Abstract
Both natural and unnatural modifications in RNA are of interest to biologists and chemists. More than 100 different analogs of the four standard RNA nucleosides have been identified in nature. Unnatural modifications are useful for structure and mechanistic studies of RNA. This Review highlights chemical, enzymatic, and combined (semisynthesis) approaches to generate site-specifically modified RNAs. The availability of these methods for site-specific modifications of RNAs of all sizes is important in order to study the relationships between RNA chemical composition, structure, and function.
RNAs undergo specific post-transcriptional modification by a wide variety of enzymes and ribonucleoprotein complexes (1, 2). More than 100 different modifications of the four standard RNA nucleosides, adenosine, cytidine, guanosine, and uridine, have been identified (3). Examples from each of the four major categories of base isomerization, base modification, sugar modification, and hypermodification are shown in Figure 1, panel a. Similarly, a large number of unnatural modifications have been synthesized and used for structure and mechanistic studies of RNA (4–6). A few examples are shown in Figure 1, panel b (7–10). Natural modified nucleotides are often found in the functionally important regions of RNA, such as the peptidyl transferase center of ribosomal RNA (rRNA), the anticodon loop of transfer RNAs, or the branch site of spliceosomal RNAs (11–13). Several modifications, such as pseudouridine (14), have been known for almost 50 years. Their locations in natural RNAs can, in some cases, be highly conserved; yet, our understanding of their biological roles is still incomplete (15). Through a combination of biological, chemical, and biophysical approaches, much can be learned regarding the roles of modified nucleotides. Similarly, the incorporation of unnatural modifications may be useful in order to study the biological roles and functions of RNA (5).
Figure 1.
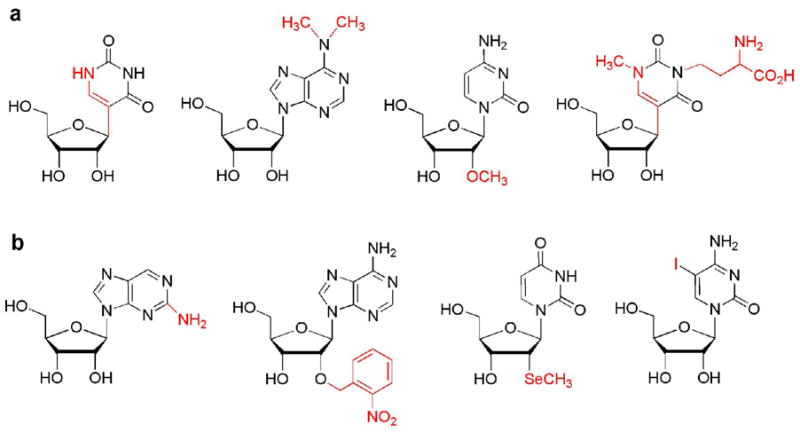
a) Four major types of natural modification are shown: isomerization of uridine to pseudouridine (Ψ) base modification of adenosine to N6, N6-dimethyladenosine (m26A), 2′-O-methylation of cytidine to 2′-O-methylcytidine (Cm), and multiple modification of pseudouridine to 1-methyl-3-(3-amino-3-carboxypropyl)pseudouridine (m1acp3Ψ). b) Four examples of unnatural modifications are shown: 2-aminopurine, 2′-O-(2-nitrobenzyl)adenine, 2′-Se-methyluridine, and 5-iodocytidine.
In a number of cases, the enzymes or small nucleolar RNAs (snoRNAs) that are responsible for site-specific RNA modification have been identified (2). Knock-out studies of the individual modifying enzymes or snoRNAs then allow the function of the modification to be deduced. In cases where the enzymes are not known, or the modification is not a natural one, alternative approaches must be considered. Over the past several decades, several chemical methods have been developed to study the effects of modifications in small RNA model systems. These studies have led to newer, more powerful approaches that allow site-specific modification of large RNAs, reconstitution into biological systems, and studies of RNA structure and function.
Chemical Synthesis
Chemical synthesis of RNA allows for site-selective incorporation of modified nucleotides. Five decades ago, Michelson and coworkers synthesized a thymidylate dinucleotide by the phosphate triester method, which served as the starting point for oligonucleotide synthesis (16). Khorana and colleagues then developed the phosphodiester method (17), which was followed by H-phosphonate and phosphoramidite approaches (18). Among the various chemical methods developed for RNA synthesis, the phosphoramidite approach is the most widely used. This method involves four repeating steps that occur on a solid support (typically CPG or polystyrene beads) following removal of the 5′-hydroxyl protecting group on the 3′ nucleotide (Figure 2): i) coupling at the 5′ site with a protected phosphoramidite, ii) capping of the unreacted 5′-hydroxyl groups, iii) oxidation of the newly formed phosphite linkage, and iv) removal of the 5′-protecting group on the newly added nucleotide. Lastly, the synthetic RNA is cleaved from the solid support, the remaining protecting groups are removed, and the final RNA product is purified (typically by HPLC or polyacrylamide gel electrophoresis).
