Abstract
Background
Boar taint is the unpleasant odour and flavour of the meat of uncastrated male pigs that is primarily caused by high levels of androstenone and skatole in adipose tissue. Androstenone is a steroid and its levels are mainly genetically determined. Studies on androstenone metabolism have, however, focused on a limited number of genes. Identification of additional genes influencing levels of androstenone may facilitate implementation of marker assisted breeding practices. In this study, microarrays were used to identify differentially expressed genes and pathways related to androstenone metabolism in the liver from boars with extreme levels of androstenone in adipose tissue.
Results
Liver tissue samples from 58 boars of the two breeds Duroc and Norwegian Landrace, 29 with extreme high and 29 with extreme low levels of androstenone, were selected from more than 2500 individuals. The samples were hybridised to porcine cDNA microarrays and the 1% most significant differentially expressed genes were considered significant. Among the differentially expressed genes were metabolic phase I related genes belonging to the cytochrome P450 family and the flavin-containing monooxygenase FMO1. Additionally, phase II conjugation genes including UDP-glucuronosyltransferases UGT1A5, UGT2A1 and UGT2B15, sulfotransferase STE, N-acetyltransferase NAT12 and glutathione S-transferase were identified. Phase I and phase II metabolic reactions increase the water solubility of steroids and play a key role in their elimination. Differential expression was also found for genes encoding 17beta-hydroxysteroid dehydrogenases (HSD17B2, HSD17B4, HSD17B11 and HSD17B13) and plasma proteins alpha-1-acid glycoprotein (AGP) and orosomucoid (ORM1). 17beta-hydroxysteroid dehydrogenases and plasma proteins regulate the availability of steroids by controlling the amount of active steroids accessible to receptors and available for metabolism. Differences in the expression of FMO1, NAT12, HSD17B2 and HSD17B13 were verified by quantitative real competitive PCR.
Conclusion
A number of genes and pathways related to metabolism of androstenone in liver were identified, including new candidate genes involved in phase I oxidation metabolism, phase II conjugation metabolism, and regulation of steroid availability. The study is a first step towards a deeper understanding of enzymes and regulators involved in pathways of androstenone metabolism and may ultimately lead to the discovery of markers to reduce boar taint.
Background
Boar taint is an off-odour and off-flavour in pig carcasses that is primarily caused by high levels of 16-androstene steroids [1] and/or skatole [2] in adipose tissue. Male pigs used for meat production are normally castrated very early in life to prevent boar taint in the meat. However, castration also removes the source of natural anabolic androgens that stimulate lean growth and, as a result, uncastrated males have improved feed efficiency and greater lean yield of the carcass compared to barrows [3]. Detection of genetic factors influencing boar taint may facilitate implementation of selective breeding practices to produce pigs with little or no taint. Because of unfavourable correlations between androstenone and other sex steroids [4], a direct selection against high levels of androstenone would result in decreased production of testosterone and estrogens, with associated negative effects on performance and sexual maturation. Therefore, a comprehensive understanding of the complex genetic system controlling boar taint is required before genetic improvement can be achieved.
Androstenone (5α-androst-16-en-3-one) is produced in the testis and is transported by blood to the salivary gland where it functions as a pheromone to stimulate mating stance in gilts [5]. It is metabolised in the liver, producing α-androstenol and β-androstenol [6,7], and deficient degradation may lead to the accumulation of androstenone in adipose tissue. Skatole (3-methylindole) is produced by the metabolism of tryptophan from the gut and is also catabolised in liver. Diaz et al. [8] identified seven metabolites of skatole in pig liver microsomes. An increase in levels of skatole in adipose tissue occurs in boars around puberty, but not in barrows or sows, indicating that skatole metabolism is regulated by testicular steroids [9,10].
Enzymes responsible for metabolism of androstenone and skatole in liver have been identified but relatively few genes have been investigated. Several studies have shown that 3β-hydroxysteroid dehydrogenase (3β-HSD) is involved in androstenone metabolism, with low mRNA, protein and enzyme expression levels correlated to high androstenone levels in adipose tissue [6,11,12]. Cytochrome P450 2E1 (CYP2E1) is involved in metabolism of skatole [13,14] and deficient CYP2E1 induction is suggested to be a major cause of high levels of skatole in adipose tissue [10]. A relationship between metabolism of androstenone and skatole in liver has been observed [9], with evidence to suggest that androstenone blocks CYP2E1 induction by skatole [10]. Sulfotransferase enzymes including SULT2A1, SULT2B1 and SULT1A1 are associated with metabolic clearance of androstenone and skatole since they are known to conjugate steroid hormones and drugs into more water soluble compounds that facilitates excretion [12,15-20].
Although both androstenone and skatole are responsible for boar taint, androstenone seems to be the biggest problem in the Duroc and Norwegian Landrace populations [21]. Levels of androstenone are also predominantly genetically determined whereas levels of skatole are more heavily influenced by feeding and environmental factors. Levels of androstenone will be a function of both production in testes and elimination in liver. We have previously studied differentially expressed genes related to the production of androstenone in boar testis [17], and the objective of this study was to investigate global gene expression profiles related to the degradation of androstenone in pig livers. Microarray technology was used to obtain gene expression profiles from liver samples of Duroc (D) and Norwegian Landrace (NL) boars with extreme high or low levels of androstenone in adipose tissue. These samples were hybridised together with a common reference sample to cDNA microarrays representing approximately 20,000 porcine gene transcripts. A total of 29 animals were selected from each group, resulting in a total of 116 arrays. A subset of the differentially expressed genes was verified using a quantitative PCR-based method.
Results
Porcine cDNA microarrays were used to obtain global gene expression profiles in liver tissues sampled from 58 D and 58 NL boars with extreme high and low levels of androstenone. Average levels of androstenone were 11.57 ± 3.2 ppm for D high (DH) and 0.37 ± 0.17 ppm for D low (DL) boars. Average levels of androstenone were 5.95 ± 2.04 ppm for NL high (NLH) and 0.14 ± 0.04 ppm for NL low (NLL) boars. Linear models were used to identify significantly differentially expressed genes and the top 1% (269) most significant genes were subsequently inspected as possible candidate genes affecting androstenone levels (See additional file 1: Microarray results for Duroc, additional file 2: Microarray results for Norwegian Landrace). The top 100 genes of D and NL are presented in table 1 and 2, respectively. Among the 269 affected genes, 25 were found to be in common for the two breeds and expression differences were generally more significant in D compared to NL.
Table 1.
Top 100 genes identified in Duroc.
| ID | Name | gene_id | FoldChange | adj.P.Val |
| 103725 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 1.227 | 8.26e-24 |
| 104194 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 0.853 | 4.98e-20 |
| 102872 | Glycine-N-acyltransferase (GLYAT) | NM_201648.1 | 1.113 | 1.27e-19 |
| 103265 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 1.480 | 4.76e-18 |
| 103234 | Hepatic flavin-containing monooxygenase (FMO) (FMO1) | NM_214064.1 | 1.179 | 2.28e-17 |
| 206700 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 1.099 | 2.71e-16 |
| 102682 | Serum amyloid P component (APCS) | NM_213887.1 | 1.294 | 2.71e-16 |
| 102284 | Sterile alpha motif domain containing 8 (Samd8) | NM_026283.2 | 0.969 | 3.44e-16 |
| 102668 | Hepatic flavin-containing monooxygenase (FMO) (FMO1) | NM_214064.1 | 1.271 | 8.01e-16 |
