Abstract
In shotgun proteomics, a complex protein mixture is digested to peptides, separated and identified by microcapillary liquid chromatography followed by tandem mass spectrometry (LC-MS-MS). In this technology, complete protein digestion is often assumed. We show that, to the contrary, modifications to a standard digestion protocol demonstrate large, reproducible improvements in protein identification, a result consistent with digestion being a limiting factor in the efficiency of protein identification.
Keywords: mass spectrometry, proteomics, digestion, protein identification
1. Introduction
In the post-genomic era of systems biology, it is becoming increasingly clear that to fully understand a protein’s function we need to examine all of its interactions simultaneously. Modulations in protein function are frequently the result of upstream, downstream, and parallel interactions, and cellular processes are mediated in part by interactions which are regulated both spatially and temporally within the cell. Measuring such phenomena requires a comprehensive analysis of proteins without bias for or against selected proteins. Unfortunately, proteins do not lend themselves well to a one-size-fits-all approach for sample detection and analysis. Proteins display a broad range of physiochemical properties and are expressed over a very large dynamic range, complicating the analysis of large numbers of proteins in parallel using a single technology.
These limitations have spurred the development of shotgun proteomics. In this technology, a complex protein mixture is digested to a peptide mixture, followed by the analysis of the peptides by microcapillary liquid chromatography-tandem mass spectrometry (μLC-MS/MS) (1; 2). The identity of the peptides present can then be deduced by correlating the respective MS/MS fragmentation spectra against theoretical spectra of peptide sequences obtained from a protein sequence database. This approach lends itself well to the analysis of proteins with a broad range of properties including extremes in molecular weight, isoelectic point and solubility because peptides are significantly less chemically diverse than the proteins they are derived from. Most shotgun proteomics methodologies assume that the digestion of all proteins in a complex mixture is driven to completion and does not limit protein identification (3). However, it is well known that many proteins are resistant to proteolysis (4; 5). Hence, incomplete digestion is one likely limitation imposed in the analysis of complex protein mixtures. Furthermore, although many experiments have examined the digestion kinetics and efficiencies of a single protease substrate (6; 7; 8), little work has examined different protocols in the context of a complex soluble protein sample, and none in the context of shotgun proteomics. Digestion of complex protein mixtures can be affected by many factors, including but not limited to poor protein solvation or denaturation, inadequate local enzyme concentration, insufficient reaction time, and even other molecular species within the mixture that can compete for protease activity. Any of these factors might interfere with the production of peptides and ultimately the identification of proteins. Thus, we modified and tested a standard digestion protocol (9) and examined the modified protocols’ effects on the identification of proteins from a complex mixture.
We demonstrate that current protein digestion protocols limit the analysis of proteins in complex mixtures. We present a comparison of four different digestion strategies for the qualitative identification of proteins by μLC-MS/MS. All analyses were performed in replicate with the same starting protein sample and analyzed at the same time using an identical experimental setup. A modified protocol using an acid-labile detergent and an immobilized protease in a microcapillary column reproducibly produced the greatest number of protein identifications. While immobilized trypsin column technology has been used in the past for digestion of a small number of proteins (10; 11; 12; 13), and for complex protein mixtures analyzed with size-exclusion chromatography (14), to the authors’ knowledge, this is the first characterization of the immobilized trypsin columns efficacy for shotgun proteomics.
Improved protein identification was indicated in the column digest by a three-fold increase in total protein identifications, as well as increased peptide sequence coverage of proteins and an almost five-fold increase in identification of low-level proteins. It is worth emphasizing that these results do not indicate improved digestion efficiency per se, but instead demonstrate improved protein identification efficiency, arguably more important in the context of proteomics method development. We thus show substantial improvement over existing methods, demonstrating that digestion variation is a significant source of inefficiency in protein identification with shotgun proteomics.