Figure 2.
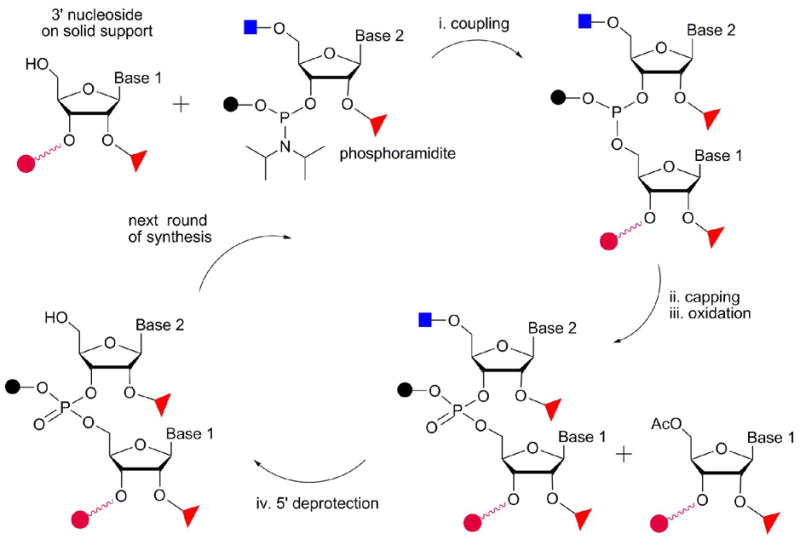
The general scheme for solid-phase RNA synthesis is shown. A number of protective groups used at the 5′ (blue squares), 2′ (open triangles), and 3′ (black circles) positions are summarized in Table 1. The solid support is represented with a red circle.
In solid-phase RNA synthesis, the choice of ribose 5′ and 2′protecting groups is important. The 2′-hydroxyl protecting group must be easy to introduce, stable during each synthetic step, and easily removed under mild conditions to generate the final RNA product. The 5′-hydroxyl and phosphoramidite protecting groups must be orthogonal to the 2′-protecting group, and be easily removed at the proper time during the synthetic cycle. Some examples include protecting groups that are fluoride-labile (e.g., TBDMS) (19), acid labile (e.g., Fpmp) (20), photolabile (e.g., Nbn) (8) or that are removed under reducing conditions (e.g., DTM) (21) or by metal catalysis (e.g., allyl) (22, 23) (Table 1). The traditional 5′-O-DMT–2′-O-TBDMS–3′-O-(2-cyanoethyl-N, N-diisopropyl) phosphoramidite chemistry (Figure 3, panel a) enables the synthesis of ~30 nucleotide (nt) RNAs on the nanomole to high micromole scale on a standard commercial DNA/RNA synthesizer using CPG supports (24).
Table 1.
Protective groups and general conditions for solid-phase oligoribonucleotide synthesis
| 5′▵ | 2

|
3′● | support

|
5′ deprotection | activator, coupling time, efficiency | oxidation | 2′ deprotection | ref |
|---|---|---|---|---|---|---|---|---|
| TBDMS | DMT | CNEt | CPG | TCA | tetrazole, 6–12 min, >95% | I2a | i) fluoride (TBAF) | 24 |
| Fpmp | DMT | CNEt | CPG | TCA | tetrazole, 3–15 min, >98% | I2a | ii) acid (NaOAc/pH 3.25) | 20 |
| Nbn | DMT | CNEt | CPG | TCA | tetrazole, 5–15 min, ~95% | I2a | iii) hν8 nm) | 8 |
| DTM | DMT | CNEt | CPG | TCA | ETT, 150 sec, >98% | I2a | iv) reduction (DTT or TCEP) | 21 |
| allyl | DMT | allyl | ref. 53 | TCA | tetrazole, 12 min, >93% | t-C4H9OOH | i) metal Pd2(dba)3 | 22 |
| TOM | DMT | CNEt | CPG | DCA | BTT, 2.5 min, >99% | I2a | i) fluoride(TBAF) | 26 |
| ACE | BzH | Me | P | HF/TEA | ETT, 90 sec, >99% | t-C4H9OOH | ii) acid (HOAc/TEMED/pH 3.8) | 27 |
I2/H2O/THF/Pyr
Figure 3.