| 100748 | Regulatory factor X domain containing 2 (RFXDC2) | NM_022841.3 | 0.594 | 7.47e-14 |
| 206864 | Similar to Alcohol dehydrogenase 6 (LOC523510) | XM_601810.2 | -0.925 | 3.49e-13 |
| 102390 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 1.152 | 5.05e-13 |
| 102098 | Carnitine O-octanoyltransferase (CROT) | NM_021151.2 | 0.886 | 5.30e-13 |
| 102991 | Cytochrome P450. family 2. subfamily E. polypeptide 1 (CYP2E1) | NM_214421.1 | -1.040 | 2.17e-12 |
| 216026 | 216026 | -0.728 | 3.72e-12 | |
| 102413 | Small nuclear RNA activating complex. polypeptide 1. 43 kDa (SNAPC1) | NM_003082.2 | 0.994 | 4.84e-12 |
| 103432 | 103432 | 0.527 | 2.07e-11 | |
| 210250 | Metallothionein (MT1A) | NM_001001266.1 | -0.965 | 3.09e-11 |
| 103062 | FKBP1A-like (LOC654323) | NM_001038000.1 | -1.078 | 3.09e-11 |
| 103279 | Orosomucoid 1 (ORM1) | NM_000607.1 | -0.947 | 3.99e-11 |
| 103021 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.869 | 6.10e-11 |
| 201488 | 201488 | 0.462 | 1.08e-10 | |
| 100575 | 17beta-estradiol dehydrogenase (HSD17B4) | NM_214306.1 | 0.452 | 1.24e-10 |
| 100723 | Aspartate aminotransferase (GOT2) | NM_213928.1 | 0.493 | 3.71e-10 |
| 103194 | Alpha-1 acid glycoprotein (AGP) | XM_585370.2 | -0.932 | 4.52e-10 |
| 202082 | Family with sequence similarity 10. member A4 (FAM10A4) | NR_002183.1 | 0.424 | 6.45e-10 |
| 100724 | Phytanoyl-CoA hydroxylase (PHYH) | NM_006214.3 | 0.518 | 8.69e-10 |
| 210077 | Cytochrome P450 2C49 (CYP2C49) | NM_214420.1 | 0.872 | 9.95e-10 |
| 101277 | Estrogen sulfotransferase (STE) | NM_213992.1 | 0.557 | 1.45e-09 |
| 216031 | Insulin-like-growth factor 2 (IGF2) | NM_213883.1 | -0.687 | 1.88e-09 |
| 210204 | Similar to HLA class I histocompatibility antigen, A-11 alpha chain precursor (LOC642049) | XM_936199.1 | -0.591 | 1.88e-09 |
| 103031 | Type I iodothyronine deiodinase (DIO1) | NM_001001627.1 | 0.499 | 2.85e-09 |
| 210252 | Sarcosine dehydrogenase (SARDH) | NM_007101.2 | -0.273 | 3.55e-09 |
| 102976 | Similar to UDP-glucuronosyltransferase 2B15 precursor (UDPGT) (HLUG4) (LOC653180) | XM_931558.1 | 0.623 | 4.69e-09 |
| 201274 | Sorbitol dehydrogenase (SORD). mRNA | NM_003104.3 | 0.523 | 5.70e-09 |
| 103801 | Similar to Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) (LOC650043) | XM_939131.1 | 0.452 | 1.83e-08 |
| 204461 | Glycine-N-acyltransferase (GLYAT). nuclear gene encoding mitochondrial protein | NM_005838.2 | 0.370 | 2.47e-08 |
| 102538 | 102538 | 0.746 | 2.65e-08 | |
| 204863 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.630 | 3.89e-08 |
| 101843 | Abhydrolase domain containing 3 (ABHD3) | NM_138340.3 | 0.370 | 9.87e-08 |
| 215880 | Transcription elongation factor B (SIII). polypeptide 3 (110 kDa. elongin A) (TCEB3) | NM_003198.1 | -0.612 | 9.87e-08 |
| 200322 | Taxilin beta (TXLNB) | NM_153235.2 | 0.385 | 9.87e-08 |
| 210206 | Metallothionein (MT1A) | NM_001001266.1 | -0.536 | 1.25e-07 |
| 201503 | Carboxylesterase (CES3) | NM_214246.1 | 0.472 | 1.44e-07 |
| 202070 | 202070 | 0.485 | 1.57e-07 | |
| 102106 | 102106 | 0.335 | 1.86e-07 | |
| 206300 | Protein tyrosine phosphatase type IVA. member 1 (PTP4A1) | NM_003463.3 | 0.337 | 2.01e-07 |
| 103047 | Similar to mouse 2310016A09Rik gene (LOC134147) | NM_138809.3 | 0.456 | 2.01e-07 |
| 204653 | FKBP1A-like (LOC654323) | NM_001038000.1 | 0.256 | 2.06e-07 |
| 103327 | UDP glucuronosyltransferase 1 family, polypeptide A5 (UGT1A5) | NM_019078.1 | 0.264 | 2.42e-07 |
| 102469 | Chromosome 1 open reading frame 128 (C1orf128) | NM_020362.2 | 0.190 | 2.42e-07 |
| 201919 | FKBP1A-like (LOC654323) | NM_001038000.1 | 0.303 | 2.53e-07 |
| 103349 | Kininogen 1 (KNG1) | NM_000893.2 | 0.428 | 3.03e-07 |
| 103940 | UDP glucuronosyltransferase 1 family, polypeptide A5 (UGT1A5) | NM_019078.1 | 0.191 | 4.03e-07 |
| 204455 | Ornithine carbamoyltransferase (OTC) | NM_000531.3 | 0.357 | 4.61e-07 |
| 212598 | L1 cell adhesion molecule (L1CAM) | NM_024003.1 | -0.167 | 4.72e-07 |
| 207639 | 207639 | 0.383 | 4.83e-07 | |
| 200534 | 200534 | 0.388 | 4.89e-07 | |
| 102183 | Protein tyrosine phosphatase type IVA. member 1 (PTP4A1) | NM_003463.3 | 0.360 | 5.72e-07 |
| 206424 | Similar to transcription elongation factor B polypeptide 3 binding protein 1 isoform 1 (LOC533427) | XM_612827.2 | 0.265 | 6.41e-07 |
| 103306 | Tryptophan 2.3-dioxygenase (TDO2) | NM_005651.1 | 0.443 | 6.41e-07 |
| 104125 | Sorbitol dehydrogenase (SORD) | NM_003104.3 | 0.428 | 7.73e-07 |
| 102709 | 102709 | 0.444 | 7.76e-07 | |
| 100472 | S-adenosylhomocysteine hydrolase (AHCY) | NM_001011727.1 | 0.321 | 8.64e-07 |
| 206129 | 206129 | -0.484 | 8.86e-07 | |
| 102624 | Ornithine carbamoyltransferase (OTC) | NM_000531.3 | 0.393 | 8.86e-07 |
| 213735 | Metallothionein (MT1A) | NM_001001266.1 | 0.422 | 1.01e-06 |
| 102836 | Carboxylesterase (CES3) | NM_214246.1 | 0.563 | 1.10e-06 |
| 201162 | FKBP1A-like (LOC654323) | NM_001038000.1 | 0.268 | 1.42e-06 |
| 201961 | 201961 | 0.293 | 1.42e-06 | |
| 216074 | Metallothionein (MT1A) | NM_001001266.1 | -0.772 | 1.64e-06 |
| 207278 | 207278 | 0.282 | 1.67e-06 | |
| 101716 | 101716 | 0.328 | 1.75e-06 | |
| 204421 | Meiosis-specific nuclear structural 1 (MNS1) | NM_018365.1 | 0.364 | 1.99e-06 |
| 103156 | 103156 | 0.333 | 2.05e-06 | |
| 103376 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.497 | 2.38e-06 |
| 100064 | Similar to UDP-galactoseN-acetylgalactosamine-alpha-R beta 1,3-galactosyltransferase (LOC645551) | XM_928571.1 | 0.159 | 2.43e-06 |
| 204950 | Abhydrolase domain containing 3 (ABHD3) | NM_138340.3 | 0.236 | 2.43e-06 |
| 204048 | Suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) (ST13) | NM_003932.3 | 0.379 | 2.74e-06 |
| 204867 | 204867 | 0.308 | 2.95e-06 | |
| 207963 | 207963 | 0.254 | 3.08e-06 | |
| 200952 | 200952 | 0.236 | 3.08e-06 | |
| 103294 | Similar to Mob4B protein (MGC124888) | NM_001033891.1 | 0.369 | 3.79e-06 |
| 208427 | RAR-related orphan receptor A (RORA) | NM_134262.1 | 0.246 | 4.25e-06 |
| 208058 | FKBP1A-like (LOC654323) | NM_001038000.1 | 0.247 | 4.48e-06 |
| 205360 | FLJ20105 protein (FLJ20105) | NM_017669.2 | 0.278 | 4.73e-06 |
| 207838 | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) (HMGCL) | NM_000191.2 | 0.156 | 4.78e-06 |
| 101413 | Replication protein A3. 14 kDa (RPA3) | NM_002947.3 | 0.253 | 5.25e-06 |
| 103180 | T-complex 1 (TCP1) | NM_001008897.1 | 0.310 | 5.69e-06 |
| 202298 | 202298 | 0.262 | 5.73e-06 | |
| 100384 | Alpha-methylacyl-CoA racemase (AMACR) | NM_014324.4 | 0.394 | 6.68e-06 |
| 103434 | Acyl-Coenzyme A oxidase 1, palmitoyl (ACOX1) | NM_004035.5 | 0.368 | 7.11e-06 |
| 202477 | Hypothetical LOC505518 (LOC505518) | XM_581816.2 | 0.240 | 7.42e-06 |
| 102858 | 102858 | 0.587 | 7.52e-06 | |
| 103058 | 103058 | 0.381 | 7.67e-06 | |
| 211612 | Gamma-aminobutyric acid (GABA) A receptor. alpha 3 (GABRA3) | NM_000808.2 | -0.135 | 8.97e-06 |
| 201347 | HRAS-like suppressor 3 (HRASLS3) | NM_007069.1 | 0.352 | 8.97e-06 |
| 102840 | Sorbitol dehydrogenase (SORD) | NM_003104.3 | 0.270 | 8.97e-06 |
| 201105 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) (COPS2) | NM_004236.1 | 0.242 | 9.06e-06 |
| 201848 | Kinesin family member C1 (KIFC1) | NM_002263.2 | 0.274 | 9.06e-06 |
Gene expression profiles were identified in Limma using empirical Bayes moderated t-statistics. Fold change and statistical significance is shown (FDR-adjusted p-values). The clone names refer to hits to pig, human, mouse or bovine sequences. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while others have no hits to the abovementioned species.
Table 2.
Top 100 genes identified in Norwegian Landrace.
| ID | Name | gene_id | FoldChange | adj.P.Val |
| 206864 | Similar to Alcohol dehydrogenase 6 (LOC523510) | XM_601810.2 | -0.730 | 1.07e-13 |
| 204853 | 204853 | -0.441 | 2.39e-13 | |
| 209551 | Isochorismatase domain containing 1 (ISOC1) | NM_016048.1 | -0.403 | 3.28e-13 |
| 103021 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.900 | 3.28e-13 |
| 204863 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.730 | 3.78e-13 |
| 209525 | Zinc finger protein 471 (ZNF471) | NM_020813.1 | -0.335 | 1.69e-12 |
| 102789 | 102789 | -0.364 | 1.69e-12 | |
| 204725 | 204725 | -0.367 | 1.69e-12 | |
| 103376 | Hydroxysteroid (17-beta) dehydrogenase 13 (HSD17B13) | NM_178135.2 | -0.589 | 2.41e-12 |
| 205190 | Similar to heterogeneous nuclear ribonucleoprotein C isoform b (LOC654074) | XM_945342.1 | 0.249 | 7.01e-12 |
