] [PubMed] [Google Scholar]
- Gallahan D., Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987 Jan;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hilgers J., Bentvelzen P. Interaction between viral and genetic factors in murine mammary cancer. Adv Cancer Res. 1978;26:143–195. doi: 10.1016/s0065-230x(08)60087-1. [DOI] [PubMed] [Google Scholar]
- Huang A. L., Ostrowski M. C., Berard D., Hager G. L. Glucocorticoid regulation of the Ha-MuSV p21 gene conferred by sequences from mouse mammary tumor virus. Cell. 1981 Dec;27(2 Pt 1):245–255. doi: 10.1016/0092-8674(81)90408-6. [DOI] [PubMed] [Google Scholar]
- Kennedy N., Knedlitschek G., Groner B., Hynes N. E., Herrlich P., Michalides R., van Ooyen A. J. Long terminal repeats of endogenous mouse mammary tumour virus contain a long open reading frame which extends into adjacent sequences. Nature. 1982 Feb 18;295(5850):622–624. doi: 10.1038/295622a0. [DOI] [PubMed] [Google Scholar]
- Knutson J. C., Yee D. Electroporation: parameters affecting transfer of DNA into mammalian cells. Anal Biochem. 1987 Jul;164(1):44–52. doi: 10.1016/0003-2697(87)90365-4. [DOI] [PubMed] [Google Scholar]
- Kuo W. L., Vilander L. R., Huang M., Peterson D. O. A transcriptionally defective long terminal repeat within an endogenous copy of mouse mammary tumor virus proviral DNA. J Virol. 1988 Jul;62(7):2394–2402. doi: 10.1128/jvi.62.7.2394-2402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B. S., Weissman S. M. Mouse mammary tumor virus-related sequences in mouse lymphocytes are inducible by 12-O-tetradecanoyl phorbol-13-acetate. J Virol. 1984 Dec;52(3):1000–1004. doi: 10.1128/jvi.52.3.1000-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Hall C. V., Ringold G. M., Dobson D. E., Luh J., Jacob P. E. Functional analysis of the steroid hormone control region of mouse mammary tumor virus. Nucleic Acids Res. 1984 May 25;12(10):4191–4206. doi: 10.1093/nar/12.10.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. T., Prakash O., Klein D., Sarkar N. H. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987 Jul;159(1):39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J., Varmus H. E. A small region of the mouse mammary tumor virus long terminal repeat confers glucocorticoid hormone regulation on a linked heterologous gene. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5866–5870. doi: 10.1073/pnas.80.19.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology. 1986 Oct 15;154(1):76–84. doi: 10.1016/0042-6822(86)90431-9. [DOI] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Weijers P. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol Cell Biol. 1985 Apr;5(4):823–830. doi: 10.1128/mcb.5.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley K. L., Toohey M. G., Peterson D. O. Transcriptional repression of a hormone-responsive promoter. Nucleic Acids Res. 1987 Sep 11;15(17):6973–6989. doi: 10.1093/nar/15.17.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Peterson D. O., Beifuss K. K., Morley K. L. Context-dependent gene expression: cis-acting negative effects of specific procaryotic plasmid sequences on eucaryotic genes. Mol Cell Biol. 1987 Apr;7(4):1563–1567. doi: 10.1128/mcb.7.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Geisse S., Westphal H. M., Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983 Aug 25;304(5928):749–752. doi: 10.1038/304749a0. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Hollingshead P. G., Pitts S. L. Multiple regulatory domains in the mouse mammary tumor virus long terminal repeat revealed by analysis of fusion genes in transgenic mice. Mol Cell Biol. 1988 Jan;8(1):473–479. doi: 10.1128/mcb.8.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wellinger R. J., Garcia M., Vessaz A., Diggelmann H. Exogenous mouse mammary tumor virus proviral DNA isolated from a kidney adenocarcinoma cell line contains alterations in the U3 region of the long terminal repeat. J Virol. 1986 Oct;60(1):1–11. doi: 10.1128/jvi.60.1.1-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]