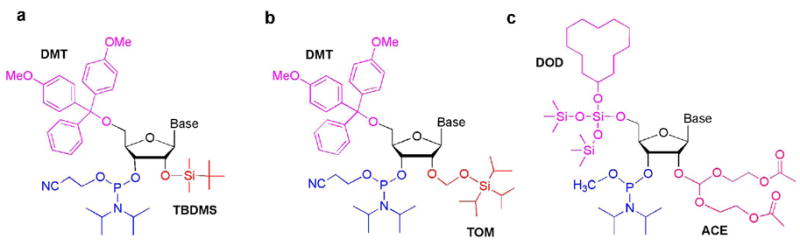
Three types of nucleoside phosphoramidites are shown: a) 5′-O-DMT–2′-O-TBDMS–3′-O-(2-cyanoethyl-NN, -diisopropyl), b) 5′-O-DMT–2′-O-TOM–3′-O-(2-cyanoethyl-NN, -diisopropyl), and c) 5′-O-DOD–2′-O-ACE–3′-O-(methyl-NN, -diisopropyl) phosphoramidites.
Several improvements to the RNA synthesis protocols have allowed for the generation of RNAs on a large scale and with longer lengths. In 1998, Pitsch and coworkers developed a novel 2′-O-protecting group, 2′-O-TOM (Figure 3, panel b), that gives high coupling yields (>99%) due to reduced steric hindrance compared to TBDMS (25, 26). With this strategy, long (up to 80 nt) RNAs with site-specific modifications can be generated. In 1998, Scaringe and coworkers developed a different strategy for RNA chemical synthesis, which employs novel 5′- and 2′-O-protecting groups. Their approach utilizes 5′-O-DOD ether and 2′-O-ACE orthoester protecting groups along with the 3′-O-(methyl-N, N-diisopropyl) phosphoramidite (Figure 3, panel c) that gives coupling yields >99% (27). Because of the presence of the silyl 5′-protecting group, which requires fluoride deprotection in each cycle, the synthesis is carried out on polystyrene (PS) supports rather than silica-based supports (e.g., CPG). An alternative to the DOD protecting group is BzH (28).
With the TOM approach, relatively large RNAs can be generated, and the chemistry is compatible with the traditional DMT/TBDMS chemistry; thus, phosphoramidites using DMT/TBDMS chemistry can be mixed with those using TOM chemistry. The DOD(BzH)/ACE amidites typically have faster coupling times (90 s) than the TOM amidites (>2 min) and also give overall higher yields and very pure RNA (few failed sequences). However, the length of RNA that can be generated using the DOD(BzH)/ACE chemistry is typically ~30–40 nt, and the DNA/RNA synthesizer has to be modified to accommodate reagents that are compatible with the DOD(BzH)/ACE protecting groups. For both methods, new phosphoramidites have to be generated for each modified nucleotide, and biologically relevant RNA lengths can often not be achieved. Furthermore, the less common base- and acid-labile modifications, such as acetyl, cannot be incorporated with either of these methods. In that case, alternative strategies must be considered (22, 23, 29), such as allyl protection and allyl linkers attached to PS/polyethylene glycol (PEG) supports (30). Some more recent strategies for RNA synthesis involve the use of CEM (31), DTM (21), and TEM (32) protection of the 2′-hydroxyl group. A fully functional 110-mer RNA has been recently synthesized using the 2′-O-CEM protection chemistry (33).
The advantages of generating site-specifically modified oligonucleotides by chemical synthesis are multifold: i) RNAs with single or multiple modifications can be generated in order to study the individual effects of the natural modified nucleotides, ii) large quantities of RNA can be generated that are 100% modified at the desired locations, and iii) the roles of specific functional groups in RNA can be studied by using synthetic analogs with only minor changes in the chemical composition. In particular, these approaches have been used to generate small, modified RNAs in high quantities for biophysical studies (28, 34–41). Similarly, unnatural modifications have allowed mechanistic studies to be carried out on RNAs such as ribozymes (5, 7–9, 42–45). The next section will focus on approaches used to generate larger, site-specifically modified RNAs using a combination of chemical and enzymatic synthesis.
Enzymatic Synthesis
The modified nucleotides cannot typically be incorporated into RNA site-specifically using T7 RNA polymerase and in vitro transcription (46). In some cases, the specific RNA modifying enzymes have been isolated that will modify either small or large RNA templates (1). Although the enzyme reactions are often highly specific for certain nucleotide sequences or structural motifs, they often do not go to completion, the modified and unmodified RNAs have to be separated for further studies, and it is difficult to incorporate more than one type of modified nucleotide into the RNA. Furthermore, the separation and analysis of mixtures can be particularly challenging if the masses of the unmodified and modified RNAs are close (e.g., the difference of a single methyl group) or identical (e.g., pseudouridine vs uridine). Some new approaches have been taken to identify and separate RNA transcripts with site-specific modifications. Certain types of base modification (e.g., m1G) interfere with Watson–Crick base pairing. These may render the RNA resistant to RNase H degradation in the presence of a complementary DNA strand (47). Similarly, RNAs containing 2′-O-Me modifications are resistant to RNase H (48, 49). RNA can be modified in a stepwise fashion at the 3′ end with a modified nucleotide 3′, 5′-bisphosphate and T4 RNA ligase (50, 51), although these methods can be time-consuming and not highly efficient.
