Abstract
Cyclization of the N-terminal glutamine residue to pyroglutamic acid in onconase, an anti-cancer chemotherapeutic agent, increases the activity and stability of the protein. Here, we examine the correlated effects of the folding/unfolding process and the formation of this N-terminal pyroglutamic acid. The results in this study indicate that cyclization of the N-terminal glutamine has no significant effect on the rate of either reductive unfolding or oxidative folding of the protein. Both the cyclized and uncyclized proteins seem to follow the same oxidative folding pathways; however, cyclization altered the relative flux of the protein in these two pathways by increasing the rate of formation of a kinetically trapped intermediate. Glutaminyl cyclase (QC) catalyzed the cyclization of the unfolded, reduced protein, but had no effect on the disulfide-intact, uncyclized, folded protein. The structured intermediates of uncyclized onconase were also resistant to QC-catalysis, consistent with their having a native-like fold. These observations suggest that, in vivo, cyclization takes place during the initial stages of oxidative folding, specifically, before the formation of structured intermediates. The competition between oxidative folding and QC-mediated cyclization suggests that QC-catalyzed cyclization of the N-terminal glutamine in onconase occurs in the endoplasmic reticulum, probably co-translationally.
The N-terminal glutamine residue of peptides and proteins undergoes non-enzymatic spontaneous cyclization resulting in the formation of pyroglutamic acid (1). This is a slow process requiring day(s) for completion, depending on the conditions (1). Examples of peptides and proteins with an N-terminal pyroglutamic acid include gonadotropin-releasing hormone, thyrotropin-releasing hormone, neurotensin, etc, for which biological activity depends on the presence of pyroglutamic acid at their N-termini (2, 3). Loss or modification of the N-terminal pyroglutamic acid residue leads to a decrease in biological activity (4, 5). This implies the existence of an enzyme that accelerates this cyclization in vivo (6-8). Indeed, enzymes with glutaminyl cyclase (QC) activity have been isolated from many sources (9-13), and the cDNA of QC was identified in many organisms (14-16).
Prior to QC catalysis, prohormone convertase must unmask the N-terminal glutamine during the secretion of several peptide hormones by removing the precursor peptide that precedes the glutamine residue on the prohormone (17). This requires that QC acts in the regulated secretory pathways, subsequently to prohormone convertase. Sub-cellular distribution studies have confirmed the presence of QC in pituitary secretory granules (18). In the case of proteins, however, in which there is no need to unmask the N-terminal glutamine, very little is known about where and when QC-mediated catalysis takes place in relation to other post-translational events such as folding and disulfide-bond formation.
Onconase (ONC; registered trademark of Alfacell Corp., Bloomfield, NJ, USA) has an N-terminal glutamine. It is a very stable 104-residue frog ribonuclease containing four disulfide bonds. Its amino acid sequence (19) and three-dimensional structure (20) are homologous to those of bovine pancreatic ribonuclease A (RNase A). The cyclized pyroglutamic acid-containing form has significantly higher ribonuclease activity (21) and is slightly more stable than its uncyclized form [unONC (22)]. ONC is toxic to tumor cells and is in Phase III clinical trials for the treatment of malignant mesothelioma, an asbestos induced lung cancer (23-25). The anti-cancer activity of ONC (due to the absence of a specific intracellular inhibitor such as one that inhibits the activity of RNase A) is related to its ribonuclease activity (19, 20, 26-29).
In this study, we examine the influence of the cyclization of N-terminal glutamine on the reductive unfolding and oxidative folding profiles (rates and pathways) in onconase. We also investigate the effect of formation of the native structure (which is coupled to disulfide-bond formation) on the QC-catalyzed cyclization of the protein.
Experimental reductive unfolding (30-39) and oxidative folding studies (40-53) of proteins, in vitro and in vivo, have contributed to a better understanding of the relationship between protein structure and folding/unfolding characteristics. The reductive unfolding and oxidative folding of ONC have been examined in detail (54-56). The reductive unfolding of ONC is a two-stage process that is initiated with the rapid reduction of the first relatively-exposed disulfide bond, resulting in the quantitative conversion of the native protein to des [30-75], an intermediate containing three native disulfide bonds (54, 56). However, the identity of the disulfide bond to be reduced in the next stage is yet to be determined.
The oxidative folding of ONC was compared to that of RNase A under several identical conditions (55). Three peaks (I1, I2 and I3) corresponding to structured intermediates of ONC were detected by HPLC (55). I1 has two disulfide bonds whereas I2 and I3 each have three (Narayan et al, in preparation). This is in contrast to RNase A which has no structured two-disulfide-bond-containing intermediate (35,41,42). The overall oxidative folding rates of ONC and RNase A do not differ significantly at 15°C and 25°C, pH 8, 25 mM DTTox (55). However, the oxidative folding rate of ONC is 100-fold faster at 37°C where the disulfide-secure intermediates of RNase A begin to melt (41). The higher rate has been attributed to the higher stability of the disulfide-secure (57) intermediate(s) of ONC (55).
Experimental Procedures
Materials
WT-ONC cDNA in a pET11 expression vector (21) was amplified by PCR and cloned to a pET 22b(+) vector in-frame with the pelB signal sequence without the starting methionine residue. WT-ONC was expressed in BL21(DE3) cells and purified as described earlier (26), except that the protein was refolded in the presence of 0.5 M arginine to prevent precipitation. The folded protein (ONC or unONC) was purified on a strong cation-exchange column after concentration, dialyzed against 20 mM acetic acid solution at 4°C and then lyophilized as described previously (54). This preparation of unONC was found to contain 10% of the cyclized species. The non-enzymatic conversion of the N-terminal glutamine to pyroglutamic acid to produce 100% cyclized protein was carried out by treating the uncyclized folded form with a 200 mM pH 7 phosphate buffer at either room temperature or 37°C for more than two days.
A model peptide corresponding to the six N-terminal residues of unONC, H-Gln-Asp-Trp-Leu-Thr-Phe-NH2, was synthesized using standard Fmoc chemistry and purified by reversed-phase HPLC.
AEMTS (> 99% pure) was purchased from Anatrace and used without further purification. DTTox and DTTred were obtained from Sigma. DTTred was used without further purification whereas DTTox was purified to remove traces of contaminating DTTred by reversed-phase HPLC (40). All other chemicals were of the highest grade commercially available.
N-terminal sequencing and mass spectrometry analyses were carried out by the Cornell Biotechnology Resource Center.
Preparation of reduced ONC/unONC
Lyophilized onconase (with a cyclized or uncyclized N-terminal Gln) was dissolved in a buffer (pH 8, 25 mM Tris-HCl, 1 mM EDTA) containing 6 M GdnHCl and 100 mM DTTred, and incubated at room temperature for 2 hours. The reducing agent and salts were removed from the reduced protein by desalting the sample using a G25 size-exclusion column (with 50 mM acetic acid as the running buffer) followed by lyophilization. The lyophilized, reduced onconase (R-ONC and R-unONC) were stored at −20°C until further use.
Reductive unfolding of ONC/unONC
Lyophilized native onconase (with a cyclized or uncyclized N-terminal Gln) was dissolved in an acetic acid buffer (50 mM) at room temperature to obtain a stock solution (5 mg/ml), and aliquots (0.2 ml each) of this solution were kept frozen at −20°C until used. Reduction experiments were initiated by introducing one such aliquot of ONC or unONC into a solution (pH 8, 100 mM Tris-HCl, 1 mM EDTA, 15°C) containing DTTred (final concentration 10 mM) such that the final protein concentration was 0.5 mg/ml.