| 204302 | Aldehyde oxidase 1 (AOX1) | NM_001159.3 | -0.481 | 9.60e-12 |
| 204466 | 204466 | -0.345 | 1.20e-11 | |
| 203752 | 203752 | -0.274 | 1.52e-11 | |
| 206901 | Non-histone protein HMG1 (LOC445521) | NM_001004034.1 | -0.286 | 5.34e-11 |
| 218781 | KIAA1344 (KIAA1344) | NM_020784.1 | 0.280 | 5.34e-11 |
| 216031 | Insulin-like-growth factor 2 (IGF2) | NM_213883.1 | -0.847 | 5.34e-11 |
| 213651 | 213651 | -0.437 | 9.01e-11 | |
| 215880 | Transcription elongation factor B (SIII). polypeptide 3 (110 kDa. elongin A) (TCEB3) | NM_003198.1 | -0.797 | 9.45e-11 |
| 204619 | Cytochrome b-5 (CYB5) | NM_001001770.1 | -0.458 | 9.45e-11 |
| 204520 | 204520 | -0.290 | 1.45e-10 | |
| 103177 | Cytochrome b-5 (CYB5) | NM_001001770.1 | -0.489 | 2.06e-10 |
| 102774 | Non-histone protein HMG1 (LOC445521) | NM_001004034.1 | -0.302 | 3.48e-10 |
| 204081 | 204081 | -0.251 | 3.48e-10 | |
| 100528 | ATP synthase. H+ transporting. mitochondrial F0 complex. subunit F6 (ATP5J) | NM_001003703.1 | 0.250 | 3.55e-10 |
| 211561 | KIAA0427 (KIAA0427) | NM_014772.1 | 0.263 | 4.63e-10 |
| 209674 | 209674 | 0.269 | 7.12e-10 | |
| 208386 | Dual specificity phosphatase 12 (DUSP12) | NM_007240.1 | -0.318 | 7.12e-10 |
| 207228 | Small nuclear ribonucleoprotein polypeptide E (SNRPE) | NM_003094.2 | 0.252 | 7.37e-10 |
| 219297 | Purine-rich element binding protein A (PURA) | NM_005859.3 | 0.499 | 8.70e-10 |
| 218205 | SMC1 structural maintenance of chromosomes 1-like 1 (yeast) (SMC1L1) | NM_006306.2 | 0.300 | 1.11e-09 |
| 103194 | Alpha-1 acid glycoprotein (AGP) | XM_585370.2 | -1.071 | 1.16e-09 |
| 219538 | 219538 | 0.352 | 1.67e-09 | |
| 201622 | Hypothetical protein LOC285016 (LOC285016) | NM_001002919.1 | -0.417 | 1.99e-09 |
| 217117 | Amyloid beta (A4) precursor protein-binding. family B. member 3 (APBB3) | NM_006051.2 | 0.294 | 2.38e-09 |
| 102677 | 4-aminobutyrate aminotransferase (ABAT) | NM_000663.2 | 0.288 | 2.38e-09 |
| 216026 | 216026 | -0.840 | 2.51e-09 | |
| 216643 | Leupaxin (Lpxn) | NM_134152.1 | 0.271 | 2.93e-09 |
| 205721 | 205721 | -0.256 | 3.02e-09 | |
| 202919 | 202919 | 0.272 | 3.02e-09 | |
| 220257 | Similar to 60S ribosomal protein L39 (LOC651724) | XM_940942.1 | 0.273 | 3.10e-09 |
| 103136 | Mitochondrial translational initiation factor 2 (MTIF2) | NM_001005369.1 | -0.250 | 3.35e-09 |
| 201942 | 201942 | -0.286 | 4.06e-09 | |
| 216638 | 216638 | 0.304 | 4.06e-09 | |
| 204605 | Similar to Maltase-glucoamylase, intestinal (LOC642103) | XM_936233.1 | -0.234 | 4.12e-09 |
| 208567 | 208567 | -0.307 | 4.71e-09 | |
| 103361 | Phenylalanine hydroxylase (PAH) | NM_000277.1 | -0.627 | 5.15e-09 |
| 202659 | FKBP1A-like (LOC654323) | NM_001038000.1 | -0.312 | 5.49e-09 |
| 207982 | La ribonucleoprotein domain family. member 4 (LARP4) | NM_052879.3 | -0.247 | 5.63e-09 |
| 204513 | Deoxyribonuclease II (DNASE2) | NM_214196.1 | -0.255 | 5.73e-09 |
| 206370 | Syndecan binding protein (syntenin) (SDCBP) | NM_001007068.1 | -0.208 | 6.33e-09 |
| 214304 | Heat shock transcription factor 1 (HSF1) | NM_005526.1 | 0.331 | 7.28e-09 |
| 103062 | FKBP1A-like (LOC654323) | NM_001038000.1 | -1.046 | 7.37e-09 |
| 200969 | Family with sequence similarity 70. member A (FAM70A) | NM_017938.2 | 0.424 | 7.37e-09 |
| 206063 | Chromosome 14 open reading frame 43 (C14orf43) | NM_194278.2 | -0.230 | 8.05e-09 |
| 206129 | 206129 | -0.447 | 1.10e-08 | |
| 206577 | Leukemia inhibitory factor receptor (LIFR) | NM_002310.3 | -0.265 | 1.39e-08 |
| 103378 | Similar to Putative steroid dehydrogenase KIK-I (LOC508455) | XM_585231.2 | -0.314 | 1.49e-08 |
| 204838 | FKBP1A-like (LOC654323) | NM_001038000.1 | -0.309 | 1.49e-08 |
| 104413 | Pyruvate carboxylase (PC) | NM_214349.1 | 0.246 | 1.49e-08 |
| 219405 | Coiled-coil domain containing 42 (CCDC42) | NM_144681.1 | 0.447 | 1.49e-08 |
| 209512 | 209512 | -0.310 | 1.72e-09 | |
| 221025 | 221025 | 0.446 | 1.74e-08 | |
| 200362 | Chromosome 3 open reading frame 43, transcript variant 1 (C3orf43) | XM_173087.5 | -0.199 | 1.88e-08 |
| 206386 | 206386 | -0.262 | 2.65e-08 | |
| 211845 | Alanyl-tRNA synthetase domain containing 1 (AARSD1) | NM_025267.2 | 0.250 | 3.31e-08 |
| 208761 | Ring finger protein 148 (RNF148) | NM_198085.1 | -0.189 | 3.31e-08 |
| 209449 | Similar to Zinc finger CCCH-type domain containing protein 11A transcript variant 3 (LOC441155) | XM_930970.1 | -0.237 | 3.31e-08 |
| 205142 | Bone morphogenetic protein receptor, type IA (BMPR1A) | NM_004329.2 | -0.340 | 3.31e-08 |
| 203722 | 203722 | -0.217 | 3.32e-08 | |
| 103279 | Orosomucoid 1 (ORM1) | NM_000607.1 | -0.999 | 3.32e-08 |
| 101898 | Bifunctional apoptosis regulator (BFAR) | NM_016561.1 | -0.189 | 3.32e-08 |
| 104972 | Cyclin-dependent kinase inhibitor 3 (CDKN3) | NM_214320.1 | 0.364 | 3.45e-08 |
| 202997 | 202997 | -0.270 | 3.45e-08 | |
| 102991 | Cytochrome P450. family 2. subfamily E. polypeptide 1 (CYP2E1) | NM_214421.1 | -1.079 | 3.68e-08 |
| 208187 | Early growth response 3 (EGR3) | NM_004430.2 | 0.378 | 3.77e-08 |
| 205347 | Histone H3.3A (H3F3A) | NM_213930.1 | -0.237 | 3.77e-08 |
| 203762 | Zinc finger protein 7 (KOX 4. clone HF.16) (ZNF7) | NM_003416.1 | -0.234 | 4.37e-08 |
| 102955 | L-kynurenine 3-monooxygenase Fpk (KMO) | NM_214076.1 | -0.294 | 4.81e-08 |
| 212532 | TBC1 domain family. member 10B (TBC1D10B) | NM_015527.2 | 0.231 | 4.99e-08 |
| 206466 | Similar to glycosyltransferase-like domain containing 1 isoform a (LOC534677) | XM_871633.1 | -0.299 | 5.15e-08 |
| 104900 | Cullin 4A (CUL4A). transcript variant 1 | NM_001008895.1 | 0.349 | 5.59e-08 |
| 208650 | FKBP1A-like (LOC654323) | NM_001038000.1 | -0.214 | 5.83e-08 |
| 205233 | 5'-3' exoribonuclease 2 (XRN2) | NM_012255.3 | 0.217 | 5.99e-08 |
| 102672 | Cytochrome b-5 (CYB5) | NM_001001770.1 | -0.528 | 6.21e-08 |
| 101034 | Programmed cell death 6 interacting protein (PDCD6IP) | NM_013374.3 | -0.247 | 6.21e-08 |
| 206253 | Cyclin-dependent kinase 6 (CDK6) | NM_001259.5 | -0.260 | 6.21e-08 |
| 209516 | 209516 | -0.208 | 6.21e-08 | |
| 100909 | Myeloid-associated differentiation marker (MYADM) | NM_001020819.1 | -0.239 | 7.42e-08 |
| 205225 | Family with sequence similarity 80, member B (FAM80B) | NM_020734.1 | 0.247 | 7.42e-08 |
| 200683 | 200683 | -0.302 | 7.64e-08 | |
| 208724 | Dickkopf-like 1 (soggy) (DKKL1) | NM_014419.3 | 0.275 | 7.64e-08 |
| 218253 | Mesoderm induction early response 1, family member 2 (MIER2) | NM_017550.1 | 0.265 | 7.97e-08 |
| 209231 | Heparin binding protein (HBP15/L22) | NM_213987.1 | -0.343 | 8.00e-08 |
| 104000 | Ribosomal protein L39 (RPL39) | NM_001000.2 | 0.264 | 8.37e-08 |
| 205725 | 205725 | -0.208 | 8.73e-08 | |
| 102726 | Complement factor I (CFI) | NM_000204.1 | -0.380 | 8.77e-08 |
| 100609 | Proline rich 13 (PRR13) | NM_001005355.1 | 0.267 | 9.03e-08 |
| 104957 | Succinyl-CoA:alpha-ketoacid coenzyme A transferase (OXCT) | NM_213938.1 | 0.392 | 9.03e-08 |
| 216727 | 216727 | -0.287 | 9.12e-08 | |
| 200320 | Ubiquitin specific peptidase 32 (USP32) | XM_939750.1 | 0.439 | 9.28e-08 |
Gene expression profiles were identified in Limma using empirical Bayes moderated t-statistics. Fold change and statistical significance is shown (FDR-adjusted p-values). The clone names refer to hits to pig, human, mouse or bovine sequences. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while others have no hits to the abovementioned species.
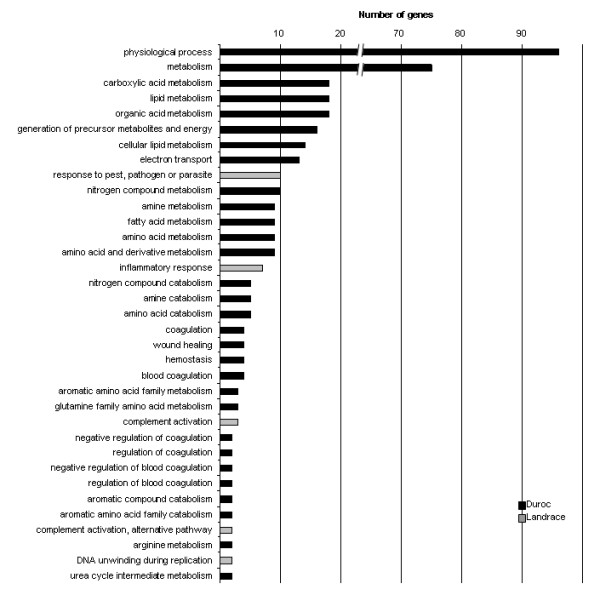
Testing for overrepresentation of gene ontology (GO) terms among the 269 affected genes was performed relative to the global representation of GO terms on the microarray. A majority of the significant genes were related to oxidoreductase activities, glucuronosyltransferase activity and the binding of iron ion, haem, DNA and oxygen (Figure 1). Additionally, cellular metabolic process pathways such as amino acid metabolism and catabolism, electron transport and inflammatory response were overrepresented (Figure 2). Furthermore, the genes were classified according to their cellular component ontology (See additional file 3: Gene ontology (GO) results for the cellular component ontology in Duroc, additional file 4: Gene ontology (GO) results for the cellular component ontology in Norwegian Landrace). For D, cytoplasm, microsomes and endoplasmatic reticulum were among the important cellular components, none of the cellular components reached the level of significance, i.e. had more than 10 significant genes related to the CC terms.
Figure 1.