Site-specific modifications can also be made to RNA by using unnatural base pair systems (52). The unique complementarity of an unnatural base pair allows one modified nucleotide to direct the incorporation of another into an RNA strand by using RNA polymerase. One early example showed that a site-specific isoC within a DNA template could direct the incorporation of the modified nucleotide isoG into an RNA transcript (53). Similarly, Hirao and coworkers have shown that the unnatural nucleotide 2-amino-6-(2-thienyl)purine can direct the site-specific incorporation of 2-oxopyridine into RNA (54). More recently, the same group has expanded the scope of the hydrophobic base pairs developed by Schweitzer and Kool (55) and Romesberg et al. (56) for the site-specific incorporation of nucleotide analogs in RNA (57). In addition, they have demonstrated site-specific incorporation of cross-linking analogs, biotinylated nucleotides, and fluorescent analogs into RNA (58–60). These systems are limited by the need to develop completely new, orthogonal base pairs; however, once these systems are established, the potential applications are numerous.
Semisynthesis Approaches
One of the major challenges in understanding the effects of the modified nucleotides in RNA is the generation of site-specifically modified RNAs that are representative of the natural RNAs, which are typically longer than is routinely achievable by direct chemical synthesis. These RNAs can be generated with internal modifications using a semisynthesis approach. In this case, the RNA is generated in two or more segments, either chemically or enzymatically, in which one or more segment contains the modification(s) of interest (5, 61). One RNA segment can be modified at the 3′ end using a bisphosphate analog of the modified nucleotide and T4 RNA ligase (51). The advantage of this approach is that any size RNA can be modified and the reaction is generally not limited by sequence or nucleoside composition. Alternatively, the modified RNA segment can be generated using available phosphoramidites and chemical synthesis. T4 RNA ligase can be used to ligate RNA fragments with a 5′ phosphate on the donor strand and a 3′ hydroxyl on the acceptor strand. One major drawback, however, is the possibility of circularization of the individual RNA fragments. Therefore, the ends of the RNAs to be joined need to be appropriately modified with phosphate, hydroxyl, or 2′, 3′-cyclic phosphate groups. The segments can instead be joined by ligation with bacteriophage T4 DNA ligase along with a bridging, complementary DNA template, or “splint” (Figure 4, panels a–c) (5, 61). This method has been used to generate successfully a wide range of modified RNAs, such as selenium-modified RNAs for X-ray crystallography studies (9); however, the ligation step often takes many hours for completion and/or suffers from poor efficiency, as well as requiring large amounts of DNA ligase. A recent modification of the method employs RNA ligase, modified RNA fragments, and DNA splint to achieve highly efficient and rapid ligation of synthetic RNA fragments (Figure 4, panel d) (62, 63). In this case, the RNA fragments are single-stranded (i.e., not paired with the DNA splint) at the ligation site in order to accommodate T4 RNA ligase, which prefers single-stranded substrates. Alternatively, long splints have been shown to increase the ligation efficiencies with T4 DNA ligase (64). The improved hybridization of the longer splint reduces the amount of RNA secondary structure formation, which could affect the ligation efficiency.
Figure 4.
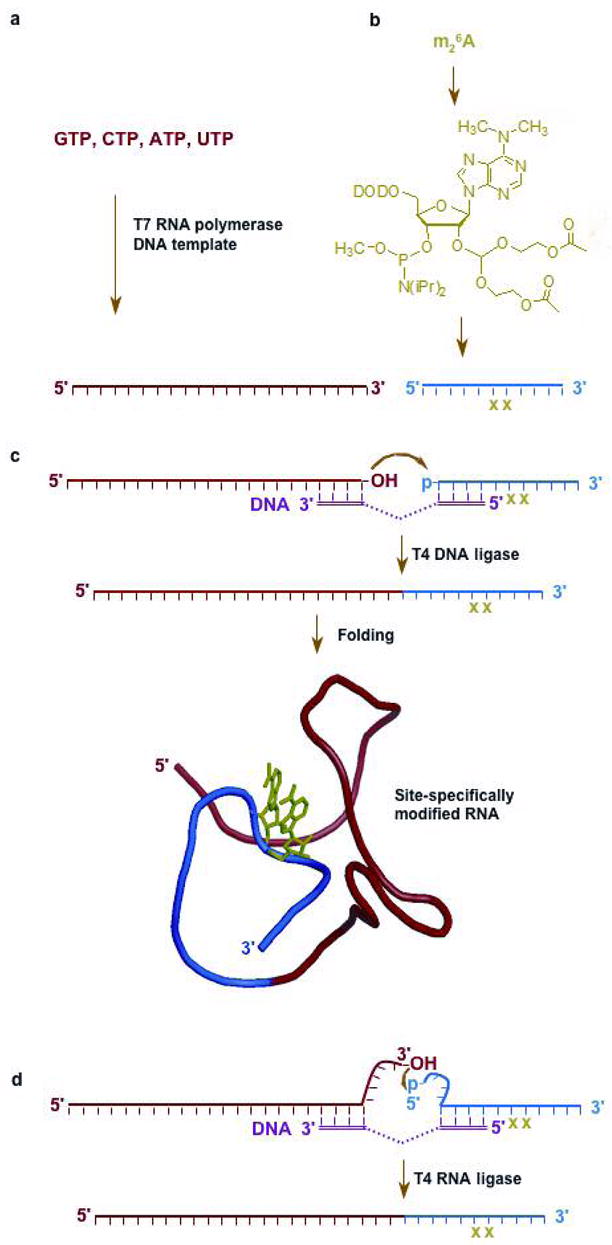
A schematic diagram shows the generation of a site-specifically modified RNA. a) The enzymatic synthesis of an unmodified RNA segment (in red) using standard ribonucleotide triphosphates, a DNA template, and T7 RNA polymerase is represented. b) Chemical synthesis can be employed to generate an RNA segment (in blue) containing modified nucleotides (in green) at specific locations using the corresponding phosphoramidites. c) DNA-dependent RNA ligation of the two RNA segments using T4 DNA ligase generates a long segment of RNA representing a natural, functional RNA from the ribosome. d) An alternative strategy shows DNA-templated RNA ligation of two RNA segments with single-stranded regions using T4 RNA ligase.