Aliquots of the reaction mixture were withdrawn periodically, and any free thiols were blocked immediately with excess AEMTS (final concentration 50 mM). After 5 minutes, the pH of the mixture was reduced to 3 by addition of 20 μl glacial acetic acid. The AEMTS-blocked, acid-quenched samples were then desalted on a Hi-Trap G25 gel filtration desalting column and analyzed by HPLC. HPLC runs were carried out on a Rainin-Hydropore strong cation-exchange column using a salt gradient (50 to 150 mM NaCl over 150 minutes).
Reduction of ONC/unONC was also carried out at a higher concentration of the reducing agent (100 mM DTTred).
Oxidative folding of ONC/unONC
Lyophilized R-ONC/R-unONC was dissolved into a 50 mM acetic acid solution to obtain a stock solution of protein (10 mg/ml). Oxidative folding of ONC/unONC was initiated by introducing an aliquot of R-ONC/R-unONC into a solution containing 25 mM DTTox (final protein concentration was 22 μM, pH 8, 100 mM Tris-HCl, 1 mM EDTA, 25°C).
Aliquots of the reaction mixture were withdrawn periodically, and any free thiols were blocked immediately with excess AEMTS (final concentration 40 to 50 mM). The AEMTS-blocked samples were desalted, prior to analysis by cation-exchange HPLC as described above. In some experiments, the blocked, desalted aliquots were divided into two halves, and incubated with and without QC, respectively, in phosphate buffered saline (PBS), pH 7.2 at room temperature and analyzed by cation-exchange chromatography.
Some aliquots were subjected to a reduction-pulse (42) by the addition of 5 mM DTTred for a period of two minutes at 25°C before being blocked with AEMTS.
Cyclization of the N-terminal glutamine
Non-enzymatic cyclization reactions of the protein and the model peptide were examined under various pH (7-8), salt concentration (10-200 mM phosphate) and temperature (4-37°C) conditions, and monitored by cation-exchange (Rainin-Hydropore strong cation-exchange column) or reversed-phase (C18 column) chromatography, respectively. The amount of the cyclized and uncyclized protein or peptide was established quantitatively by measuring the areas of the corresponding peaks on the chromatogram.
QC-catalyzed cyclization of the protein and the model peptide was carried out at pH 7, 37C° and analyzed as above. Human pituitary glutaminyl cyclase was kindly provided by Prof. Robert C. Bateman, Jr. (13).
Results
Frog onconase is expressed in E. coli as a mixture of the cyclized and uncyclized forms with about 50 % being cyclized. Since cyclization of the N-terminal glutamine results in a change in the pI of the protein, the retention times on a cation-exchange column of the native (as well as of the intermediate and reduced) forms of the cyclized and uncyclized protein species differ, facilitating their isolation. The cyclized and uncyclized forms of the expressed native protein, found in the earlier and later eluting peaks, respectively, from a strong cation exchange column, were identified by mass spectrometry. Cyclization of the N-terminal glutamine of ONC was further verified by N-terminal sequencing of both protein species as follows. The sequence (QD-LTFQKKHITNT, where the dash stands for W, which is modified under the conditions of the Edman-degradation, and thus, cannot be identified), found in the later-eluting peak, agrees with the expected sequence of expressed unONC. By contrast, the protein that is eluted earlier on the cation-exchange column was resistant to Edman degradation, which is consistent with an N-terminal cyclization of the glutamine (19).
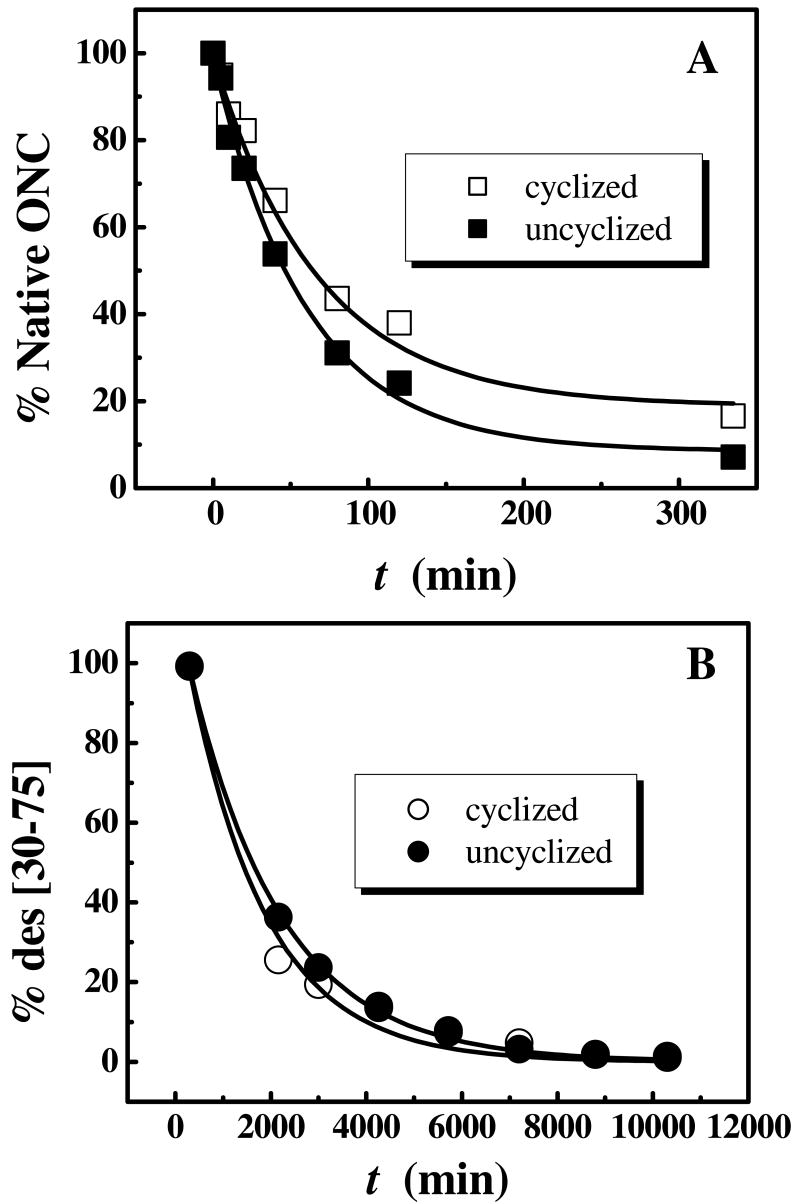
Reductive unfolding of unONC was carried out on uncyclized onconase containing 10% cyclized protein that was used as an internal control for direct comparison. The (30-75) disulfide bond of ONC is susceptible to reduction under mildly reducing conditions resulting in the full conversion of the native protein to des [30-75] without detectable accumulation of any other species (54, 56). In the second stage, des [30-75] is converted to the fully-reduced protein without significant accumulation of any other intermediates (54, 56). The two stages of the reduction of native ONC have to be examined in separate experiments. If examined in one experiment, under mildly reducing conditions, the reduction of des [30-75] (the second stage) to R-ONC takes a very long time whereas, if strongly reducing conditions are applied, the reduction of the (30-75) disulfide bond in the native protein (the first stage) takes place very rapidly not enabling one to make precise measurements of reduction rates under either reducing condition. Hence, the first stage in the reduction process of native ONC/unONC is studied under mild reducing conditions and the second stage is carried out on isolated des [30-75] under strongly reducing conditions. The cyclization of N-terminal glutamine did not alter the reduction rate of either stage of the reduction significantly although, in the first stage, a slight increase of the reduction rate of unONC was observed (Figure 1 A and B).