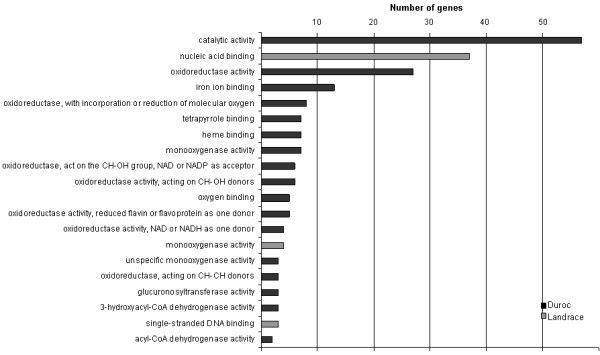
Figure 2.
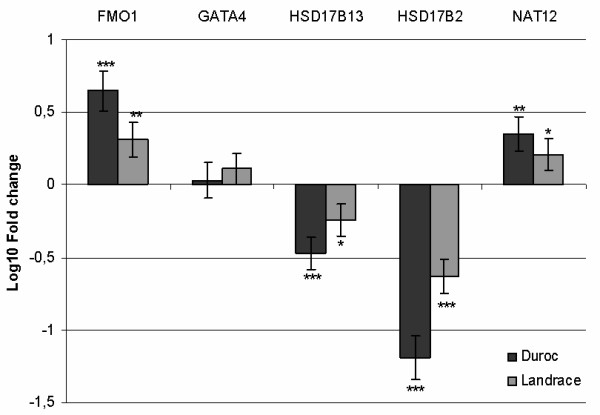
To validate the microarray results, a real competitive PCR (rcPCR) approach was applied to five selected genes: Flavin-containing monooxygenase 1 (FMO1), transcription factor GATA-4 (GATA4), 17β-hydroxysteroid dehydrogenase 13 (HSD17B13), 17β-hydroxysteroid dehydrogenase 2 (HSD17B2) and N-acetyltransferase 12 (NAT12). Their expression levels were normalised to the housekeeping gene transferrin receptor (TFRC). Differential expression of FMO1, HSD17B13, HSD17B2 and NAT12 was confirmed in both D and NL breeds (p < 0.05) while altered expression of GATA4 was not found in either of the two breeds (Figure 3).
Figure 3.
Discussion
Elevated levels of androstenone in adipose tissue can result from both increased biosynthesis in the testes and deficient metabolism in liver. We have previously conducted a microarray experiment investigating gene expression profiles in the testes of boars with extreme levels of androstenone, and in this study we have examined gene expression profiles in the liver of some representatives from that study along with new individuals. Liver metabolism can be divided into phase I and phase II reactions [22]. Phase I metabolism involves oxidation and hydroxylation reactions that make the substrate more water soluble. Phase II conjugation reactions further increase hydrophilicity by adding polar groups. As a result of these metabolic reactions, compounds including endogenous steroids, fatty acids and drugs are inactivated and eliminated. We have identified a number of differentially expressed genes that function in pathways affecting both phase I and phase II reactions involved in metabolism of androstenone in the liver.
Phase I metabolism
The most significantly differentially expressed gene involved in phase I oxidation reactions was cytochrome P450 2C49 (CYP2C49). This gene is a member of the CYP2C family [23] and genes of this family encode monooxygenases that catalyse several reactions involved in the metabolism of drugs and endogenous compounds. Different CYP2C isoforms show some cross reactivity towards substrates which makes it difficult to differentiate CYP2C activities, and substrate specificity for CYP2C49 is not known [24]. In our study, CYP2C49 transcripts were shown to have a significant up-regulation in boars with high levels of androstenone in both D and NL lines (DH and NLH) but differences were clearly most prominent in the D breed.
Another member of the cytochrome P450 superfamily, that was differentially expressed in our study, is cytochrome P450 2E1 (CYP2E1). In contrast to expression of the CYP2C49 gene, CYP2E1 showed a down-regulation in DH and NLH boars. CYP2E1 encodes an enzyme that metabolises many endogenous and exogenous substrates such as alcohols, ketones and drugs [24]. Androstenone has been shown to block skatole induced expression of CYP2E1 in pig hepatocytes and to inhibit activity of CYP2E1 in porcine liver microsomes [10,25]. Liver metabolism of skatole also involves this gene and enzyme [13,14,26-28], and the observed pubertal increase in skatole levels has been attributed to the inhibition of CYP2E1 by the sex steroids androstenone and 17β-estradiol [25]. This is the first study, however, to show gene expression differences for CYP2E1 in relation to androstenone.
Cytochrome P450 member 2A19 (CYP2A19), which is the pig ortholog of human CYP2A6, catalyses 7-hydroxylation of coumarin in pigs [26], in addition to being involved in skatole metabolism [29]. In this study, CYP2A19 was significantly down-regulated in DH boars. Gene expression differences in animals with high and low androstenone levels have not previously been reported for porcine CYP2A19, however its protein expression has been shown to be inhibited by androstenone [30]. Diaz and Squires [26] found that both CYP2A6 protein content and its enzymatic activity were negatively correlated with skatole levels in adipose tissue. In contrast, results presented by Terner et al. [28] indicate that the CYP2A6 enzyme is not important for metabolism of skatole in primary cultured porcine hepatocytes.
Two other members of the cytochrome P450 family, CYP27A1 and CYP2C33, were also up-regulated in DH. CYP27A1 is a multifunctional enzyme that oxygenates cholesterol, bile acids and vitamin D. It has not previously been identified as a candidate gene associated with androstenone, but it is regulated by other sex steroids in human cells [31]. CYP2C33 belongs to the same sub-family as CYP2C49, but its function has yet to be described in the literature. Like the CYP2C49 gene, CYP2C33 was found to be up-regulated in DH and NLH boars. The differential expression of CYP2A19, CYP27A1 and CYP2C33 in D and not NL boars might suggest that more genes are involved in phase I reactions in D. This is supported by a high number of oxidoreductase pathways significant in D (figure 1). Monooxygenase reactions are, however, clearly important in both breeds according to our gene ontology results (figure 1). Down-regulation of CYP2E1 and CYP2A19 might suggest a different role for these genes compared to the up-regulated CYP2C18, CYP2C33 and CYP27A1.
Our study implicates another class of genes involved in phase I oxidation reactions, namely the flavin-containing monooxygenases (FMOs). The FMO family of enzymes converts lipophilic compounds into more polar metabolites and decreases activity of the compounds, a similar activity to that of the cytochrome P450s [32]. Microarray expression results show that flavin-containing monooxygenase 1 (FMO1) was significantly up-regulated in DH in the microarray results. The expression pattern in DH boars was confirmed by rcPCR and the rcPCR analysis also revealed that FMO1 was up-regulated in NLH boars. As shown in figure 2, the fold change in NLH was only half of DH, which might explain why this gene was not found to be significantly differentially expressed in the microarray results. Regulation of FMO involves sex steroids. In male mice, castration was reported to increase FMO1 expression [33], while in rats the opposite effect has been shown, with positive regulation of FMO by testosterone and negative by estradiol [34]. The monooxygenase activity of FMO1 in addition to its regulation by steroid hormones makes it an interesting candidate gene for boar taint.
Phase II metabolism
Phase I oxidation reactions of androstenone by enzymes including cytochrome P450s and FMOs are often followed by phase II conjugation reactions that are catalysed by glucuronosyltransferases, sulfotransferases, acetyltransferases and glutathione S-transferases [22]. Through the addition of polar moieties, these enzymes increase substrate solubility. Conjugation reactions are an important means of excreting steroid hormones and other compounds, these reactions have also been proposed to keep inactive steroids easily available in cells. The hydroxysteroid sulfotransferases SULT2A1 and SULT2B1 have been associated with androstenone levels in previous studies [12,15-17]. These genes were not found to be significantly differentially expressed in our study, however the estrogen sulfotransferase STE (SULT2E1) was found to be up-regulated in DH boars and down-regulated in NLH boars. A previous microarray study performed by our group showed an up-regulation of CYP19 gene expression in the testis of D and NL boars with high androstenone levels [17]. These genes encode enzymes biosynthesising estrogens and might explain why an estrogen conjugation enzyme is differentially expressed in livers of the same pigs. Furthermore, boars secrete large quantities of estrogens from the testis [35], and both fat and plasma estrogen levels have previously been shown to be highly correlated with levels of androstenone in adipose tissue [36,37].
Phase II conjugation by glucuronidation is another major pathway of liver elimination of endogenous and exogenous compounds, and in D this molecular function was significantly overrepresented. It is catalysed by uridine diphospho-glucuronosyltransferases (UGTs) which transfer glucuronic acid to substrates to increase solubility [38]. The UGTs have been divided into two subfamilies, the UGT1s and the UGT2s, and in this study the family member UGT1A5 was found to be up-regulated in DH boars. Furthermore, a transcript similar to UGT2B15 was up-regulated in D and a transcript similar to UGT2A1 was down-regulated in NLH. UGT2B15 has been shown to conjugate several androgens in humans [38], whereas UGT1A5 has been found to be catalytically active against some exogenous compounds [39]. UGT2A1 contributes to glucuronidation of steroids and phenolic compounds in olfactory tissue and has been found to eliminate odourants [40]. The UGT family of conjugation enzymes have previously been associated with the androstenone metabolites α-androstenol and β-androstenol [41,42].
N-acetyltransferases (NATs) are another family of conjugation enzymes and act by transferring the acetyl group of acetyl coenzyme A to aromatic amines to increase their water solubility [22]. NAT type 12 (NAT12) was found to be up-regulated in NLH based on the microarray results and this was verified by rcPCR. The rcPCR results also revealed an up-regulation in DH boars. Androgens have been shown to increase expression of NAT type 1 in human cancer cells [43], but the enzyme family has not previously been associated with levels of androstenone. Tryptophan, the precursor of skatole, contains an aromatic, suggesting a role for this family of conjugation enzymes in the control of skatole levels, possibly through regulation by steroid hormones.
Glutathione S-transferases (GSTs) are functionally diverse enzymes mostly known to catalyse conjugation reactions of endogenous substances, haem, fatty acids, xenobiotics and products of oxidative processes [44]. They have also been implicated in the intracellular transport of steroids to their site of action [44]. In this study, a glutathione S-transferase gene was found to be down-regulated in NLH. The gene is described in the database as LOC396850. We have previously described up-regulation of two GST genes, GSTO1 and MGST1, in association with testicular androstenone levels [17], and here we additionally propose a role for glucuronidation in liver metabolism. Expression of phase II metabolic genes in D and NL pigs suggest breed specific mechanisms. We found differential expression of a GST variant (LOC396850) in NL but not D boars, up-regulation of STE in DH but down-regulation in NLH, and up-regulation of two UGT genes in DH but down-regulation of another family member in NLH. Breed specific phase II mechanisms for these breeds are in agreement with our previous finding for the protein SULT2B1 [12].