2 Experimental Section
2.1 Materials
RapiGest SF acid labile surfactant was purchased from Waters Corporation (Milford, MA). The proteases endoproteinase Lys-C and trypsin (modified, sequencing grade) were obtained from Roche. Poroszyme immobilized trypsin enzyme was purchased from Applied Biosystems (Foster City, CA). All other laboratory reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless noted otherwise.
2.2 Sample Preparation
Bacteria culture
Two 5mL test tubes of OP media were inoculated from single colonies of Escherichia coli (strain OP50) and incubated overnight at 37°C. Both 5mL cultures were added to 1L OP media for a 1:100 dilution and incubated for 5h at 37°C to an OD600nm of approximately 0.6. This culture was chilled for 5 min on ice and spun down at 3000 rpm for 5 min at 4°C. The resulting bacterial pellets were then lysed using the French press twice at 1000psi, and spun at 4000 rpm for 30 min at 4°C. The resulting supernatant was then spun at 14000 rpm for 10 min at 4°C to remove insoluble proteins. The final supernatant was collected and assayed for protein concentration using RC DC Protein Assay Kit (Bio Rad, 500–0122) at 8μg/μL.
Digestion protocols
Four digestion protocols were tested in quadruplicate for a total of 16 samples as described below. Protocol short names are shown in parentheses.
Lys-C/Trypsin digestion (Lys-C/Tryp)
Four samples each containing 60μg protein were solubilized in 100mM Tris pH 8.5, 8M urea. The sample was reduced in 5mM dithio-theitol (DTT) for 30 min at 60°C to reduce disulfide bridges; cysteines were then alkylated in the dark in 25mM iodoacetic acid (IAA) for 30 min at 25°C. Endoproteinase Lys-C was added to each of four samples for ratio of 1:100 enzyme:protein and incubated overnight with shaking at 37°C. Sample was diluted 1:1 in 100mM Tris, pH 8.5 for a final concentration of 4M urea and Trypsin enzyme was added for a ratio of 1:30 enzyme:protein along with 2mM CaCl2, and then incubated overnight with shaking at 37°C.
Twenty-four hour solution-phase trypsin digestion (24-Hour-Solution)
Four samples each containing 60μg protein were solubilized in 100mM Tris pH 8.5; 0.1% RapiGest SF. The sample was reduced with DTT and alkylated with IAA, as above. Trypsin enzyme was added for a ratio of 1:30 enzyme:protein along with 2mM CaCl2, and then incubated overnight (approximately 24-hour) with shaking at 37°C.
One-hour solution-phase trypsin digestion (1-Hour-Solution)
Four samples each containing 60μg protein were solubilized in 100mM Tris pH 8.5; 0.1% RapiGest SF, reduced with DTT and alkylated with IAA, as above. Trypsin enzyme was added for a ratio of 1:30 enzyme:protein with 2mM CaCl2, and then incubated for one-hour with shaking at 37°C.
Immobilized trypsin column digestion (1-Hour-Column)
Four samples each containing 60μg protein were solubilized in 100mM Tris pH 8.5; 0.1% RapiGest SF, reduced with DTT and alkylated with IAA, as above. Each 50μL of 1.2μg/μL sample was digested by using an immobilized trypsin column.
The trypsin column was constructed by attaching 50cm of 250μm ID fused silica (Polymicro Tech, Phoenix, AZ) to a MicroTight inline MicroFilter (UpChurch Scientific, Oak Harbor, WA). Poroszyme immobilized trypsin was slurry packed using a pressure bomb into the fused silica behind the inline MicroFilter to create a 7cm trypsin column. The resulting column was flushed with 25% MeOH, 0.01% formic acid and then looped with ends sealed to retain moisture and stored at 4°C until immediately prior to use.
In preparation for sample digestion, the column was equilibrated at 37°C and washed with 100 mM Tris pH 8.5, 1 mM CaCl2, and 5% acetonitrile for 5 min. The 50 μL sample was then passed over the column at 1 μL per min using an HPLC pump. The sample was collected for a total of 90 min (90μL) to ensure that all the sample was retained and any dead volume and flow inaccuracy was accounted for.