Figure 1.
The reductive unfolding of cyclized and uncyclized onconase. A. Percentage of native ONC/unONC in the presence of 10 mM DTTred, starting from the native protein (first stage). B. Percentage of des [30-75] in the presence of 100 mM DTTred starting the reduction from isolated des [30-75] (second stage).
The oxidative folding of ONC/unONC was carried out at pH 8, 25 mM DTTox, 25°C on a mixture containing 10% cyclized and 90% uncyclized protein, facilitating the direct comparison of the rates. Aliquots were taken at different times and any further rearrangements of the disulfides were prevented by adding excess AEMTS as described in the Experimental Procedures section. No significant difference in the overall oxidative folding rates of the two forms was observed in these experiments.
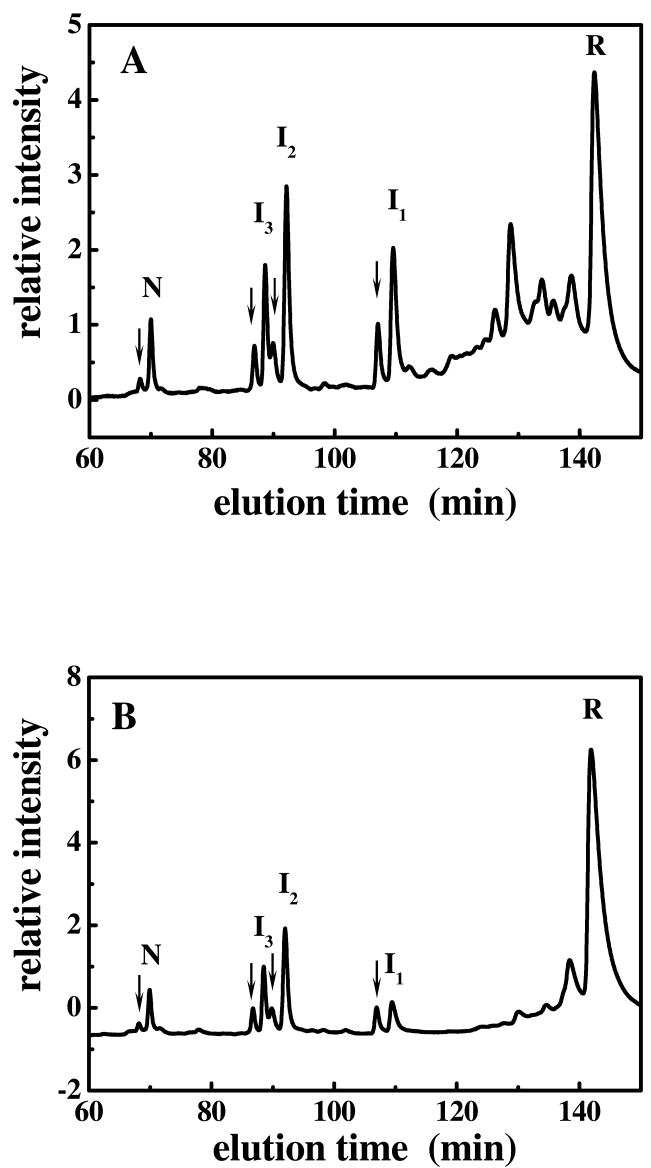
However, further examination revealed some differences in the distribution of the oxidative folding intermediates in ONC versus unONC. A reduction pulse (a short mild reduction), which is sufficient to reduce unstructured intermediates (in which the disulfide bonds are exposed) to the fully-reduced protein without affecting the structured ones (in which the three-dimensional structure protects the disulfide bonds from easy reduction), is a useful tool to distinguish structured intermediates from unstructured ones (42, 55, 58). We applied a reduction pulse to one-half of the aliquots of the regeneration mixture for both ONC and unONC, but no reduction pulse to the other half of the aliquots, before processing all the samples for HPLC analysis. Figure 2A shows the chromatogram for the two-hour oxidation of R-unONC (containing 10% R-ONC) without a reduction pulse. A reduction pulse revealed the presence of three peaks for ONC corresponding to structured intermediates (I1, I2, and I3,) in an earlier study (55). I1 includes two-disulfide-containing species whereas I2 and I3 correspond to three-disulfide-containing intermediates (as concluded from their masses after-AEMTS blocking). The corresponding three peaks for unONC are apparent, after the reduction pulse, in the chromatogram in Figure 2B. (The earlier-eluting smaller peaks are indicated by arrows in the Figure, and correspond to the N, I3, I2, I1 species of ONC; unpublished data.) However, the relative proportion of these structured intermediates is different for the oxidative folding of ONC and unONC. Relative to I2, the I1 and I3 species, respectively, are more abundant in the oxidative folding of the cyclized protein.
Figure 2.
Chromatogram of the oxidative folding of R-unONC (containing 10% cyclized R-ONC) at pH 8, 25 mM DTTox and 25°C at two hours. A. Without and, B. With a reduction pulse. The earlier-eluting smaller peaks (indicated by arrows) corresponding to N, I3, I2, I1 are for the cyclized protein. All free thiols in each sample were blocked with AEMTS; the samples were then desalted, and analyzed using cation-exchange HPLC analysis.
We also examined the effect of the three-dimensional structure on the non-enzymatic cyclization of the N-terminal glutamine of the protein. Native uncyclized ONC and its six-residue N-terminal peptide fragment were incubated under various conditions described under Experimental Procedures. The two forms (cyclized and uncyclized) were separated from one another using reversed-phase HPLC and cation-exchange HPLC for the peptide and the protein, respectively. It takes less than two days for the non-enzymatic cyclization to be completed at 37°C for both the peptide and the protein, in agreement with the literature (10, 59).