Engineering of Natural, Modified RNAs
The site-specific modification of an RNA containing thousands of nucleotides is a daunting task; however, Polacek and coworkers have recently succeeded in engineering 23S rRNA (>2500 nucleotides in length) from the large ribosomal subunit by a method called gapped-circularly permutated-reconstitution (65, 66). Rather than piece together the entire 23S rRNA, a circularly permuted 23S rRNA was generated in which the 5′ and 3′ ends of the RNA were covalently connected and new ends were introduced at the site of modification. The circularly permuted RNA was created by a series of polymerase chain reaction, enzyme digestion, ligation, and cloning steps, starting with a plasmid carrying a tandem repeat of the 23S rRNA gene. When the 23S rRNA was created, a gap was left in which a modified synthetic fragment was placed in trans (65). In the second case, ligation of the modified fragment into the gapped site was necessary to generate a functional RNA (66). The ribosome can then be reconstituted with the modified rRNA (circularly permuted RNA and synthetic fragment) and remaining components (5S rRNA and proteins). The advantage of the gapped-cp-reconstitution method is that it allows the incorporation of non-natural nucleotides into the desired location on very large RNAs, although further improvements are still needed in order to improve the efficiency of the reconstituted system. This newly created ribosome system was used to understand the role of specific nucleotides in the peptidyl transferase mechanism (65, 66), and nicely highlights the use of combined chemical and enzymatic approaches to generate site-specifically modified RNAs that are able to carry out biological functions.
Detection of Site-Specific RNA Modifications
A number of approaches have been taken to detect and locate modified nucleotides in RNA. Most involve the use of enzyme digestion followed by chromatography and/or mass spectrometry, or the use of chemical modification and reverse transcriptase inhibition. The use of these methods is particularly important for the characterization of site-specifically modified RNAs that are engineered through chemical synthesis, enzymatic synthesis, or semisynthesis approaches described above.
Typically, small synthetic RNAs can be characterized by matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI) mass spectrometry (MS) (67) to provide information about the mass and purity of the RNA product. Complete P1 nuclease digestion of the modified RNA followed by treatment with alkaline phosphatase will generate a mixture of free nucleosides that can be separated by reverse-phase high-performance liquid chromatography (HPLC) (68, 69). The retention times of the digested products are then compared to authentic standards. The identity of the digestion products can be further confirmed by using ESI MS. This method allows the composition (number and types of modified nucleosides) of the RNA to be confirmed, but not the site or position within the RNA strand.
The position of modified nucleotides in a given RNA oligonucleotide can be identified by nucleotide-specific RNase digestion and subsequent analysis of the resulting small oligonucleotide fragments by MALDI MS or by coupled liquid chromatography/ESI MS (67). Tandem mass spectrometry on the individual oligonucleotide fragments can be used to further analyze and identify the position of modification (67). Mass silent modifications, such as pseudouridine, require modification by chemical reagents (e.g., cyanoethylation) (70) for further verification. Another approach for determining the position of modified nucleotides in larger RNAs (>100 nucleotides) is the primer extension-based method, in which reverse transcriptase is stopped or paused because of the presence of modified nucleotides (71); however, some modifications cannot be detected by this approach, and some stops are caused by RNA secondary structure. In this case, the RNA should be fragmented and analyzed by combined digestion/HPLC/MS methods.