Regulation of steroid availability
17β-hydroxysteroid dehydrogenases (17β-HSDs) regulate the availability of androgens and estrogens in tissues by catalysing interconvertion of active and inactive forms of steroids [45]. They do so by regulating the occupancy of steroid nuclear hormone receptors like androgen receptor, estrogen receptor and progesterone receptor [46]. Gene expression of 17β-HSD in porcine liver has been shown to be negatively associated with levels of androstenone in adipose tissue [37]. In a previous study, we found that the HSD17B4 gene was significantly up-regulated in testicle samples from DH and NLH [17], and in this study HSD17B4 was up-regulated in the liver of DH boars. Additionally, we found that HSD17B2 was down-regulated in DH and that the isoform HSD17B13 was down-regulated in both DH and NLH. Differential expression of both HSD17B2 and HSD17B13 was verified by rcPCR, which also showed significant down-regulation of HSD17B2 in NLH boars. The HSD17B2 enzyme catalyses the interconversion of testosterone and androstenedione, as well as estradiol and estrone [47]. The function of isoform HSD17B13, also known as short-chain dehydrogenase/reductase 9, has yet to be described but its expression has been characterised in human liver [48]. The short-chain dehydrogenase/reductase 8 gene (DHRS8), also known as HSD17B11, was found to be down-regulated in NLH boars. HSD17B11 is involved in androgen metabolism [49] and we previously found this gene to be down-regulated in the testis of DH boars [17]. The HSDs belong to two superfamilies: the short-chain dehydrogenase/reductases (SDRs) and the aldo-keto reductases (AKRs) [46]. The four HSDs described above all belong to the SDR superfamily but a gene from the AKRs was also found to be differentially expressed. The aldo-keto reductase family member 1D1 (AKR1D1) was down-regulated in NLH animals. AKR1D1 is a liver specific enzyme that regulates the hormone levels of several steroids [46]. The enzyme has a 5β-reductase activity and is necessary for the catabolism of aldosterone, cortisol and androgens [50]. Aldosterone, cortisol and androgens are three of four metabolic compounds formed from pregnenolone, with the fourth compound being androstenone [5]. A role for AKR1D1 in androstenone catabolism might therefore be possible. We previously reported up-regulation of the family member AKR1C4 in the testis of DH and NLH boars [17], but AKR1D1 has not formerly been associated with porcine androstenone levels.
Another class of enzymes that regulate the availability of steroids is the plasma proteins. Binding of plasma protein is a reversible reaction that has no physiological effect, but it controls the amount of free drugs and hormones available to tissues by protecting them from metabolism [51]. Alpha-1-acid glycoprotein (AGP) is an acute phase serum protein synthesised in the liver and secreted to plasma where it binds and carries drugs and steroid hormones [51]. The protein has been shown to interact with CYP3A and CYP2C19 in humans and to inhibit cytochrome P450 activity [52,53]. It is suggested that serum protein interaction is an important factor for cytochrome P450 mediated metabolism, and that the interaction is isoform specific [53]. AGP has also been studied in pigs as a binding factor of pheromones, but has not been found to bind progesterone in pig nasal mucosa [54]. AGP has two variants in humans: orosomucoid 1 and 2 (ORM1 and ORM2) [51]. In this study we found that porcine transcript homologous to bovine AGP and human ORM1 were significantly down-regulated in DH and NLH boars. Significant expression of these genes might explain the inflammatory response ontology term in figure 2 as these genes have mostly been described in relation to inflammatory response. AGP has not been investigated as a binding protein and transporter for androstenone, but different levels of androstenone could be related to differences in its availability through plasma binding by AGP.
Regulatory factors
One of the most significant molecular function terms in NL was nucleic acid binding. This term includes transcription factor binding and several transcription factors were found to be differentially expressed in this study. The GATA factor 4 (GATA4) was found to be down-regulated based on the microarray data, but we were not able to confirm this result with rcPCR. Other transcription factors identified in this study should be further investigated, including RAR-related orphan receptor A (RORA), transcription elongation factor B3 (TCEB3), nuclear factor I/X (NFIX), transcription factor 8 (TCF8) and heat shock transcription factor 1 (HSF1). We have previously identified iron ion binding, ferric ion binding and electron transport as being associated with levels of androstenone [17], possibly through interaction with the haem-containing cytochrome P450s. In this study we also found iron ion binding and electron transport, in addition to haem binding, to be important pathways, supporting our previous findings. Cytochrome b5 (CYB5) is involved in electron transfer to cytochrome P450s and has been proposed as a candidate gene for androstenone through its interaction with cytochrome P450 c17 (CYP17) [55]. In this study we found CYB5 to be down-regulated in NLH boars, however we previously identified CYB5 as being up-regulated in the testis of both DH and NLH [17]. Both CYP2E1 and CYP2A6 have been found to be activated by CYB5 in humans [56], supporting the down-regulation of CYB5 together with CYP2E1 and CYP2A6 in this study. Up-regulation in testis and down-regulation in liver might also support a key regulatory role of CYB5 in both tissues.
Breed differences
Breed differences in boar taint candidate genes have previously been shown between D and NL [12,17]. Consistent with these findings, we found breed differences in the expression profiles of genes involved in both phase I metabolism and phase II metabolism. However, rcPCR results might suggest that genes appearing in one breed can be differentially expressed also in the other breed. Additional and more comprehensive rcPCR studies are planned to clarify this. In general, the D breed showed higher levels of significance compared to the NL breed, potentially due to the larger contrasts between high and low androstenone groups in this breed. A number of new candidate genes have been identified in this study, however, we do not know their effect on performance and sexual maturation in pig. Additional studies are needed to investigate potential roles of the genes identified for phenotypes related to fertility.
Relationship with skatole
Pigs in this study were selected based on extreme androstenone values. However, levels of androstenone and skatole have previously been found to be highly correlated, showing genetic correlations of 0.62 for D and 0.36 for NL [21]. Some of the genes found differentially expressed in this study have previously been found to be associated with tryptophan or skatole and this could be explained by high correlations with androstenone. This might also be the reason for the significance of GO terms such as aromatic amino acid metabolism and catabolism, since the skatole precursor tryptophan belongs to this family of amino acids. The tryptophan 2, 3-dioxygenase gene (TDO2), which oxidises tryptophan, was differentially expressed in DH. A gene of the FMO family, kynurenine 3-monooxygenase (KMO), was found to be down-regulated in NLH. KMO is involved in tryptophan degradation and might therefore be interesting in regards to skatole [57]. The aldehyde oxidase (AOX1) gene was significantly down-regulated in high androstenone animals of both breeds and this gene has not previously been associated with androstenone levels. It has, however, been shown to play an important role in skatole metabolism in several species including pigs [58-60].
Conclusion
In this study we compared global gene expression profiles from the livers of boars with extreme high and low levels of androstenone from two breeds, Duroc and Norwegian Landrace. Breed differences are evident for molecular functions and biological processes involved in metabolism of androstenone, however many of the same genes are differentially expressed in the two breeds as well. Genes encoding different oxidising enzymes including the cytochrome P450 family (CYP2E1, CYP2A19, CYP2C49, CYP27A1 and CYP2C33) and the flavin-containing monooxygenase family (FMO1 and KMO) were significantly differentially expressed. Furthermore, genes involved in conjugation reactions, including the UDP-glucuronosyltransferases (UGT1A5, UGT2A1 and UGT2B15), sulfotransferases (STE), N-acetyltransferases (NAT12) and glutathione S-transferase were significant, in addition to genes of the 17β-hydroxysteroid dehydrogenase family (HSD17B2, HSD17B4, HSD17B11 and HSD17B13), which are known to regulate availability of active steroids. We suggest a novel role for plasma proteins including AGP and ORM1 in regulating availability of androstenone in pigs. This is the first published microarray experiment describing liver metabolism of androstenone. A number of new candidate genes have been identified, both from phase I and phase II metabolism as well as pathways regulating steroid availability.
Materials and methods
Animals
Animals used in this study were Duroc (D) and Norwegian Landrace (NL) boars from NORSVIN's three boar testing stations. The D boars were on average 156 days old at 100 kg live weight compared to the NL boars that were on average 143 days old at 100 kg live weight. The boars were slaughtered on average 14 days later. Tissue samples from liver were frozen in liquid N2 immediately after slaughter and stored at -80°C until used for RNA extraction as described below. Samples from adipose tissue were collected from the neck at slaughter and stored at -20°C until used for androstenone measurements. Androstenone levels were measured by a modified time-resolved fluoroimmunoassay at the hormone laboratory, Norwegian School of Veterinary Science (NVH) [61] using an antiserum produced at NVH [62]. The androstenone measurements were performed on more than 2500 boars, and statistical power calculations showed that selecting animals from each tail of the androstenone distribution would yield sufficient power to detect differentially expressed genes with a limited number of arrays. The 30 most extreme boars from each tail of the androstenone level distribution were therefore selected. Due to poor RNA quality for two of the samples, 29 samples were subsequently used from each group. 42 of the animals used were the same individuals as those used in our previous study examining gene expression in boar testis [17], with 16 being new animals as liver samples were not available for all of the previously studied animals. Average androstenone levels for the selected boars were 1.17 ppm and 3.22 ppm for NL and D, respectively. (See additional file 5: Androstenone levels). Average values for the groups were 5.95 ± 2.04 ppm for NL high (NLH), 0.14 ± 0.04 ppm for NL low (NLL), 11.57 ± 3.2 ppm for D high (DH) and 0.37 ± 0.17 ppm for D low (DL). In order to reduce family effects, a maximum of two and three half sibs were chosen from NL and D, respectively. The selected animals were used for expression profiling by microarrays and for the following verification of selected genes by rcPCR.
Expression profiling using microarrays
The present work utilises and extends methods described in our previous microarray experiment [17]. The porcine cDNA microarrays were produced at the Faculty of Agricultural Sciences, University of Aarhus and contained 27,774 features printed in duplicates. 26,877 features were PCR products amplified from cDNA clones produced by the Sino-Danish Porcine Genome Sequencing project [63,64], and 867 were control features. The 26,877 features represent approximately 20K gene transcripts. Additional information about the porcine cDNA microarray can be found at NCBIs Gene Expression Omnibus (GEO, [65]) using the platform accession number GPL3585. This is a different batch of microarrays compared to the one we used in the testis experiment.