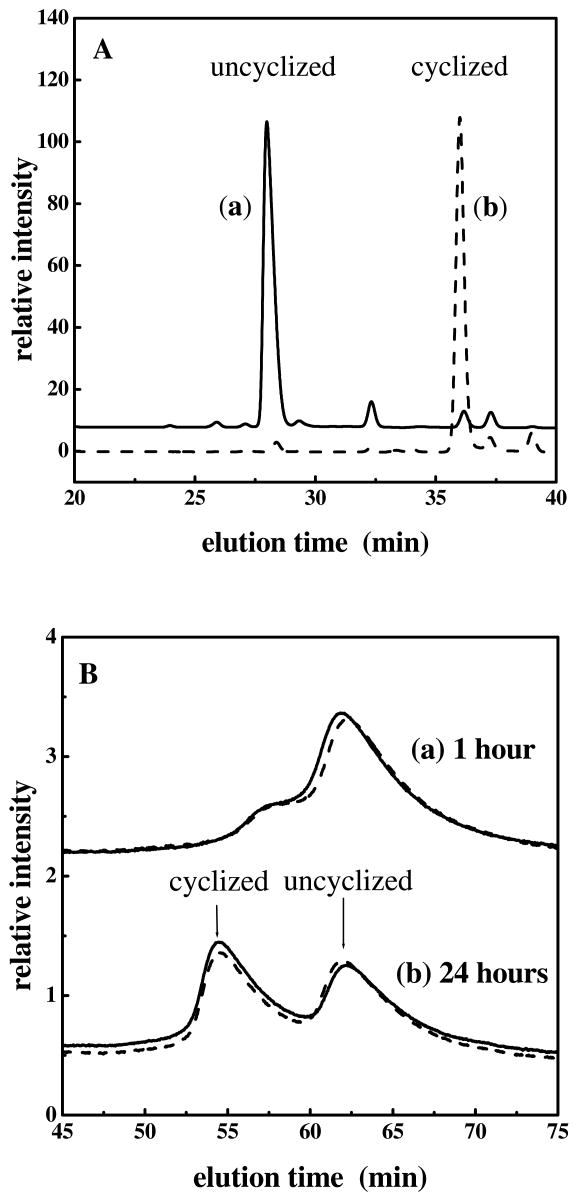
The cyclization of N-terminal glutamine in peptides and proteins is accelerated by an enzyme, glutaminyl cyclase (QC) in vivo (9-13). We examined the effect of QC on the cyclization of native unONC, and of the N-terminal six-residue peptide under identical conditions of enzyme and substrate concentration, (pH 7; 37°C). Whereas QC dramatically accelerated the cyclization of the peptide (the cyclization is basically completed in one minute, as evident from the different elution times of the peaks for one-minute incubation with and without QC in Figure 3A), it had no effect on the cyclization of the native, folded unONC (Fig. 3B). In Figure 3B, there is one dominant peak at one-hour reaction times (corresponding to the uncyclized protein) with or without QC indicating that very little (enzymatic or non-enzymatic) cyclization of native unONC occurs in 1 hour. In 24 hours, equal fractions of cyclized and uncyclized proteins appear. The cyclized material resulted from slow non-enzymatic cyclization of unONC, and the solid and dashed curves overlap because no QC-catalyzed cyclization appeared in 24 hours under these conditions.
Figure 3.
A. Overlaid reversed-phase HPLC chromatograms of (a) the uncyclized model peptide (—) and (b) the QC-cyclized model peptide (---), at 1 min. B. Chromatograms of native unONC after incubating with (—) and without (---) QC at (a) 1 hour and (b) 24 hours. Experiments were carried out at pH 7, 37°C.
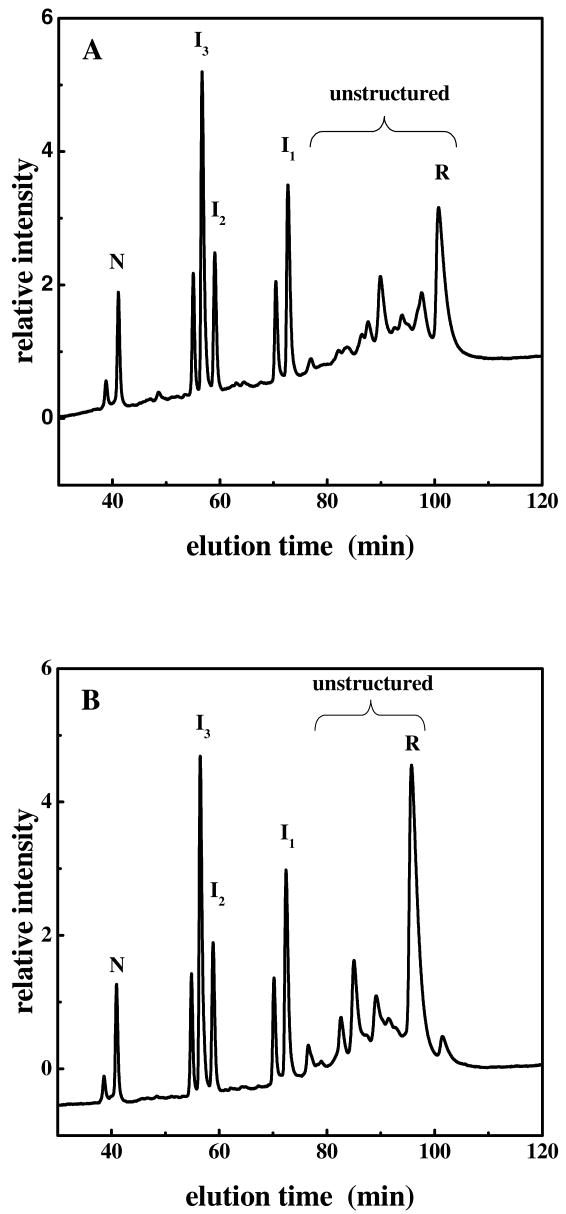
Subsequently, we examined the reduced form and the oxidative folding intermediates of unONC to determine whether they were also resistant to QC catalysis. Aliquots of the oxidative folding mixture of unONC (at four hours) were withdrawn, and the thiols were blocked with AEMTS, followed by desalting and incubation with and without QC before HPLC analyses. A comparison of the chromatograms in Figure 4, reveals that, whereas the structured species, N, I1, I2 and I3, were resistant to QC-catalyses (shown as the same elution pattern between 40 and 75 min. in Fig. 4A & B), the unstructured ensemble of unONC including the reduced protein (indicated by the peaks under the braces in Figures 4A and 4B) shifts to earlier elution times, indicating that QC is effective in catalyzing cyclization in unstructured species.
Figure 4.
Chromatogram of an oxidative folding mixture of unONC (containing 10% cyclized ONC) at 4 hours at pH 8, 25 mM DTTox. All free thiols in each sample were blocked with AEMTS, and the samples were desalted prior to analysis using cation-exchange HPLC. After desalting, the blocked protein was incubated without QC (A) and with QC (B) at pH 7.2, room temperature. The unstructured species are indicated with braces.
Discussion
Although the non-enzymatic cyclization cannot be stopped/quenched, the cyclization rate is significantly slowed down at low temperatures (and is in fact relatively slow even at room temperature); thus, the non-enzymatic cyclization did not cause detectable error during the processing of the samples to determine the rates of oxidative folding and reductive unfolding. On the other hand, this non-enzymatic cyclization of the intermediates of reductive unfolding and oxidative folding, could be exploited for the identification of the HPLC peaks of the corresponding cyclized and uncyclized intermediate pairs, when they are kept above 10°C. This could be achieved by the isolation of peaks on a chromatogram of unONC intermediates and by re-injecting the isolated peaks after allowing cyclization to proceed.
Effect of local unfolding on the reduction of disulfide bonds during reductive unfolding
The X-ray structure of native ONC in Figure 5 reveals a hydrogen bond between the N-terminal pyroglutamic acid and Lys 9 (20). The absence of this hydrogen bond decreases the stability of the protein (22). This decreased stability is not evident in the reductive unfolding experiments (Fig. 1), since the reduction rates are not significantly accelerated in unONC. This supports a previous contention that both the reduction of the first disulfide bond (30-75) and that of the second disulfide bond in des [30-75] are mediated by a local unfolding event (54, 56). This explanation is plausible because a similar phenomenon is observed in a mutant variant of RNase A, a homolog of ONC. The Pro93Ala mutation in RNase A, which greatly destabilizes the protein and increases the rate of reduction of the first, nearby 40-95 disulfide bond 120-fold, had apparently little effect on the reduction of the remote (65-72) disulfide bond; both disulfide bonds in RNase A are exposed for reduction by a local unfolding event (35, 60).