Post-digestion
Following digestion, each of the 16 reactions were acidified to 50mM HCl to hydrolyze the RapiGest SF in the 12 samples that contained it and to inhibit any remaining active enzyme. Samples were then incubated for 45 min at 37°C and centrifuged for 4 min at 4000rpm. The supernatant was collected and diluted with MilliQ purified water to a total volume of 120μL for consistent peptide concentrations.
2.3 Mass spectrometry and database searches
Each of the 16 trypsin digests were analyzed by automated microcapillary liquid chromatography tandem mass spectrometry. A 100μm ID fused silica capillary (Polymicro Tech, Phoenix, AZ) was manually pulled to a 5μm tip and slurry packed with 12cm 5μm Aqua C18 material (Phenomenex, Ventura, CA) using a pressure bomb. The column was placed inline with an Agilent 1100 Binary HPLC and Autosampler (Palo Alto, CA) and interfaced with a LTQ linear ion-trap mass spectrometer (ThermoElectron, San Jose, CA) using a home built microspray ion source.
The sample was loaded onto the microcapillary column using a conventional binary HPLC and autosampler. This system uses two different split flow configurations: a sample loading configuration and a sample running configuration (Supp. Figure 2). In both states, the HPLC pump flows at 150μL/min, and the flow rate to the electrospray tip is reduced through the use of a split. The main difference between the two states is the position and flow resistance provided by length of split capillary. In the loading configuration, a relatively long split occurs between the HPLC pump and the autosampler, avoiding sample loss, while maintained output flow rate at 5μL/min (diverting 145μL/min to waste). In the running configuration, a relatively short restriction is placed immediately prior to the column, reducing the effect of the dead volume between the autosampler and column while reducing the flow rate to 200nL/min (diverting 149.8μL/min to waste). This configuration allows rapid sample loading while facilitating nanoliter flow rates for high sensitivity during peptide separation and analysis. All splits were controlled using the divert valve of the mass spectrometer.
The HPLC separation was provided by a gradient between Buffer A (95% water, 5% acetonitrile and 0.1% formic acid) and Buffer B (20% water, 80% acetonitrile, and 0.1% formic acid). The HPLC gradient was held constant for 20-min at 100% A during the loading of the sample from the autosampler to the microcapillary column. The buffer conditions then changed from 0% B to 15% B over 5 min, followed by a 70-min gradient from 15% B to 40% B, and a 5-min gradient from 40% B to 85% B. The column was then re-equilibrated in 100% A for 20 min. As peptides eluted from the microcapillary column, they electrosprayed directly into the mass spectrometer. A cycle of one full-scan mass spectrum (400–2000m/z) followed by five data-dependent MS/MS spectra at a 20% normalized collision energy was repeated continuously throughout each step of the separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system.
Sequence database search
Each MS/MS spectrum was searched against a protein database containing the E. coli open reading frames (described below) using a normalized implementation of the SEQUEST algorithm (15; 16). All database searches were performed with no enzyme specificity. Multiply charged spectra were searched twice, once as +2 and once as +3.
The spectra-peptide associations generated by SEQUEST were then filtered using DTAS-elect (17). Peptides were filtered by requiring a normalized XCorr of 0.3 for peptides of charge state +1, +2 or +3. The minimum DeltCN was 0.1 and the minimum proportion of fragment ions observed was 0.1. Peptides were required to have a minimum sequence length of seven amino acids and to be fully tryptic with internal missed cleavage sites allowed. Proteins with at least one peptide passing these criteria were accepted as an identification.