Total RNA was extracted from liver tissue using Qiagen's RNeasy midi kit according to manufacturer's instruction (Qiagen, CA, USA). RNA quantity was measured on a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, DE, USA) and RNA quality was evaluated by the 28S:18S rRNA ratio using a RNA 6000 Nano LabChip® Kit on 2100 Bioanalyzer (Agilent Technologies, CA, USA). The microarray experiment was conducted as a common reference design using RNA purified from liver tissue sampled from an unrelated Danish Landrace × Hampshire pig as a reference. Aminoallyl-cDNA was synthesised from 15 μg of total RNA using the SuperScript indirect cDNA labelling system (Invitrogen Corporation, CA, USA) and labelled using ARES Alexa Fluor labelling kits (Molecular Probes, OR, USA). Amino-modified and fluorescently labelled cDNA was purified using NucleoSpin 96 Extract II PCR Clean-up kits (Macherey-Nagel, Düren, Germany). The individual samples were labelled with Alexa Fluor 647 and the reference was labelled with Alexa Fluor 555. "Green" spike-in RNA from the Lucidea Universal ScoreCard (Amersham Biosciences) was added to the individuals and "red" spike-in RNA was added to the reference. Hybridisation was performed in a Discovery XT hybridisation station (Ventana Discovery Systems, AZ, USA), followed by manual washing and drying by centrifugation. The microarrays were scanned using a ScanArray Express scanner (Perkin Elmer Inc., MA, USA) and image analyses were conducted using GenePix Pro 6 software (version 6.0.1.26, Molecular Devices Corp., CA, USA). Statistical analyses were carried out in R version 2.3.1 [66] using the software package Linear Models for Microarray Analysis (Limma version 2.7.2) [67-69] which is part of the Bioconductor package [70]. Log-transformed ratios of mean foreground intensities (not background corrected) were print tip loess normalised. The duplicate correlation function in Limma was used to consider duplicate printing of each feature. To evaluate the analyses, MA-plots (M = log2594/log2488, A = (log2594 + log2488)/2), image plots and box plots were constructed using the Limma tools for visualisation both before and after normalisation (see additional file 6: Boxplots). To assess differential expression, Limma uses linear models in combination with an empirical Bayes method to moderate the standard errors of the estimated log-fold changes [68]. The nominal p-values were corrected for multiple testing by false discovery rates (FDR) using Benjamini and Hochberg approach [71]. Each of the groups DH, DL, NLH and NLL animals were hybridised in individual batches, causing some confounding between androstenone levels and hybridisation batch. However, as neither boxplots nor MA-plots were found to differ between hybridisation batches, it was assumed that the impact of hybridisation batch on the obtained data can be ignored. The top 1% of the genes was considered for further analyses. The array features were mapped to a LocusLink identifier and an annotation package was built using the Bioconductor package AnnBuilder (version 1.9.14). GO terms (p < 0.01 and more than 10 significant genes) were analysed for overrepresentation using the GOHyperG function of the Bioconductor package GOstats (version 1.6.0) [72,73]. More detailed descriptions of the microarray experiments are available at the GEO database through the series accession number GSE 11073.
Quantitative real competitive PCR analysis
A real competitive PCR (rcPCR) gene expression analysis was used to verify a subset of the results from the microarray study. Quantitative Gene Expression (QGE) was performed using MassARRAY methodology and the iPLEX protocol (Sequenom, CA, USA). Total RNA was isolated from liver using an automatic DNA/RNA extractor (BioRobot M48 workstation; Qiagen; CA, USA). Total RNA was treated with TURBO DNA-free™ (Ambion, Huntingdon, UK) for removal of contaminating DNA and first strand cDNA synthesis was conducted on 0.5 μg total RNA using SuperScript™-II Rnase H- Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Assays for the genes included in this study (See additional file 7: Gene transcripts included in the rcPCR analyses) were designed and multiplexed into a single reaction using MassARRAY QGE Assay Design software (Sequenom, CA, USA). The competitor, a synthetic DNA molecule matching the targeted cDNA sequence at all positions except for one single base, served as an internal standard for each transcript. A 10-fold competitor dilution was initially used over a wide range of concentrations to determine an approximate equivalence point. Following this, a 7-fold dilution of competitor from 4.04 × 10-11 to 1.43 × 10-19 was used to achieve accurate quantification. The cDNA and competitor were co-amplified in the same PCR reaction with the following conditions: 95°C for 15 minutes, 45 cycles each of 95°C for 20 second, 56°C for 30 seconds and 72°C for 1 minute, and 72°C for 3 minutes. A clean-up step was performed to remove unincorporated nucleotides. The iPLEX reaction cocktail mix and PCR conditions were according to manufacturer's instructions [74]. Parallel PCR reactions were performed for all samples and the products were printed with two replicates on a SpectroCHIP. The primer extension reaction uses PCR products as templates and generates distinct mass signals for competitor and cDNA-derived products. Mass spectrometric analysis generated signals from which peak areas were calculated. Gene expression levels were analysed using TITAN software version 1.0–13 [75,76] that runs in the R statistical environment. Raw data from the Genotype Analyzer Software (Sequenom) were imported into TITAN and analysed using the default values of linear least squares polynomial regression and 4000 bootstrap replicates. cDNA concentrations were corrected with respect to the housekeeping gene (TFRC), and p-values and confidence intervals for fold changes were calculated.
List of abbreviations
D: Duroc; NL: Norwegian Landrace; DH: Duroc high androstenone; NLH: Norwegian Landrace high androstenone; DL: Duroc low androstenone; NLL: Norwegian Landrace high androstenone; 17β-HSD: 17β-hydroxysteroid dehydrogenase; 3β-HSD: 3β-hydroxysteroid dehydrogenase; AGP: alpha-1-acid glycoprotein; AKR: aldo-keto reductase; AKR1C4: aldo-keto reductase family member 1C4; AKR1D1: aldo-keto reductase family member 1D1; AOX1: aldehyde oxidase; CYB5: cytochrome b5; CYP17: cytochrome P450 family 17; CYP19: cytochrome P450 family 19; CYP27A1: cytochrome P450 family member 27A1; CYP2A: cytochrome P450 family 2A; CYP2A19: cytochrome P450 family member 2A19; CYP2A6: cytochrome P450 family member 2A6; CYP2C19: cytochrome P450 family member 2C19; CYP2C33: cytochrome P450 family member 2C33; CYP2C49: cytochrome P450 family member 2C49; CYP2E1: cytochrome P450 family member 2E1; CYP3A: cytochrome P450 family member 3A; DHRS8: short-chain dehydrogenase/reductase 8; FMO: flavin-containing monooxygenase; FMO1: flavin-containing monooxygenase; GATA4: transcription factor GATA-4; GO: gene ontology; GST: glutathione S-transferase; GSTO1: glutathione S-transferase omega; HSDs: hydroxysteroid dehydrogenases; HSD17B2: hydroxysteroid 17β dehydrogenase 2; HSD17B4: hydroxysteroid 17β dehydrogenase 4; HSD17B11: hydroxysteroid 17β dehydrogenase 11; HSD17B13: hydroxysteroid 17β dehydrogenase 13; HSF1: heat shock transcription factor 1; KMO: kynurenine 3-monooxygenase; MGST1: glutathione S-transferase; NAT: N-acetyltransferase; NAT12: N-acetyltransferase family member 12; NFIX: nuclear factor I/X; ORM1: orosomucoid 1; ORM2: orosomucoid 2; QGE: quantitative gene expression; RORA: RAR-related orphan receptor A; rcPCR: real competitive PCR; SDR: short-chain dehydrogenase/reductase; STE: estrogen sulfotransferase; SULT1A1: sulfotransferase family member 1A1; SULT2E1: sulfotransferase family member 1E1; SULT2A1: sulfotransferase family member 2A1; SULT2B1: sulfotransferase family member 2B1; TCEB3: transcription elongation factor B3; TCF8: transcription factor 8; TDO2: tryptophan 2,3-dioxygenase; TFRC: transferrin receptor; UGT: uridine diphospho-glucuronosyltransferase; UGT1A5: UDP-glucuronosyltransferase family member 1A5; UGT2A1: UDP-glucuronosyltransferase family member 2A1; UGT2B15: UDP-glucuronosyltransferase family member 2B15
Authors' contributions
MM carried out the microarray experimental work, performed statistical analysis and drafted the paper. SL was involved in planning the project, provided laboratory facilities for rcPCR work and took part in writing the paper. CB was involved in planning the project and was in charge of lab facilities for microarray studies. JH was involved in microarray experimental work and took part in writing the paper. HH carried out bioinformatics work. IB was involved in statistical analysis. TM was involved in power calculations and statistical supervision. EG coordinated the study, was involved in planning the project, carried out rcPCR molecular work and took part in writing the paper.
Supplementary Material
Microarray results for Duroc. Gene expression profiling was performed using 58 arrays and the 1% most differentially expressed genes were considered significant. The clone names are sequences with a hit to pig, human, mouse or bovine genes. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while some have no hits to the abovementioned species. The ID column gives the feature ID on the microarray, M value indicates fold change, t gives the t-statistics, P.value is the nominal p-value and adj.P.value is the FDR corrected p-value.
Microarray results for Norwegian Landrace. Gene expression profiling was performed using 58 arrays and the 1% most differentially expressed genes were considered significant. The clone names are sequences with a hit to pig, human, mouse or bovine genes. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while some have no hits to the abovementioned species. The ID column gives the feature ID on the microarray, M value indicates fold change, t gives the t-statistics, P.value is the nominal p-value and adj.P.value is the FDR corrected p-value.
Gene ontology (GO) results for the cellular component ontology in Duroc. The top 1% differentially expressed genes in Duroc were analysed for over-represented (p < 0.01) GO terms in the cellular component ontology.
Gene ontology (GO) results for the cellular component ontology in Norwegian Landrace. The top 1% differentially expressed genes in Norwegian Landrace were analysed for over-represented (p < 0.01) GO terms in the cellular component ontology.
Androstenone values. The androstenone values (ppm) in Duroc high (DH), Duroc low (DL), Landrace high (NLH) and Landrace low (LL) animals used in this study.
Boxplots. Boxplots of normalised arrays for Duroc (D) and Norwegian Landrace (L).
Gene transcripts included in the rcPCR analyses.
Acknowledgments
Acknowledgements
This study was financed by the Norwegian pig breeders association (NORSVIN) and The Research Council of Norway. The work conducted at University of Aarhus was supported by the EU-project SABRE. We want to thank Ellen Dahl and Øystein Andresen for being in charge of the androstenone analyses performed at the hormone laboratory at the Norwegian School of Veterinary Science (NVH). We also thank Elin Bergseth at NORSVIN for collecting samples, Dr. Peter Sørensen and Helle Jensen at the microarray platform at University of Aarhus for kind assistance, Dr. Paul Oeth at Sequenom for carrying out the rcPCR primer design and Dr. Matthew Baranski for valuable help with English grammar.
Contributor Information
Maren Moe, Email: maren.moe@umb.no.
Sigbjørn Lien, Email: sigbjorn.lien@umb.no.
Christian Bendixen, Email: christian.bendixen@agrsci.dk.
Jakob Hedegaard, Email: jakob.hedegaard@agrsci.dk.
Henrik Hornshøj, Email: henrikh.jensen@agrsci.dk.
Ingunn Berget, Email: ingunn.berget@umb.no.
Theo HE Meuwissen, Email: theo.meuwissen@umb.no.
Eli Grindflek, Email: eli.grindflek@umb.no.
References
- Patterson RLS. 5alpha-androst-16-ene-3-one: – Compound responsible for taint in boar fat. Journal of the science of food and agriculture. 1968. p. 19.