Figure 5.
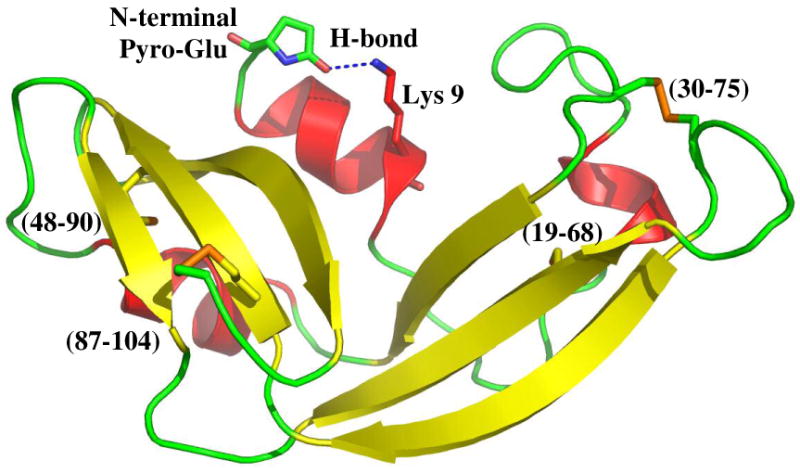
The X-ray structure of native ONC (20). The four disulfide bonds and the hydrogen bond between the γ-carboxyl oxygen of the N-terminal pyroglutamic acid and the ε -amino hydrogen of Lys 9 are indicated.
The N-terminus of unONC is not-accessible in the folded protein
Application of the reduction-pulse technique led to the detection of structured intermediates corresponding to peaks I1, I2 and I3 in the oxidative folding of both the cyclized and uncyclized forms. This is because the reduction-pulse technique effectively screens for stable three-dimensional folds that provide protection to the disulfide bonds against the reducing agent (42). Such protection has been demonstrated only in structured intermediate species which have a native-like fold (42, 61, 62). It is known that disulfide-bond-containing proteins frequently retain their native structure in the absence of either one or even more disulfide bonds (57, 63-72). Considering the extremely high stability of ONC (Tm = 85.4°C at pH 8) (28), it is expected that reduction of one or even two disulfide bonds in the protein might not cause unfolding at room temperature (27, 54, 72). Thus, it is plausible to propose that I2 and I3 may have the overall native fold of the protein since they possess three of the four native disulfide bonds, and survive a reduction pulse (Fig. 2). I1, which also survives a reduction pulse, may also have the overall native fold but further structural characterization is necessary to determine the effect of the absence of two disulfide bonds on the native three-dimensional structure of ONC in this species. Interestingly, the stability of I1 of unONC is lower than that of I1 of the cyclized species; this is evident from the effect of a reduction pulse (Fig 2B). The same phenomenon was observed at 37°C with I1 of ONC in an earlier study (55), suggesting that the effect of heat destabilization is similar to that due to the absence of the native-like interactions of the pyroglutamyl moiety in this intermediate.
We found that QC does not catalyze cyclization of the native uncyclized protein and its structured intermediates, although it effectively catalyzes cyclization of the fully-reduced uncyclized protein, the unstructured uncyclized intermediates and the N-terminal peptide. These results indicate that the folded structure hinders the catalysis by making the N-terminus of the protein non-accessible for the enzymatic action of QC. A similar hindrance was revealed in an earlier study of a bacterially-expressed variant of onconase (73). In that study, the in vitro use of methionine amino peptidase (MAP) was attempted for the removal of the N-terminal methionine, but the results indicated that the enzyme had no access to the N-terminus of the protein; the first methionine residue could be removed from the polypeptide chain by the specific action of MAP only after denaturing the structure of the protein (73). In our bacterial expression system, used for both RNase A and ONC, the starting methionine is cleaved off together with the signal peptide.
Cyclization changes the rate of formation of I3
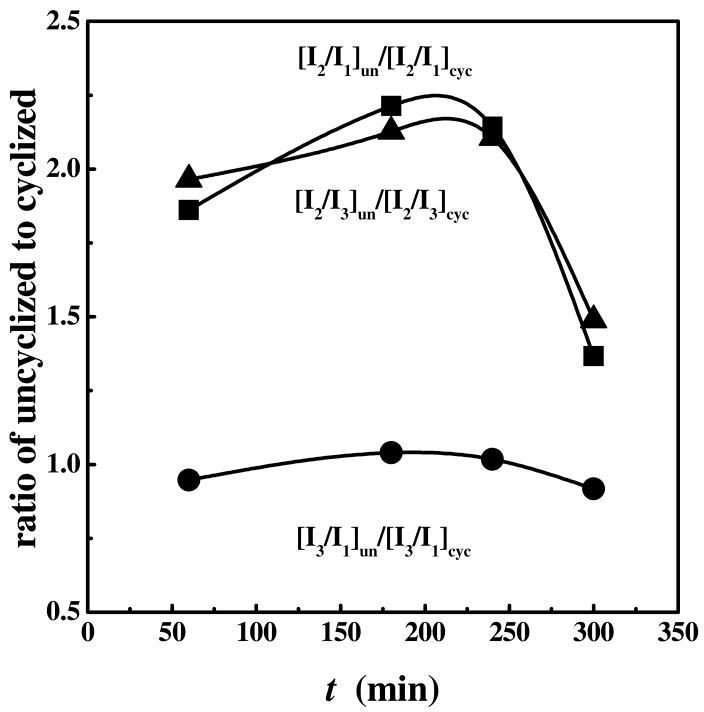
Comparison of the oxidative folding of ONC and unONC revealed further important features of the oxidative folding process of this protein. Both the cyclized and uncyclized proteins seem to follow the same oxidative folding pathways, through I2 or I3. However, from chromatograms (such as Fig. 2), it is apparent that the ratios between the intermediates are different for the oxidative folding of the cyclized and uncyclized forms (Fig. 6). Specifically, the concentrations of I3 (and I1) relative to I2 are consistently lower in the uncyclized form than in the cyclized one throughout the course of the experiments. By contrast, the ratio of I3 to I1 is nearly identical in both forms of onconase at all times examined (Fig. 6). Thus, cyclization altered the relative flux of the protein in these pathways by increasing the concentration of I3, a kinetically trapped intermediate (55).
Figure 6.
The ratios of fractions of the intermediates during the time course of oxidative folding of the uncyclized form relative to the cyclized form of onconase. ▲: [I2/I3]un/[I2/I3]cyc ■: [I2/I1]un/ [I2/I1]cyc and, ●: [I3/I1]un/ [I3/I1]cyc.
In theory, the concentration of I3 could be changed by decreasing the rate of its unfolding or by increasing the rate of its formation or by both. We argue here that it is less likely that its unfolding rate (i.e., by reduction and/or reshuffling) is substantially changed, based on the following facts. ONC is an extremely stable protein, with a Tm of 85°C (pH 8) under the conditions of the experiments (28). I3, which has three disulfide bonds, is plausibly a des species as we have shown above. The absence of a disulfide bond, as in I3, destabilizes the protein, primarily, by increasing the entropy of the unfolded state compared to that of the four-disulfide native protein. The distances between the cysteines of the native disulfide bonds in the amino acid sequence of ONC are similar to or less than those in RNase A that has an analogous fold as ONC. Thus, it is not expected that the stability of ONC would decrease by ≥ 40°C due to the absence of one disulfide bond. This argument is supported by the melting temperatures of two des species of ONC. [Tm of des [30-75] is decreased by ∼30°C, and Tm of mutant analogs of des[87-104] is decreased by ∼20°C (27,28,74)]. Therefore, I3 is plausibly stable at 37°C where its reshuffling rate or reduction remains very slow. Since the effect of the absence of the native-like interaction of the pyroglutamic moiety on the oxidative folding intermediates of unONC is very similar to those of increased temperature (37°C) for both intermediates of ONC, the stability of I3-unONC at 25°C should not be altered too much by the absence of one disulfide bond. The resistance of I3 to a reduction pulse under these conditions (in contrast to I1 in Fig. 2 and in ref. 55) further supports these arguments.