Estimation of False Discovery and True Discovery Rates
We only wish to count the number of true positive protein identifications under each digestion protocol. This requires an estimate of the rate of false positive assignment of peptides to spectra, which we obtain with the following procedure: Spectra are searched against a FASTA database consisting of the NCBI 2004-May-02 E. coli protein database concatenated with a database of common contaminant proteins and a decoy database consisting of randomized sequences of the same length and amino acid distributions as the original protein database. The number of protein hits to the randomized database is used as a proxy measure of the number of false protein hits to the E. coli database. The number of true positive protein hits is then determined by subtracting this estimated number of false-discoveries from the number of hits to the normal (unrandomized) sequences in the concatenated database.
2.4 Estimate of mRNA transcript abundance
Expression-level information about the E. coli strain K12 MG1655 was downloaded from http://asap.ahabs.wisc.edu/annotation/php/ASAP1.htm. Calibrated expression levels were obtained for microarray experiment EXPSET0003 data set PALSP49:1 (18). All gene IDs from the NC000913 genome were queried from the web interface to obtain calibrated expression levels for all E. coli proteins. Expression values of mRNA for 4017 genes in log phase E. coli (18) were ranked and used to divide the genes into three groups of approximately 1006 proteins each: those proteins with mRNA expression values in the lowest 33rd percentile; those between the 34th and 66th percentiles; and those between the 67th and 100th percentiles. The number of genes identified in each group for the three non-Lys-C digestion protocols are divided by the number of genes identified in that group in the Lys-C/Trypsin digest. Expression is used as a rough proxy for protein-level.
3 Results
The four digestion protocols are compared using three methods, all of which measure the protocols’ effects on protein identification. The first method measures effects on total proteins identified. The second method measures the extent to which identified peptides cover their respective protein sequences. The third method measures amount of low-level proteins identified by examining the number of proteins identified at low, medium and high levels, as indicated by each protein’s mRNA expression levels.
Total number of identified proteins
The number of proteins identified under each of the four digestion protocols in four replicates is shown in Table 1. All three modified digestion protocols show increased protein identification compared to the standard 48-hour Lys-C/Trypsin digestion protocol, with the highest number of proteins identified using the 1-Hour-Column digestion protocols, followed by the 1-Hour-Solution and finally the 24-Hour-Solution protocols. The four replicates for each protocol show low standard deviation of less than 30 proteins. These trends are maintained across a wide variety of search parameters.
Table 1.
Number of proteins identified under four digestion protocols
| Protein IDs | ||||||
|---|---|---|---|---|---|---|
| Condition1 | Replicate | MS/MS Scans | Forward | Random2 | Mean34 | Std. Dev.5 |
| Lys-C/Trypsin | 1 | 51742 | 172 | 4 | 200 | 22.1 |
| 2 | 56172 | 210 | 5 | |||
| 3 | 55806 | 224 | 5 | |||
| 4 | 56914 | 210 | 3 | |||
|
| ||||||
| 24-Hour-Solution | 1 | 50159 | 352 | 5 | 368 | 20.2 |
| 2 | 51048 | 397 | 4 | |||
| 3 | 50979 | 375 | 2 | |||
| 4 | 50315 | 363 | 6 | |||
|
| ||||||
| 1-Hour-Solution | 1 | 50587 | 519 | 10 | 517 | 15.2 |
| 2 | 49113 | 519 | 6 | |||
| 3 | 49075 | 497 | 8 | |||
| 4 | 50100 | 534 | 6 | |||
|
| ||||||
| 1-Hour-Column | 1 | 55236 | 591 | 4 | 581 | 22.9 |
| 2 | 50789 | 589 | 2 | |||
| 3 | 52808 | 560 | 3 | |||
| 4 | 52344 | 614 | 2 | |||
See text for description of each experiment.
Hits to the random DB are a rough measure of false positive rate.
Mean of Forward Protein IDs minus Random Protein IDs.
All differences pairwise significant with a p-value of less than 0.01.
Standard deviation is inferential as opposed to statistical.