Conclusions
This Review highlights the methods for site-specific incorporation of modified nucleotides into RNA. RNAs with a wide range of sizes and functions can now be generated with one or more natural or unnatural modified nucleotides at defined locations. Further design, synthesis, analysis, and biochemical or biophysical experiments on site-specifically modified rRNAs will continue to lead us to a better understanding of their natural, biological functions. Similarly, the generation and use of non-natural modifications will allow further structural and biological studies of functionally important RNAs, as well as large ribonucleoprotein complexes.
Acknowledgments
We are grateful for support from the National Institutes of Health (Grants GM054632 and AI061192).
ABBREVIATIONS
- ACE
bis(2-acetoxyethoxy)methyl
- BTT
5-(benzoylthio)tetrazole
- BzH
benzhydryloxy-bis(trimethylsilyloxy)silyl
- CEM
2-cyanoethoxymethyl
- CNEt
2-cyanoethyl
- CPG
controlled pore glass
- DCA
dichloroacetic acid
- DMT
4, 4′oxytrityl
- DOD
[bis(trimethylsilyl)oxy]cyclo-dodecyloxysilyl
- DTM
tert-butyldithiomethyl
- DTT
1, 4-dithiothreitol
- ESI
electrospray ionization
- ETT
5-(ethylthio)tetrazole
- Fpmp
1-(2-fluorophenyl)-4-methoxypiperidin-4-yl
- HPLC
high-performance liquid chromatography
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
- Nbn
2-nitrobenzyl
- PEG
polyethylene glycol
- PS
polystyrene
- Pd2(dba)3
tris(dibenzylideneacetone)dipalladium(0)
- Pyr
pyridine
- t-C4H9OOH
tert-butylhydroperoxide
- TBAF
tetrabutylammoniumfluoride
- TBDMS
tert-butyldimethylsilyl
- TCA
trichloroacetic acid
- TCEP
tris(2-carboxyethyl)phosphine
- TEA
triethylamine
- TEM
4-tolylsulfonylethoxymethyl
- TEMED
N, N, N′, N′-tetramethyethylenediamine
- THF
tetrahydrofuran
- TOM
[(triisopropylsilyl)oxy]methyl
References
- 1.Grosjean H. Modification and editing of RNA: historical overview and important facts to remember. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. 1. Springer-Verlag; New York: 2005. pp. 1–16. [Google Scholar]
- 2.Ye K. H/ACA guide RNAs, proteins and complexes. Curr Op Struct Biol. 2007;17:287–292. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallo M, Montserrat JM, Iribarren AM. Design and applications of modified oligonucleotides. Braz J Med Biol Res. 2003;36:143–151. doi: 10.1590/s0100-879x2003000200001. [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann RA, Gait MJ, Moore MJ. Incorporation of modified nucleotides into RNA for studies on RNA structure, function, and intermolecular interactions. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. ASM Press; Washington, DC: 1998. pp. 59–84. [Google Scholar]
- 6.Cobb AJA. Recent highlights in modified oligonucleotide chemistry. Org Biomol Chem. 2007;5:3260–3275. doi: 10.1039/b709797m. [DOI] [PubMed] [Google Scholar]
- 7.Doudna JA, Szostak JW, Rich A, Usman N. Chemical synthesis of oligoribonucleotides containing 2-aminopurine: substrates for the investigation of ribozyme function. J Org Chem. 1990;55:5547–5549. doi: 10.1021/jo00308a003. [DOI] [PubMed] [Google Scholar]
- 8.Chaulk SG, MacMillan AM. Caged RNA: photo-control of a ribozyme reaction. Nucleic Acids Res. 1998;26:3173–3178. doi: 10.1093/nar/26.13.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobärtner C, Micura R. Chemical synthesis of selenium-modified oligoribonucleotides and their enzymatic ligation leading to an U6 snRNA stem-loop segment. J Am Chem Soc. 2004;126:1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- 10.Irani RJ, SantaLucia J., Jr The synthesis of 5-iodocytidine phosphoramidite for heavy atom derivatization of RNA. Tet Lett. 1999;40:8961–8964. [Google Scholar]
- 11.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 12.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem. 1960;235:1488–1498. [PubMed] [Google Scholar]
- 15.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michelson AM, Todd AR. Nucleotides part XXXII. Synthesis of a dithymidine dinucleotide containing a 3′:5′-internucleotidic linkage. J Chem Soc. 1955;1955:2632–2638. [Google Scholar]
- 17.Khorana HG. Nucleic acid synthesis. Pure Appl Chem. 1968;17:349–381. [Google Scholar]
- 18.Gait MJ. An introduction to modern methods of DNA synthesis. In: Gait MJ, editor. Oligonucleotide Synthesis: A Practical Approach. IRL Press Limited; Washington, DC: 1984. pp. 1–22. [Google Scholar]
- 19.Ogilvie KK, Theriault N, Sadana KL. Synthesis of oligoribonucleotides. J Am Chem Soc. 1977;99:7741–7743. doi: 10.1021/ja00465a073. [DOI] [PubMed] [Google Scholar]