The presence of kinetically trapped species in the oxidative folding of Onconase
The destabilization (melting) of disulfide-secure intermediates in oxidative folding was shown to alter the folding rate by two orders of magnitude (55). By contrast, the presence of a kinetically-trapped species, such as I3 usually has little effect on the overall oxidative folding rate in the first stage of the oxidative folding process, in general, when folding can proceed by other pathways (55). The rate will drop more significantly only in the second stage of the process when most of the precursor intermediates are already converted to either the native or the kinetically-trapped species. Since we measured the oxidative folding rates during the first 5 hours of the process, it is understandable that a change in the concentrations of I3 did not change the overall rate. The same consideration applies to I1, and, thus, I1 must also lie on a kinetically trapped pathway. The alternative explanation, that the effect of the cyclization on I1, which lies on a direct pathway, is compensated exactly by an opposite effect on this or on another direct pathway, is not very likely.
Support for an I1-to-I3 pathway
If we consider the parallel effects of both the absence of cyclization and of an increase in temperature (to 37°C) on I1 and I3 during oxidative folding [Fig. 6 (55)], the most plausible explanation is that I1 and I3 lie on a common pathway where I3 is a kinetically trapped species. A des species of ONC could form either by reshuffling from the 3S ensemble or by oxidation of precursor species of the 2S ensemble. Since an earlier study showed no formation of I3 by reshuffling from the 3S species (54), I3 must form by oxidation. There are three possible 2S precursor species of I3; of them I1 is the only one which accumulates to a considerable extent supporting the hypothesis of an I1-to-I3 pathway. This interpretation is consistent with data found by the disulfide mapping of these two species (R. Gahl and H.A.S., unpublished results). However, the alternative explanation cannot be excluded with certainty, namely, that I1 is also a kinetically trapped species, and the real 2S precursor of I3 does not accumulate to a detectable level because its oxidation (to I3) is extremely rapid.
Sub-cellular localization of QC catalysis
Our results provide further insights into the subcellular localization of in vivo QC catalysis. QC catalysis has been localized mainly in the secretory granules (18). By contrast, the resistance of folded unONC against QC-catalyzed cyclization that we revealed here suggests that in vivo QC catalysis of proteins may occur much earlier on the secretory pathways than it is observed with some hormone peptides. The activity of ONC depends on cyclization of the N-terminal glutamine (21). Since the non-enzymatic cyclization is very slow, catalysis by QC is necessary to produce fully functional ONC in vivo. Given that the native but uncyclized onconase is resistant to QC-catalysis as demonstrated by our studies, cyclization plausibly occurs in the ER, before the native three-dimensional structure is formed. Since early-forming structured intermediates of unONC are also resistant to QC catalysis, cyclization seems to take place very early during the oxidative folding process, probably co-translationally.
Acknowledgments
We are grateful to Dr. Vaclav Čeřovský for helping with the synthesis of the model peptide and to Professor Robert C. Batemann, University of Southern Mississippi, for kindly providing a sample of QC and for helpful discussions.
Abbreviations
- ONC
frog onconase (Rana pipiens)
- N-ONC
native onconase
- R-ONC
fully-reduced onconase
- unONC
uncyclized onconase
- I1, I2, and I3
intermediates with two, three, and three native disulfides, respectively, formed during the oxidative folding of ONC
- RNase A
bovine pancreatic ribonuclease A
- des [30-75]
an ONC intermediate having all native disulfide bonds but lacking the (30-75) disulfide bond
- QC
glutaminyl cyclase
- ER
endoplasmic reticulum
- WT
wild-type
- AEMTS
2-aminoethylmethylthiosulfonate
- DTTred and DTTox
reduced and oxidized dithiothreitol, respectively
- GdnHCl
guanidine hydrochloride
- EDTA
ethylenediamine-tetra-acetic acid
- Tris-HCl
tris(hydroxymethyl) aminomethane hydrochloride
- HPLC
high performance liquid chromatography
- MALDI-TOF
Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight
- Fmoc
9-fluorenylmethyloxycarbonyl
- MAP
methionine amino peptidase
- PBS
phosphate buffered saline
Footnotes
This research was supported by NIH grant GM-24893, by HSRF grant NF61431, by Marie Curie IRG-04515 and EIF-011636, and by the National Office for Research and Technology Hungary RET-08/2004. E.W is an HHMI international scholar.
References
- 1.Richter K, Kawashima E, Egger R, Kreil G. Biosynthesis of thyrotropin releasing hormone in the skin of Xenopus laevis: partial sequence of the precursor deduced from cloned cDNA. EMBO J. 1984;3:617–621. doi: 10.1002/j.1460-2075.1984.tb01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awade AC, Cleuziat P, Gonzales T, Robert-Baudouy J. Pyrrolidone carboxyl peptidase (Pcp): an enzyme that removes pyroglutamic acid (pGlu) from pGlu-peptides and pGlu-proteins. Proteins. 1994;20:34–51. doi: 10.1002/prot.340200106. [DOI] [PubMed] [Google Scholar]
- 3.Garavelli JS. The RESID database of protein structure modifications: 2000 update. Nucleic Acids Res. 2000;28:209–211. doi: 10.1093/nar/28.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham GN, Podell DN. Pyroglutamic acid. Non-metabolic formation, function in proteins and peptides, and characteristics of the enzymes effecting its removal. Mol Cell Biochem. 1981;38(Spec No Pt 1):181–190. doi: 10.1007/BF00235695. [DOI] [PubMed] [Google Scholar]
- 5.Van Coillie E, Proost P, Van Aelst I, Struyf S, Polfliet M, De Meester I, Harvey DJ, Van Damme J, Opdenakker G. Functional comparison of two human monocyte chemotactic protein-2 isoforms, role of the amino-terminal pyroglutamic acid and processing by CD26/dipeptidyl peptidase IV. Biochemistry. 1998;37:12672–12680. doi: 10.1021/bi980497d. [DOI] [PubMed] [Google Scholar]
- 6.Gololobov MY, Wang W, Bateman RC., Jr Substrate and inhibitor specificity of glutamine cyclotransferase (QC) Biol Chem Hoppe Seyler. 1996;377:395–398. [PubMed] [Google Scholar]
- 7.Bateman RC, Jr, Temple JS, Misquitta SA, Booth RE. Evidence for essential histidines in human pituitary glutaminyl cyclase. Biochemistry. 2001;40:11246–11250. doi: 10.1021/bi011177o. [DOI] [PubMed] [Google Scholar]