The fact that the 1-Hour-Solution protocol identifies more proteins than the 24-Hour-Solution protocol is unexpected, and deserves additional comment. It is generally assumed that longer digestion duration will result in more complete digestion, and hence in increased protein identification. There are several possible explanations for why this is not the case in these experiments. Endogenous proteases could be degrading proteins below the detectable molecular weight range in the 24-Hour-Solution protocol. Alternatively, peptides might remain intact, but be modified chemically. To address this possibility, we re-searched spectra allowing for carbamylation of Lysine or oxidation of Met, with no significant change in results (the carbamylated LysC/Tryp had 201 true positives with a standard deviation of 19.8). Additional experiments will need to be performed to determine the exact cause of this unexpected result.
Sequence coverage of identified proteins
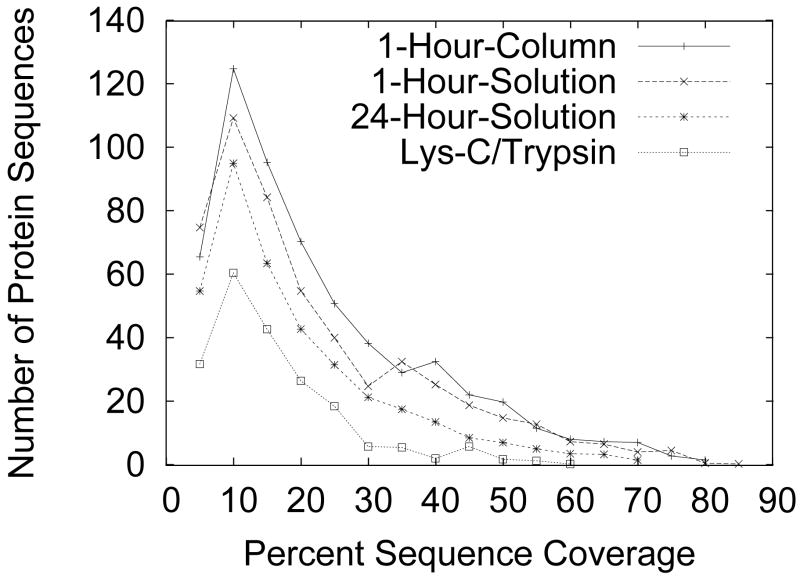
It is rare that all peptides from the entire protein sequence are identified by mass spectrometry—usually only a fraction of the protein’s peptides are recovered. While many factors (e.g. peptide fragmentation efficiency), contribute to this incomplete detection, sample digestion almost certainly plays a role. Increased sequence coverage indicates increased confidence in a protein identification; thus, an ideal shotgun proteomics digestion protocol will result in high sequence coverage across all proteins. The distribution of sequence coverage for all proteins identified in each of the four digestion protocols is shown in Figure 1. Sequence coverage for the 1-Hour-Column and 1-Hour-Solution digestion protocols are the highest, followed by the 24-Hour-Solution and finally the standard Lys-C/Trypsin protocol.
Figure 1.
Distribution of percent sequence coverage of identified proteins by identified tryptic peptides. It is rare that peptides from the entire protein sequence are identified by mass spectrometry; usually only a fraction of the protein’s peptides are recovered. Increased sequence coverage of a protein by its identified peptides increases confidence of the protein identification.
Identification of proteins at low, medium and high levels
It is possible that the difference in total number of protein identifications noted in Table 1 is due only to improved digestion of highly-abundant proteins. Thus, we sought to determine the extent to which proteins occurring at different levels within the cell are identified using each digestion protocol. The 4017 proteins in the E. coli proteome were divided into three equal groups based on the relative abundance of their respective mRNA transcripts: high-level, medium-level and low-level proteins. Table 2 shows the ratio of the number of proteins identified in each of these three groups relative to the number of proteins identified in that group by the standard Lys-C/Trypsin protocol. The 1-Hour-Column, 1-Hour-Solution and 24-Hour-Solution digestion protocols all show increased identification of high-, mid- and low-level proteins relative to the Lys-C/Trypsin protocol, with the largest difference being four-fold increase for low-level proteins identified by the 1-Hour-Column digestion.