- 20.Capaldi DC, Reese CB. Use of the 1-(2-fluorophenyl)-4-methoxypiperidin-4-yl (Fpmp) and related groups in oligoribonucleotide synthesis: stability of internucleotide linkages to aqueous acid. Nucleic Acids Res. 1994;22:2209–2216. doi: 10.1093/nar/22.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenyuk A, Földesi A, Johansson T, Estmer-Nilsson C, Blomgren P, Brännvall M, Kirsebom LA, Kwiatkowski M. Synthesis of RNA using 2′-O-DTM protection. J Am Chem Soc. 2006;128:12356–12357. doi: 10.1021/ja0636587. [DOI] [PubMed] [Google Scholar]
- 22.Heidenhain SB, Hayakawa Y. Improved methods for the preparation of 2′-deoxyribonucleoside and ribonucleoside 3′-phosphoramidites with allylic protectors. Nucleosides Nucleotides. 1999;18:1771–1787. [Google Scholar]
- 23.Sproat BS, Iribarren AM, Guimil Garcia R, Beijer B. New synthetic routes to synthons suitable for 2′-O-allyl oligoribonucleotide assembly. Nucleic Acids Res. 1991;19:733–738. doi: 10.1093/nar/19.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scaringe SA, Francklyn C, Usman N. Chemical synthesis of biologically active oligoribonucleotides using ′-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990;18:5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Pitsch S. Synthesis and pairing properties of oligoribonucleotide analogues containing a metal-binding site attached to a ′-D-allofuranosyl cytosine. Nucleic Acids Res. 1998;26:4315–4323. doi: 10.1093/nar/26.19.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl(2′-O-tom)-protected phosphoramidites. Helv Chim Acta. 2001;84:3773–3795. [Google Scholar]
- 27.Scaringe SA, Wincott FE, Caruthers MH. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J Am Chem Soc. 1998;120:11820–11821. [Google Scholar]
- 28.Meroueh M, Grohar PG, Qiu J, SantaLucia J, Jr, Scaringe SA, Chow CS. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdan FM, Chow CS. The synthesis of allyl- and allyloxycarbonyl-protected RNA phosphoramidites. Useful reagents for solid-phase synthesis of RNAs with base-labile modifications. Tetrahedron Lett. 1998;39:1897–1900. [Google Scholar]
- 30.Zhang X, Gaffney BL, Jones RA. RNA synthesis using a universal basestable allyl linker. Nucleic Acids Res. 1997;25:3980–3983. doi: 10.1093/nar/25.20.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohgi T, Masutomi Y, Ishiyama K, Kitagawa H, Shiba Y, Yano J. A new RNA synthetic method with a 2′-O-(2-cyanoethoxymethyl) protecting group. Org Lett. 2005;7:3477–3480. doi: 10.1021/ol051151f. [DOI] [PubMed] [Google Scholar]
- 32.Zhou C, Honcharenko D, Chattopadhyaya J. 2-(4-Tolylsulfonyl)ethoxymethyl (TEM)–a new 2′-OH protecting group for solid-supported RNA synthesis. Org Biomol Chem. 2007;5:333–343. doi: 10.1039/b614210a. [DOI] [PubMed] [Google Scholar]
- 33.Shiba Y, Masuda H, Watanabe N, Ego T, Takagaki K, Ishiyama K, Ohgi T, Yano J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2′-O-protecting group: structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007;35:3287–3296. doi: 10.1093/nar/gkm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agris PF, Malkiewicz A, Kraszewski A, Everett K, Nawrot B, Sochacka E, Jankowska J, Guenther R. Site-selected introduction of modified purine and pyrimidine ribonucleosides into RNA by automated phosphoramidite chemistry. Biochimie. 1995;77:125–134. doi: 10.1016/0300-9084(96)88115-6. [DOI] [PubMed] [Google Scholar]
- 35.Bajji AC, Davis DR. Synthesis of the tRNALys, 3 anticodon stem-loop domain containing the hypermodified ms2t6A nucleoside. J Org Chem. 2002;67:5352–5358. doi: 10.1021/jo025826t. [DOI] [PubMed] [Google Scholar]
- 36.Chui HMP, Desaulniers JP, Scaringe SA, Chow CS. Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m3′ and ′. J Org Chem. 2002;67:8847–8854. doi: 10.1021/jo026364m. [DOI] [PubMed] [Google Scholar]
- 37.Höbartner C, Kreutz C, Flecker E, Ottenschläger E, Pils W, Grubmayr K, Micura R. The synthesis of 2′-O-[(triisopropylsilyl)oxy]methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m22G, m1I, m3U, m4C, m6A, m62A) for use in automated RNA solid-phase synthesis. Monat Für Chemie. 2003;134:851–873. [Google Scholar]
- 38.Hall KB, McLaughlin LW. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry. 1991;30:1795–1801. doi: 10.1021/bi00221a010. [DOI] [PubMed] [Google Scholar]