- 8.Schilling S, Hoffmann T, Rosche F, Manhart S, Wasternack C, Demuth HU. Heterologous expression and characterization of human glutaminyl cyclase: evidence for a disulfide bond with importance for catalytic activity. Biochemistry. 2002;41:10849–10857. doi: 10.1021/bi0260381. [DOI] [PubMed] [Google Scholar]
- 9.Messer M. Enzymatic cyclization of L-glutamine and L-glutaminyl peptides. Nature. 1963;197:1299. doi: 10.1038/1971299a0. [DOI] [PubMed] [Google Scholar]
- 10.Busby WH, Quackenbush GE, Humm J, Youngblood WW, Kizer JS. An enzyme(s) that converts glutaminyl-peptides into pyroglutamyl-peptides. Presence in pituitary, brain, adrenal medulla, and lymphocytes. J Biol Chem. 1987;262:8532–8536. [PubMed] [Google Scholar]
- 11.Fischer WH, Spiess J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc Natl Acad Sci USA. 1987;84:3628–3632. doi: 10.1073/pnas.84.11.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen GJ, Russell JB. Transport of glutamine by Streptococcus bovis and conversion of glutamine to pyroglutamic acid and ammonia. J Bacteriol. 1989;171:2981–2985. doi: 10.1128/jb.171.6.2981-2985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sykes PA, Watson SJ, Temple JS, Bateman RC., Jr Evidence for tissue-specific forms of glutaminyl cyclase. FEBS Lett. 1999;455:159–161. doi: 10.1016/s0014-5793(99)00872-8. [DOI] [PubMed] [Google Scholar]
- 14.Pohl T, Zimmer M, Mugele K, Spiess J. Primary structure and functional expression of a glutaminyl cyclase. Proc Natl Acad Sci USA. 1991;88:10059–10063. doi: 10.1073/pnas.88.22.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song I, Chuang CZ, Bateman RC., Jr Molecular cloning, sequence analysis and expression of human pituitary glutaminyl cyclase. J Mol Endocrinol. 1994;13:77–86. doi: 10.1677/jme.0.0130077. [DOI] [PubMed] [Google Scholar]
- 16.Dahl SW, Slaughter C, Lauritzen C, Bateman RC, Connerton I, Pedersen J. Carica papaya glutamine cyclotransferase belongs to a novel plant enzyme subfamily: cloning and characterization of the recombinant enzyme. Protein Expr Purif. 2000;20:27–36. doi: 10.1006/prep.2000.1273. [DOI] [PubMed] [Google Scholar]
- 17.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 18.Bockers TM, Kreutz MR, Pohl T. Glutaminyl-cyclase expression in the bovine/porcine hypothalamus and pituitary. J Neuroendocrinol. 1995;7:445–453. doi: 10.1111/j.1365-2826.1995.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 19.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an antitumor protein from Rana pipiens oocytes and early embryos-homology to pancreatic ribonucleases. J Biol Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- 20.Mosimann SC, Ardelt W, James MNG. Refined 1.7 Å X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with antitumor activity. J Mol Biol. 1994;236:1141–1153. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 21.Boix E, Wu YN, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. Role of the N terminus in RNase A homologues: differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J Mol Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 22.Notomista E, Catanzano F, Graziano G, Di Gaetano S, Barone G, Di Donato A. Contribution of chain termini to the conformational stability and biological activity of onconase. Biochemistry. 2001;40:9097–9103. doi: 10.1021/bi010741s. [DOI] [PubMed] [Google Scholar]
- 23.Irie M, Nitta K, Nonaka T. Biochemistry of frog ribonucleases. Cell Mol Life Sci. 1998;54:775–784. doi: 10.1007/s000180050206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov AA, Ilinskaya ON. Cytotoxic ribonucleases: molecular weapons and their targets. FEBS Lett. 2003;540:15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 25.Matousek J, Soucek J, Slavik T, Tomanek M, Lee JE, Raines RT. Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comp Biochem Physiol C Toxicol Pharmacol. 2003;136:343–356. doi: 10.1016/j.cca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Leland PA, Schultz LW, Kim BM, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;95:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leland PA, Staniszewski KE, Kim BM, Raines RT. A synapomorphic disulfide bond is critical for the conformational stability and cytotoxicity of an amphibian ribonuclease. FEBS Lett. 2000;477:203–207. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- 28.Notomista E, Catanzano F, Graziano G, Dal Piaz F, Barone G, D'Alessio G, Di Donato A. Onconase: an unusually stable protein. Biochemistry. 2000;39:8711–8718. doi: 10.1021/bi000415x. [DOI] [PubMed] [Google Scholar]
- 29.Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. Entry into cells and selective degradation of tRNAs by a cytotoxic member of the RNase A family. J Biol Chem. 2002;277:15142–15146. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- 30.Kuwajima K, Ikeguchi M, Sugawara T, Hiraoka Y, Sugai S. Kinetics of disulfide bond reduction in α-lactalbumin by dithiothreitol and molecular basis of superreactivity of the Cys6-Cys120 disulfide bond. Biochemistry. 1990;29:8240–8249. doi: 10.1021/bi00488a007. [DOI] [PubMed] [Google Scholar]
- 31.Ewbank JJ, Creighton TE. Pathway of disulfide-coupled unfolding and refolding of bovine α-lactalbumin. Biochemistry. 1993;32:3677–3693. doi: 10.1021/bi00065a022. [DOI] [PubMed] [Google Scholar]
- 32.Mendoza JA, Jarstfer MB, Goldenberg DP. Effects of amino acid replacements on the reductive unfolding kinetics of pancreatic trypsin inhibitor. Biochemistry. 1994;33:1143–1148. doi: 10.1021/bi00171a013. [DOI] [PubMed] [Google Scholar]
- 33.Creighton TE. The protein folding problem. In: Pain RH, editor. Mechanisms of Protein Folding. Oxford University Press; New York: 1994. pp. 1–25. [Google Scholar]
- 34.Yamashita H, Nakatsuka T, Hirose M. Structural and functional characteristics of partially disulfide-reduced intermediates ovotransferrin N lobe. Cystine localization by indirect end-labeling approach and implications for the reduction pathway. J Biol Chem. 1995;270:29806–29812. doi: 10.1074/jbc.270.50.29806. [DOI] [PubMed] [Google Scholar]
- 35.Li YJ, Rothwarf DM, Scheraga HA. Mechanism of reductive protein unfolding. Nature Struct Biol. 1995;2:489–494. doi: 10.1038/nsb0695-489. [DOI] [PubMed] [Google Scholar]
- 36.Ma LC, Anderson S. Correlation between disulfide reduction and conformational unfolding in bovine pancreatic trypsin inhibitor. Biochemistry. 1997;36:3728–3736. doi: 10.1021/bi962310t. [DOI] [PubMed] [Google Scholar]
- 37.Chang JY. A two-stage mechanism for the reductive unfolding of disulfide-containing proteins. J Biol Chem. 1997;272:69–75. [PubMed] [Google Scholar]
- 38.Singh RR, Rao AGA. Reductive unfolding and oxidative refolding of a Bowman Birk inhibitor from horsegram seeds (Dolichos biflorus): evidence for “hyperreactive” disulfide bonds and rate-limiting nature of disulfide isomerization in folding. Biochim Biophys Acta. 2002;1597:280–291. doi: 10.1016/s0167-4838(02)00301-1. [DOI] [PubMed] [Google Scholar]