- Vold E. Fleischproduktionseigenschaften bei ebern und kastraten. Meldinger fra Norges Landbrukshøgskole. 1970;49:1–25. [Google Scholar]
- Babol J, Squires EJ. Quality of meat from entire male pigs. Food research international. 1995;28:201–212. [Google Scholar]
- Sinclair PA, Squires EJ, Raeside JI. Early postnatal plasma concentrations of testicular steroid hormones, pubertal development, and carcass leanness as potential indicators of taint in market weight intact male pigs. Journal of animal science. 2001;79:1868–1876. doi: 10.2527/2001.7971868x. [DOI] [PubMed] [Google Scholar]
- Brooks RI, Pearson AM. Steroid hormone pathways in the pig, with special emphasis on boar odor: a review. Journal of animal science. 1986;62:632–645. doi: 10.2527/jas1986.623632x. [DOI] [PubMed] [Google Scholar]
- Doran E, Whittington FM, Wood JD, McGivan JD. Characterisation of androstenone metabolism in pig liver microsomes. Chemico-biological interactions. 2004;147:141–149. doi: 10.1016/j.cbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Bonneau M, Terqui M. A note on the metabolism of 5alpha-androst-16-en-3-one in the young boar in vivo. Reproduction, nutrition, development. 1983;23:899–905. doi: 10.1051/rnd:19830610. [DOI] [PubMed] [Google Scholar]
- Diaz GJ, Skordos KW, Yost GS, Squires EJ. Identification of phase I metabolites of 3-methylindole produced by pig liver microsomes. Drug metabolism and disposition. 1999;27:1150–1156. [PubMed] [Google Scholar]
- Babol J, Squires EJ, Lundstrom K. Relationship between metabolism of androstenone and skatole in intact male pigs. J Anim Sci. 1999;77:84–92. doi: 10.2527/1999.77184x. [DOI] [PubMed] [Google Scholar]
- Doran E, Whittington FM, Wood JD, McGivan JD. Cytochrome P450IIE1 (CYP2E1) is induced by skatole and this induction is blocked by androstenone in isolated pig hepatocytes. Chemico-biological interactions. 2002;140:81–92. doi: 10.1016/s0009-2797(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Nicolau-Solano SI, McGivan JD, Whittington FM, Nieuwhof GJ, Wood JD, Doran O. Relationship between the expression of hepatic but not testicular 3β-hydroxysteroid dehydrogenase with androstenone deposition in pig adipose tissue. Journal of animal science. 2006;84:2809–2817. doi: 10.2527/jas.2005-595. [DOI] [PubMed] [Google Scholar]
- Moe M, Grindflek E, Doran O. Expression of 3β-hydroxysteroid dehydrogenase, cytochrome P450c17 and sulfotransferase 2B1 proteins in liver and testis of pigs of two breeds: relationship with adipose tissue androstenone level. Journal of animal science. 2007;85:2924–2931. doi: 10.2527/jas.2007-0283. [DOI] [PubMed] [Google Scholar]
- Babol J, Squires E, Lundstrom K. Hepatic metabolism of skatole in pigs by cytochrome P4502E1. Journal of animal science. 1998;76:822–828. doi: 10.2527/1998.763822x. [DOI] [PubMed] [Google Scholar]
- Squires EJ, Lundstrom K. Relationship between cytochrome P450IIE1 in liver and levels of skatole and its metabolites in intact male pigs. J Anim Sci. 1997;75:2506–2511. doi: 10.2527/1997.7592506x. [DOI] [PubMed] [Google Scholar]
- Sinclair PA, Gilmore WJ, Lin Z, Lou Y, Squires EJ. Molecular cloning and regulation of porcine SULT2A1: relationship between SULT2A1 expression and sulfoconjugation of androstenone. Journal of Molecular Endocrinology. 2006;36:301–311. doi: 10.1677/jme.1.01847. [DOI] [PubMed] [Google Scholar]
- Sinclair PA, Squires EJ. Testicular sulfoconjugation of the 16-androstene steroids by hydroxysteroid sulfotransferase: its effect on the concentrations of 5alpha-androstenone in plasma and fat of the mature domestic boar. J Anim Sci. 2005;83:358–365. doi: 10.2527/2005.832358x. [DOI] [PubMed] [Google Scholar]
- Moe M, Meuwissen T, Lien S, Bendixen C, Wang X, Conley LN, Berget I, Tajet H, Grindflek E. Gene expression profiles in testis of pigs with extreme high and low levels of androstenone. BMC Genomics. 2007:8. doi: 10.1186/1471-2164-8-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Lou Y, Squires JE. Molecular cloning and functional analysis of porcine SULT1A1 gene and its variant: a single mutation SULT1A1 causes a significant decrease in sulfation activity. Mammalian genome. 2004;15:218–226. doi: 10.1007/s00335-002-2318-4. [DOI] [PubMed] [Google Scholar]
- Babol J, Squires EJ, Lundström K. Relationship between oxidation and conjugation metabolism of skatole in pig liver and concentrations of skatole in fat. Journal of animal science. 1998;76:829–838. doi: 10.2527/1998.763829x. [DOI] [PubMed] [Google Scholar]
- Diaz GJ, Squires EJ. Phase II in vitro metabolism of 3-methylindole metabolites in porcine liver. Xenobiotica. 2003;33:485–498. doi: 10.1080/0049825031000076186. [DOI] [PubMed] [Google Scholar]
- Tajet H, Andresen Ø, Meuwissen THE. Prevention of boar taint in pig production: the 19th symposium of the nordic committee for veterinary scientific cooperation gardermoen, norway. 21–22 november 2005. Abstracts. Acta Veterinaria Scandinavica Supplement. 2005;48:22–23. doi: 10.1186/1751-0147-48-S1-P1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JA, Faletto MB. Advances in mechanisms of activation and deactivation of environmental chemicals. Environmental Health Perspectives. 1993;100:169–176. doi: 10.1289/ehp.93100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Journal of clinical pharmacology. 2001;52:349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaanild MT. Porcine cytochrome P450 and metabolism. Current Pharmaceutical Design. 2006;12:1421–1427. doi: 10.2174/138161206776361183. [DOI] [PubMed] [Google Scholar]
- Zamaratskaia G, Gilmore WJ, Lundstrom K, Squires EJ. Effect of testicular steroids on catalytic activities of cytochrome P450 enzymes in porcine liver microsomes. Food and Chemical Toxicology. 2007;45:676–681. doi: 10.1016/j.fct.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Diaz GJ, Squires EJ. Metabolism of 3-methylindole by porcine liver microsomes: responsible cytochrome P450 enzymes. Toxicology science. 2000;55:284–292. doi: 10.1093/toxsci/55.2.284. [DOI] [PubMed] [Google Scholar]
- Skinner TM, Doran E, McGivan JD, Haley CS, Archibald AL. Cloning and mapping of the porcine cytochrome-p450 2E1 gene and its association with skatole levels in the domestic pig. Animal genetics. 2005;36:417–422. doi: 10.1111/j.1365-2052.2005.01342.x. [DOI] [PubMed] [Google Scholar]
- Terner MA, Gilmore WJ, Lou Y, Squires EJ. The role of CYP2A and CYP2E1 in the metabolism of 3-methylindole in primary cultured porcine hepatocytes. Drug metabolism and disposition: the biological fate of chemicals. 2006;34:848–854. doi: 10.1124/dmd.105.008128. [DOI] [PubMed] [Google Scholar]
- Skaanild MT, Friis Ch. Porcine CYP2A polymorphisms and activity. Basic and Clinical Pharmacology and Toxicology. 2005;97:115–121. doi: 10.1111/j.1742-7843.2005.pto_148.x. [DOI] [PubMed] [Google Scholar]
- Chen G, Cue R-A, Lundstrom K, Wood JD, Doran O. Regulation of CYP2A6 protein expression by skatole, indole and testicular steroids in primary cultured pig hepatocytes. Drug metabolism and disposition. 2008;36:56–60. doi: 10.1124/dmd.107.017285. [DOI] [PubMed] [Google Scholar]
- Tang W, Norlin M, Wikvall K. Regulation of human CYP27A1 by estrogens and androgens in HepG2 and prostate cells. Archives of biochemistry and biophysics. 2007;462:13–20. doi: 10.1016/j.abb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Cashman JR. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochemical and biophysical research communications. 2005;338:599–604. doi: 10.1016/j.bbrc.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Falls JG, Ryu D-Y, Cao Y, Levi PE, Hodgson E. Regulation of mouse liver flavin-containing monooxygenases 1 and 3 by sex steroids. Archives of biochemistry and biophysics. 1997;342:212–223. doi: 10.1006/abbi.1997.9965. [DOI] [PubMed] [Google Scholar]
- Dannan GA, Guengerich FP, Waxman DJ. Hormonal regulation of rat liver microsomal enzymes: Role of gonadal steroids in programming, maintenance, and suppression of delta 4-steroid 5 alpha-reductase, flavin-containing monooxygenase, and sex-specific cytochromes P-450. The Journal of Biological Chemistry. 1986;261:10728–10735. [PubMed] [Google Scholar]
- Claus R, Hoffman B. Oestrogens, compared to other steroids of testicular origin, in bloodplasma of boars. Acta Endocrinologica. 1980;94:404–411. doi: 10.1530/acta.0.0940404. [DOI] [PubMed] [Google Scholar]
- Zamaratskaia G, Rydhmer L, Chen G, Madej A, Andersson HK, Lundstrom K. Boar taint is related to endocrine and anatomical changes at puberty but not to aggressive behaviour in entire male pigs. Reproduction in domestic animals. 2005;40:500–506. doi: 10.1111/j.1439-0531.2005.00613.x. [DOI] [PubMed] [Google Scholar]
- Chen G, Bourneuf E, Marklund S, Zamaratskaia G, Madej A, Lundstrom K. Gene expression of 3β-hydroxysteroid dehydrogenase and 17β-hydroxysteroid dehydrogenase in relation to androstenone, testosterone, and estrone sulphate in gonadally intact male and castrated pigs. Journal of animal science. 2007;85:2457–2463. doi: 10.2527/jas.2007-0087. [DOI] [PubMed] [Google Scholar]
- Hum DW, Bèlager A, Lèvesque È, Barbier O, Beaulieu M, Albert C, Vallèe M, Guillemette C, Tchernof A, Turgeon D, Dubois S. Characterization of UDP-glucuronosyltransferases active on steroid hormones. Journal of steroid biochemistry and molecular biology. 1999;69:413–423. doi: 10.1016/s0960-0760(99)00061-8. [DOI] [PubMed] [Google Scholar]
- Finel M, Li X, Gardner-Stephen D, Bratton S, Mackenzie PI, Radominska-Pandya A. Human UDP-Glucuronosyltransferase 1A5: Identification, expression, and activity. Journal of Pharmacology and Experimental Therapeutics. 2005;315:1143–1149. doi: 10.1124/jpet.105.091900. [DOI] [PubMed] [Google Scholar]
- Jedlitschky G, Cassidy AJ, Sales M, Norman P, Burchell B. Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochemical journal. 1999;340:837–843. [PMC free article] [PubMed] [Google Scholar]