Table 2.
Ratios of number of proteins identified in three digestion protocols relative to the standard Lys-C/Trypsin digestion protocol. The first value in each cell is the mean across four samples; the second is the standard deviation.
| Percentile | |||
|---|---|---|---|
| Condition1 | 0–33 | 34–66 | 67–100 |
| 1-Hour-Column | 4.46 ± 0.39 | 3.67 ± 0.19 | 2.25 ± 0.10 |
| 1-Hour-Solution | 3.98 ± 0.50 | 3.17 ± 0.18 | 2.02 ± 0.06 |
| 24-Hour-Solution | 2.31 ± 0.39 | 2.02 ± 0.12 | 1.64 ± 0.12 |
See text for detailed description of digestion conditions.
Additional experiments
To account for the effect of different protein denaturants, enzyme form (e.g. immobilized versus soluble), endogenous proteases, and digestion time, a separate set of experiments were performed to provide a more thorough set of controls. In the supplementary experiments, we repeated the serial endoproteinase Lys-C/trypsin digestion using urea in one experiment and RapiGest in a second (Supp. Figure 4). The samples containing RapiGest (ER) had an average 33% greater number of peptides (p-value < 0.001) and 25% greater number of protein identifications (p-value < 0.100) compared to those with urea (EU).
To confirm that the improvement in the column digestion was actually a function of the column and not the difference between the modified trypsin and the Poroszyme immobilized trypsin, we performed a subsequent experiment (Supp. Figure 4) comparing the following conditions: first, immobilized trypsin packed into a microcapillary column; second, immobilized trypsin in suspension in a microfuge tube; third, soluble trypsin in a microfuge tube. During these three additional conditions, all the non-tested conditions were maintained the same: the buffer, temperature, and length of digestion. In all of the replicates, the Poroszyme column digested sample outperformed the digestion in free solution and the digestion performed using the Poroszyme trypsin used in a suspension. Furthermore, the soluble trypsin digest gave superior results when compared to the identical microfuge based digestion using immobilized trypsin in suspension, suggesting that the digestion improvement is a result of the column and not the immobilized trypsin.
4 Discussion
In this section we point out and speculate on the implications of the main results regarding digestion conditions with respect to peptide and protein identifications: first, digestion protocols using RapiGest have improved protein identifications when compared with those that use urea; second, longer digestion times are not necessarily better; and third, digestions on a column result in more identifications than digestions in microfuge tubes.
Digestion protocols using RapiGest have improved protein identifications when compared with urea
In all of our experiments, we observed a significant increase in protein identifications in the presence of RapiGest when compared to urea. It is likely that this improvement in peptide and protein identifications is a result of improved solubilization and denaturation of the protein without reduction in the protease activity.
RapiGest is an acid labile surfactant that is commercially available from Waters. This surfactant is anionic and has been reported to improve the solubilization/denaturation of proteins while maintaining the activity of trypsin. Trypsin maintains 100% of its activity in 0.1% RapiGest and improves the digestion of proteins commonly resistant to proteoly-sis (19). A secondary advantage of RapiGest is that upon acid treatment it is cleaved into two components, a small ionic moiety and a hydrophobic, aqueous insoluble group. Centrifugation removes this insoluble group, allowing mass spectrometry analysis without the interference caused by surfactants used during most other biochemical protein preparations.
Urea is a common denaturant used in trypsin digestions. While urea can be an effective denaturant, more than 50% of trypsin activity is lost with urea concentrations greater than 4M. Thus, a common procedure is to first denature the protein mixture in 8M urea and perform an initial digestion using endoproteinase Lys-C, which maintains 60% of its activity at this urea concentration, followed by dilution to 4M and an additional digestion in trypsin (9) This serial digestion maintains the same cleavage specificity (i.e. Lys-C cleaves at K and trypsin cleaves at K and R) as using trypsin alone but uses the enhanced activity of Lys-C in 8M urea to improve the overall digestion efficiency. However, our results suggest that the use of a denaturant (such as RapiGest) that can maintain the activity of the protease is important to comprehensive peptide identification.