- 39.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rife JP, Cheng CS, Moore PB, Strobel SA. N2-Methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res. 1998;26:3640–3644. doi: 10.1093/nar/26.16.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of ′ at tRNA’s position 39. Nucleic Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beigelman L, McSwiggen JA, Draper KG, Gonzalez C, Jensen K, Karpeisky AM, Modak AS, Matulic-Adamic J, DiRenzo AB, Haeberli P, Sweedler D, Tracz D, Grimm S, Wincott FE, Thackray VG, Usman N. Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J Biol Chem. 1995;270:25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- 43.Gordon PM, Fong R, Deb SK, Li NS, Schwans JP, Ye JD, Piccirilli JA. New strategies for exploring RNA’s 2′-OH expose the importance of solvent during group II intron catalysis. Chem Biol. 2004;11:237–246. doi: 10.1016/j.chembiol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Carriero S, Damha MJ. Inhibition of pre-mRNA splicing by synthetic branched nucleic acids. Nucleic Acids Res. 2003;31:6157–6167. doi: 10.1093/nar/gkg824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye JD, Li NS, Dai Q, Piccirilli JA. The mechanism of RNA strand scission: an experimental measure of the Brønsted coefficient, ′nuc. Angew Chem Int Ed. 2007;46:3714–3717. doi: 10.1002/anie.200605124. [DOI] [PubMed] [Google Scholar]
- 46.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou YM, Li Z, Gamper H. Isolation of a site-specifically modified RNA from an unmodified transcript. Nucleic Acids Res. 2006;34:e21. doi: 10.1093/nar/gnj018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y-T, Shu M-D, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 49.Lapham J, Crothers DM. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA. 1996;2:289–296. [PMC free article] [PubMed] [Google Scholar]
- 50.Beckett D, Uhlenbeck OC. Enzymatic synthesis of oligoribonucleotides. In: Gait MJ, editor. Oligonucleotide Synthesis: A Practical Approach. IRL Press Limited; Washington, DC: 1984. pp. 185–197. [Google Scholar]
- 51.England TE, Uhlenbeck OC. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978;17:2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- 52.Piccirilli JA, Krauch T, Morony SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 53.Tor Y, Dervan PB. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J Am Chem Soc. 1993;115:4461–4467. [Google Scholar]
- 54.Fujiwara T, Kimoto M, Sugiyama H, Hirao I, Yokoyama S. Synthesis of 6-(2-thienyl)purine nucleoside derivatives that form unnatural base pairs with pyridin-2-one nucleosides. Bioorg Med Chem Lett. 2001;11:2221–2223. doi: 10.1016/s0960-894x(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 55.Schweitzer BA, Kool ET. Hydrophobic, non-hydrogen-bonding bases and base pairs in DNA. J Am Chem Soc. 1995;117:1863–1872. doi: 10.1021/ja00112a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry AA, Olsen AG, Matsuda S, Yu C, Geierstanger BH, Romesberg FE. Efforts to expand the genetic alphabet: identification of a replicable unnatural DNA self-pair. J Am Chem Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
- 57.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 58.Kimoto M, Endo M, Mitsui T, Okuni T, Hirao I, Yokoyama S. Site-specific incorporation of a photo-crosslinking component into RNA by T7 transcription mediated by unnatural base pairs. Chem Biol. 2004;11:47–55. doi: 10.1016/j.chembiol.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 59.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005;33:e129. doi: 10.1093/nar/gni128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J Am Chem Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 61.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 62.Höbartner C, Rieder R, Kreutz C, Puffer B, Lang K, Polonskaia A, Serganov A, Micura R. Syntheses of RNAs with up to 100 nucleotides containing site-specific 2′-methylseleno labels for use in x-ray crystallography. J Am Chem Soc. 2005;127:12035–12045. doi: 10.1021/ja051694k. [DOI] [PubMed] [Google Scholar]
- 63.Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA. 2006;12:2014–2019. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurschat WC, Müller J, Wombacher R, Helm M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erlacher MD, Lang K, Shankaran N, Wotzel B, Hüttenhofer A, Micura R, Mankin AS, Polacek N. Chemical engineering of the peptidyl transferase center reveals an important role of the 2′-hydroxyl group of A2451. Nucleic Acids Res. 2005;33:1618–1627. doi: 10.1093/nar/gki308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amort M, Wotzel B, Bakowska-Zywicka K, Erlacher MD, Micura R, Polacek N. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5140. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:2–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 68.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4785. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pomerantz SC, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 70.Mengel-Jørgensen J, Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 2002;30:e135. doi: 10.1093/nar/gnf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]