- Sinclair PA, Hancock S, Gilmore WJ, Squires EJ. Metabolism of the 16-androstene steroids in primary cultured porcine hepatocytes. J Steroid Biochem Mol Biol. 2005. [DOI] [PubMed]
- Saat YA, Gower DB, Harrison FA, Heap RB. Studies on the metabolism of 5α-androst-16-en-3-one in boar testis in vivo. Biochemical journal. 1974;144:347–352. doi: 10.1042/bj1440347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher NJ, Tetlow NL, Cheung C, Broadhurst GM, Minchin RF. Induction of human arylamine N-acetyltransferase type I by androgens in human prostate cancer cells. Cancer Research. 2007;67:85–92. doi: 10.1158/0008-5472.CAN-06-2635. [DOI] [PubMed] [Google Scholar]
- Listowsky I, Abramovitz M, Homma H, Niitsu Y. Intracellular binding and transport of hormones and xenobiotics by glutathione S-transferase. Drug metabolism reviews. 1988;19:305–318. doi: 10.3109/03602538808994138. [DOI] [PubMed] [Google Scholar]
- Baker ME. Evolution of 17beta-hydroxysteroid dehydrogenases and their role in androgen, estrogen and retinoid action. Molecular and cellular endocrinology. 2001;171:211–215. doi: 10.1016/s0303-7207(00)00414-7. [DOI] [PubMed] [Google Scholar]
- Bauman DR, Steckelbroeck S, Penning TM. The roles of aldo-keto reductases in steroid hormone action. Drug News Perspect. 2004;17:563–578. doi: 10.1358/dnp.2004.17.9.872570. [DOI] [PubMed] [Google Scholar]
- Wu L, Einstein M, Geissler WM, Chan HK, Elliston KO, Andersson S. Expression cloning and characterization of human 17beta-hydroxysteroid dehydrogenase type 2, a microsomal enzyme possessing 20alpha-hydroxysteroid dehydrogenase activity. Journal of biological chemistry. 1993;268:12964–12969. [PubMed] [Google Scholar]
- Liu S, Huang C, Li D, Ren W, Zhang H, Qi M, Li X, Yu L. Molecular cloning and expression analysis of a new gene for short-chain dehydrogenase/reductase 9. Acta Biochimica Polonica. 2007;54:213–218. [PubMed] [Google Scholar]
- Chai Z, Brereton P, Suzuki T, Sasano H, Obeyesekere V, Escher G, Saffery R, Fuller P, Enriquez C, Krozowski Z. 17β-hydroxysteroid dehydrogenase type XI localizes to human steroidogenic cells. Endocrinology. 2003;144:2084–2091. doi: 10.1210/en.2002-221030. [DOI] [PubMed] [Google Scholar]
- Palermo M, Marazzi MG, Hughes BA, Stewart PM, Clayton PT, Shackleton CHL. Human Δ4-3-oxosteroid 5β-reductase (AKR1D1) deficiency and steroid metabolism. Steroids. 2008;73:417–423. doi: 10.1016/j.steroids.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Fournier T, Medjoubi-N N, Porquet D. Alpha-1-acid glycoprotein. Biochimica et Biophysica Acta. 2000;1482:157–171. doi: 10.1016/s0167-4838(00)00153-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ding LR, Ishii M, Fischer NE, Inaba T. The interaction of human and bovine serum proteins with CYP3A in human liver microsomes: inhibition of testosterone 6beta-hydroxylation by albumin, alpha-globulins, alpha-1-acid glycoprotein and gamma-globulins. Toxicology letters. 2002;136:33–41. doi: 10.1016/s0378-4274(02)00285-0. [DOI] [PubMed] [Google Scholar]
- Xu BQ, Ishii M, Ding LR, Fischer NE, Inaba T. Interaction of serum proteins with CYP isoforms in human liver microsomes: inhibitory effects of human and bovine albumin, alpha-globulins, alpha-1-acid glycoproteins and gamma-globulins on CYP2C19 and CYP2D6. Life Sciences. 2003;72:1953–1962. doi: 10.1016/s0024-3205(03)00093-6. [DOI] [PubMed] [Google Scholar]
- Guiraudie G, Pageat P, Cain A-H, Madec I, Meillour PNL. Functional characterization of olfactory binding proteins for appeasing compounds and molecular cloning in the vomeronasal organ of pre-pubertal pigs. Chemical senses. 2003;28:609–619. doi: 10.1093/chemse/bjg052. [DOI] [PubMed] [Google Scholar]
- Meadus WJ, Mason JI, Squires EJ. Cytochrome P450c17 from porcine and bovine adrenal catalyses the formation of 5,16-androstadien-3 beta-ol from pregnenolone in the presence of cytochrome b5. Journal of steroid biochemistry and molecular biology. 1993;46:565–572. doi: 10.1016/0960-0760(93)90183-w. [DOI] [PubMed] [Google Scholar]
- Duarte MP, Palma BB, Gilep AA, Laires A, Oliveira JS, Usanov SA, Rueff J, Kranendonk M. The stimulatory role of human cytochrome b5 in the bioactivation activities of human CYP1A2, 2A6 and 2E1: a new cell expression system to study cytochrome P450 mediated biotransformation. Mutagenesis. 2005;20:93–100. doi: 10.1093/mutage/gei012. [DOI] [PubMed] [Google Scholar]
- Crozier KR, Moran GR. Heterologous expression and purification of kynurenine-3-monooxygenase from Pseudomonas fluorescens strain 17400. Protein Expression and Purification. 2007;51:324–333. doi: 10.1016/j.pep.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Diaz GJ, Squires EJ. Role of aldehyde oxidase in the hepatic in vitro metabolism of 3-methylindole in pigs. Journal of agricultur food chemistry. 2000;48:833–837. doi: 10.1021/jf990805u. [DOI] [PubMed] [Google Scholar]
- Skordos KW, Skiles GL, Laycock JD, Lanza DL, Yost GS. Evidence supporting the formation of 2,3-epoxy-3-methylindoline: a reactive intermediate of the pneumotoxin 3-methylindole. Chemical research in toxicology. 1998;11:741–749. doi: 10.1021/tx9702087. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Appleton ML, Carlson JR, Yost GS. Identification of β-glucuronidase-resistant diastereomeric glucuronides of 3-hydroxy-3-methyloxindole formed during 3-methylindole metabolism in goats. Drug metabolism and disposition. 1996;24:119–125. [PubMed] [Google Scholar]
- Tuomola M, Harpio R, Knuuttila P, Mikola H, Løvgren T. Time-resolved fluoroimmunoassay for the measurement of androstenone in porcine serum and fat samples. Journal of agricultural food chemistry. 1997;45:3529–3534. [Google Scholar]
- Andresen Ø. Development of radioimunoassay for 5alpha-adrost-16-en-3-one in pig peripheral plasma. Acta endochrinologia. 1974;76:377–387. doi: 10.1530/acta.0.0760377. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Cirera S, Hedegaard J, Gilchrist MJ, Panitz F, Jørgensen C, Scheibye-Knudsen K, Arvin T, Lumholdt S, Sawera M, Green T, Nielsen BJ, Havgaard JH, Rosenkilde C, Wang J, Li H, Li R, Liu B, Hu S, Dong W, Li W, Yu J, Wang J, Stærfeldt H-H, Wernersson R, Madsen LB, Thompsen B, Hornshøj H, Bujie Z, Wang X, Wang X, Bolund L, Brunak S, Yang H, Bendixen C, Fredholm M. Porcine transcriptome analysis based on 97 non-normalized cDNA libraries and assembly of 1,021,891 expressed sequence tags. Genome biology. 2007;8:R45. doi: 10.1186/gb-2007-8-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Schierup RH, Jørgensen FG, Gorodkin J, Panitz F, Staerfeldt HH, Christensen OF, Mailund T, Hornshøj H, Klein A, Wang J, Liu B, Hu S, Dong W, Li W, Wong GKS, Yu J, Wang J, Bendixen C, Fredholm M, Brunak S, Yang H, Bolund L. Pigs in sequence space: a 0.66X pig genome survey based on shotgun sequencing. BMC Genomics. 2005:6. doi: 10.1186/1471-2164-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2007. http://www.ncbi.nlm.nih.gov/geo/Ref Type: Electronic Citation.
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of computational and graphical statistics. 1996;5:299–314. [Google Scholar]
- Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004. p. 3. [DOI] [PubMed]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editor. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal Of The Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2006;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Hornshøj H, Conley LN, Hedegaard J, Sørensen P, Panitz F, Bendixen C. Microarray expression profiles of 20.000 genes across 23 healthy porcine tissues. PLoS ONE. 2007;2:e1203. doi: 10.1371/journal.pone.0001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2007. http://www.sequenom.com
- Elvidge GP, Price TS, Glenny L, Ragoussis J. Development and evaluation of real competitive PCR for high-throughput quantitative applications. Analytical Biochemistry. 2005;339:231–241. doi: 10.1016/j.ab.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 2007. http://itmat.upenn.edu/~tsprice/software.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray results for Duroc. Gene expression profiling was performed using 58 arrays and the 1% most differentially expressed genes were considered significant. The clone names are sequences with a hit to pig, human, mouse or bovine genes. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while some have no hits to the abovementioned species. The ID column gives the feature ID on the microarray, M value indicates fold change, t gives the t-statistics, P.value is the nominal p-value and adj.P.value is the FDR corrected p-value.
Microarray results for Norwegian Landrace. Gene expression profiling was performed using 58 arrays and the 1% most differentially expressed genes were considered significant. The clone names are sequences with a hit to pig, human, mouse or bovine genes. Some genes are represented by several different clones on the array and may therefore show up more than once in the table, while some have no hits to the abovementioned species. The ID column gives the feature ID on the microarray, M value indicates fold change, t gives the t-statistics, P.value is the nominal p-value and adj.P.value is the FDR corrected p-value.
Gene ontology (GO) results for the cellular component ontology in Duroc. The top 1% differentially expressed genes in Duroc were analysed for over-represented (p < 0.01) GO terms in the cellular component ontology.
Gene ontology (GO) results for the cellular component ontology in Norwegian Landrace. The top 1% differentially expressed genes in Norwegian Landrace were analysed for over-represented (p < 0.01) GO terms in the cellular component ontology.
Androstenone values. The androstenone values (ppm) in Duroc high (DH), Duroc low (DL), Landrace high (NLH) and Landrace low (LL) animals used in this study.
Boxplots. Boxplots of normalised arrays for Duroc (D) and Norwegian Landrace (L).
Gene transcripts included in the rcPCR analyses.