Another potential drawback to the use of urea is that it contains varying amounts of cyanate that can result in artifactual carbamylation of lysine and/or cysteine residues during the sample preparation. Although we were not able to detect an increase in protein carbamylation sites in the sample preparations with urea (data not shown), carbamylation leading to reduced peptide identification is nonetheless a concern in any preparation that uses urea.
During these experiments, it has become clear that a major limiting factor for protein identification is the lack of mass spectrometry compatible surfactants that can be used to ensure protein solubility while maintaining trypsin activity. Although the use of RapiGest shows significant improvement over urea, the availability of alternative (nonionic or zwitterionic) surfactants will likely prove more useful than others for selected classes of proteins within complex mixtures. Over the last couple years alternative surfactants have been developed (20; 21; 22); however, only after commercialization will these products truly benefit the greater proteomics community.
Longer digestion times do not necessarily result in more identifications
In every digestion performed we found that shorter digestion times worked as well or better than longer digestions for peptide identification. While the mechanism for the improved protein and peptide identification with shorter digestion times is not clear, the risk of asparagine and glutamine deamidation increases substantially with time under pH and temperature conditions optimal for trypsin activity (23). This increased deamidation will shift the mass of the peptides by 1 Da and complicate the characterization of peptide tandem mass spectra by database searching. Trypsin also undergoes autolysis, reducing enzyme activity and site-specificity over time. Given these concerns along with our experimental results, we recommend limiting digestion time to less than 24 hours, although an exact digestion-time recommendation would require additional experiments.
Digestions on-column yield more identifications than digestions in microfuge tubes
These data show that driving a protein mixture through a microcapillary immobilized trypsin column results in significantly more efficient protein identification by μLC-MS/MS of the resulting peptides than any other approach. Although other laboratories have reported similar improvement in tryptic digestion efficiencies of simple mixtures using column digestions (10; 11; 12; 13), the exact mechanism for the improved on-column digestion is not known. However, it is likely that the enzyme substrate ratio on the column is very high, creating a microenvironment that proceeds at a higher rate than immobilized trypsin suspended in a large volume or trypsin in solution. Additionally, the chymotryptic-like activities of the 26S proteasome are often inhibited by allosteric and competitive regulation by peptides mimicking cleavage products (24). Although to our knowledge this has not been reported for trypsin, it is not unlikely that some of the thousands of tryptic peptides that are produced in these digestions might have an inhibitory effect on the trypsin activity. Using trypsin in a column, the cleavage products are removed from the protease and replenished with new substrate as the material is pushed through the column. While we do not have data to conclusively prove this mechanism, our data support this hypothesis.
5 Conclusions
Even slight modifications to a standard digestion protocol can have a significant effect on the qualitative identification of proteins in complex mixtures. Whereas most proteomics research has focused on the implementation of improved chromatographic separation strategies and database searching algorithms, these data suggest that subtle changes in digestion and sample preparation protocols might have as great—if not greater—impact on protein and peptide identification by shotgun proteomics technologies. While we have shown a significant improvement over prior protocols, none of the digestion protocols presented here should be considered fully optimized. Furthermore, this analysis is limited to a single E. coli sample; it is entirely possible that different samples might show different results. Nonetheless, these results leave little doubt that digestion protocols are a limiting factor in peptide identification using shotgun proteomics.
Supplementary Material
Acknowledgments
The authors would like to thank Gennifer Merrihew of the MacCoss lab for general laboratory pedagogy. The authors would also like to thank anonymous reviewers for their helpful comments on the manuscript. Financial support for this research was provided in part by an American Society for Mass Spectrometry Research Award, a Basic Research Grant from Merck Research Laboratories, and National Institutes of Health Grant P41-RR11823. A. A. Klammer acknowledges support from National Institute for General Medical Sciences training grant T32-GM007735.
References
- 1.McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates JR., III Analytical Chemistry. 1997;69(4):767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 2.Yates JR., III Analytical Chemistry. 1998;33:1–19. [Google Scholar]
- 3.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fountoulakis M. Journal of Chemical Technology and Biotechnology. 1995;62:81–90. [Google Scholar]
- 5.Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, Ochsner UA. Journal of Bacteriology. 2000;182:4557–4563. doi: 10.1128/jb.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koeppe RE, Krieger M, Stroud RM. Biochimica et Biophysica Acta. 1977;481(2):617–621. doi: 10.1016/0005-2744(77)90294-7. [DOI] [PubMed] [Google Scholar]
- 7.Casteneda-Agullo M, del Castillo LM. Journal of General Physiology. 1959;42(3):617. doi: 10.1085/jgp.43.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halsey JF, Harrington WF. Biochemistry. 1973;12(4):693–701. doi: 10.1021/bi00728a019. [DOI] [PubMed] [Google Scholar]
- 9.Washburn MP, Wolters D, Yates JR., III Nature Biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Blackburn RK, Anderegg RJ. American Society for Mass Spectrometry. 1997;8:483–494. [Google Scholar]
- 11.Nadler T, Blackburn C, Mark J, Gordon N, Regnier FE, Vella G. Journal of Chromatography A. 1996;743(1):91–98. [Google Scholar]
- 12.Lei J, Chen DA, Regnier FE. Journal of Chromatography A. 1998;808:121–131. doi: 10.1016/s0021-9673(98)00098-3. [DOI] [PubMed] [Google Scholar]
- 13.Slysz GW, Schriemer DC. Rapid communications in mass spectrometry. 2003;17:1044–1050. doi: 10.1002/rcm.1022. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Regnier FE. Journal of Chromatography A. 2001;913:429–436. doi: 10.1016/s0021-9673(01)00604-5. [DOI] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, Yates JR., III Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.MacCoss MJ, Wu CC, Yates JR., III Analytical Chemistry. 2002;74(21):5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 17.Tabb DL, McDonald WH, Yates JR., III Journal of Proteome Research. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen TE, Herrgård MJ, Liu M, Qiu Y, Glasner JD, Blattner FR, Palsson BØ. Journal of Bacteriology. 2003;185:6392–6399. doi: 10.1128/JB.185.21.6392-6399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suder P, Bierczynska A, Konig S, Silberring J. Rapid Communications in Mass Spectrometry. 2004;18:822–824. doi: 10.1002/rcm.1411. [DOI] [PubMed] [Google Scholar]
- 20.Norris JL, Porter NA, Caprioli RM. Analytical Chemistry. 2003;75:6642–6647. doi: 10.1021/ac034802z. [DOI] [PubMed] [Google Scholar]
- 21.Norris JL, Hangauer MJ, Porter NA, Caprioli RM. Journal of the American Society for Mass Spectrometry. 2005;40:1319–1326. doi: 10.1002/jms.914. [DOI] [PubMed] [Google Scholar]
- 22.Norris JL, Porter NA, Caprioli RM. Analytical Chemistry. 2005;77:5036–5040. doi: 10.1021/ac050460g. [DOI] [PubMed] [Google Scholar]
- 23.Lundell N, Schreitmuller T. Analytical Biochemistry. 1999;266:31–47. doi: 10.1006/abio.1998.2919. [DOI] [PubMed] [Google Scholar]
- 24.Papapostolou D, Coux O, Reboud-Ravaux M. Biochemical and Biophysical Research Communications. 2002;295:1090–1095. doi: 10.1016/s0006-291x(02)00803-3. [DOI] [PubMed] [Google Scholar]
- 25.Venn J. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 1880;9:1–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.