- 39.Yan YB, Zhang RQ, Zhou HM. Biphasic reductive unfolding of ribonuclease A is temperature dependent. Eur J Biochem. 2002;269:5314–5322. doi: 10.1046/j.1432-1033.2002.03251.x. [DOI] [PubMed] [Google Scholar]
- 40.Rothwarf DM, Scheraga HA. Equilibrium and kinetic constants for the thiol-disulfide interchange reaction between glutathione and dithiothreitol. Proc Natl Acad Sci USA. 1992;89:7944–7948. doi: 10.1073/pnas.89.17.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothwarf DM, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A. 4. Temperature dependence of the regeneration rate. Biochemistry. 1993;32:2698–2703. doi: 10.1021/bi00061a030. [DOI] [PubMed] [Google Scholar]
- 42.Rothwarf DM, Li YJ, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A: identification of two nativelike three-disulfide intermediates involved in separate pathways. Biochemistry. 1998;37:3760–3766. doi: 10.1021/bi972822n. [DOI] [PubMed] [Google Scholar]
- 43.Welker E, Wedemeyer WJ, Narayan M, Scheraga HA. Coupling of conformational folding and disulfide-bond reactions in oxidative folding of proteins. Biochemistry. 2001;40:9059–9064. doi: 10.1021/bi010409g. [DOI] [PubMed] [Google Scholar]
- 44.Maskos K, Huber-Wunderlich M, Glockshuber R. DsbA and DsbC-catalyzed oxidative folding of proteins with complex disulfide bridge patterns in vitro and in vivo. J Mol Biol. 2003;325:495–513. doi: 10.1016/s0022-2836(02)01248-2. [DOI] [PubMed] [Google Scholar]
- 45.Buczek P, Buczek O, Bulaj G. Total chemical synthesis and oxidative folding of δ-conotoxin PVIA containing an N-terminal propeptide. Bioploymers. 2004;80:50–57. doi: 10.1002/bip.20211. [DOI] [PubMed] [Google Scholar]
- 46.Bulaj G, Koehn RE, Goldenberg DP. Alteration of the disulfide-coupled folding pathway of BPTI by circular permutation. Protein Sci. 2004;13:1182–1196. doi: 10.1110/ps.03563704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cemazar M, Zahariev S, Pongor S, Hore PJ. Oxidative folding of Amaranthus α-amylase inhibitor: disulfide bond formation and conformational folding. J Biol Chem. 2004;279:16697–16705. doi: 10.1074/jbc.M312328200. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Jin R, Dong HY, Feng YM. In vitro refolding/unfolding pathways of amphioxus insulin-like peptide: implications for folding behavior of insulin family proteins. J Biol Chem. 2004;279:55224–55233. doi: 10.1074/jbc.M409030200. [DOI] [PubMed] [Google Scholar]
- 49.Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, Glockshuber R, Ellgaard L. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- 50.Gent J, Braakman I. Low-density lipoprotein receptor structure and folding. Cell Mol Life Sci. 2004;61:2461–2470. doi: 10.1007/s00018-004-4090-3. [DOI] [PubMed] [Google Scholar]
- 51.Shioi S, Imoto T, Ueda T. Analysis of the early stage of the folding process of reduced lysozyme using all lysozyme variants containing a pair of cysteines. Biochemistry. 2004;43:5488–5493. doi: 10.1021/bi036048h. [DOI] [PubMed] [Google Scholar]
- 52.Varsanyi M, Szarka A, Papp E, Makai D, Nardai G, Fulceri R, Csermely P, Mandl J, Benedetti A, Banhegyi G. FAD transport and FAD-dependent protein thiol oxidation in rat liver microsomes. J Biol Chem. 2004;279:3370–3374. doi: 10.1074/jbc.M307783200. [DOI] [PubMed] [Google Scholar]
- 53.Arolas JL, D'Silva L, Popowicz GM, Aviles FX, Holak TA, Ventura S. NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor. Structure. 2005;13:1193–1202. doi: 10.1016/j.str.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Xu G, Narayan M, Welker E, Scheraga HA. Characterization of the fast-forming intermediate, des [30-75], in the reductive unfolding of onconase. Biochemistry. 2004;43:3246–3254. doi: 10.1021/bi036215d. [DOI] [PubMed] [Google Scholar]
- 55.Welker E, Hathaway L, Scheraga HA. A new method for rapid characterization of the folding pathways of multidisulfide-containing proteins. J Am Chem Soc. 2004;126:3720–3721. doi: 10.1021/ja031658q. [DOI] [PubMed] [Google Scholar]
- 56.Narayan M, Xu G, Ripoll DR, Zhai HL, Breuker K, Wanjalla C, Leung HJ, Navon A, Welker E, McLafferty FW, Scheraga HA. Dissimilarity in the reductive unfolding pathways of two ribonuclease homologues. J Mol Biol. 2004;338:795–809. doi: 10.1016/j.jmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 57.Welker E, Narayan M, Volles MJ, Scheraga HA. Two new structured intermediates in the oxidative folding of RNase A. FEBS Lett. 1999;460:477–479. doi: 10.1016/s0014-5793(99)01391-5. [DOI] [PubMed] [Google Scholar]
- 58.Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Oxidative folding of proteins. Acc Chem Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- 59.Ribo M, Bosch M, Torrent G, Benito A, Beaumelle B, Vilanova M. Quantitative analysis, using MALDI-TOF mass spectrometry, of the N-terminal hydrolysis and cyclization reactions of the activation process of onconase. Eur J Biochem. 2004;271:1163–1171. doi: 10.1111/j.1432-1033.2004.04020.x. [DOI] [PubMed] [Google Scholar]
- 60.Cao A, Welker E, Scheraga HA. Effect of mutation of proline 93 on redox unfolding/folding of bovine pancreatic ribonuclease A. Biochemistry. 2001;40:8536–8541. doi: 10.1021/bi010692j. [DOI] [PubMed] [Google Scholar]
- 61.Welker E, Narayan M, Wedemeyer WJ, Scheraga HA. Structural determinants of oxidative folding in proteins. Proc Natl Acad Sci USA. 2001;98:2312–2316. doi: 10.1073/pnas.041615798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. Disulfide bonds and protein folding. Biochemistry. 2000;39:4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- 63.Forman-Kay JD, Clore GM, Stahl SJ, Gronenborn AM. 1H and 15N resonance assignments and secondary structure of the human thioredoxin C62A, C69A, C73A mutant. J Biomol NMR. 1992;2:431–445. doi: 10.1007/BF02192807. [DOI] [PubMed] [Google Scholar]
- 64.Stone MJ, Chandrasekhar K, Holmgren A, Wright PE, Dyson HJ. Comparison of backbone and tryptophan side-chain dynamics of reduced and oxidized Escherichia coli thioredoxin using 15N NMR relaxation measurements. Biochemistry. 1993;32:426–435. doi: 10.1021/bi00053a007. [DOI] [PubMed] [Google Scholar]
- 65.Vogl T, Brengelmann R, Hinz HJ, Scharf M, Lotzbeyer M, Engels JW. Mechanism of protein stabilization by disulfide bridges: calorimetric unfolding studies on disulfide-deficient mutants of the α-amylase inhibitor tendamistat. J Mol Biol. 1995;254:481–496. doi: 10.1006/jmbi.1995.0632. [DOI] [PubMed] [Google Scholar]
- 66.Kelley JJ, III, Caputo TM, Eaton SF, Laue TM, Bushweller JH. Comparison of backbone dynamics of reduced and oxidized Escherichia coli glutaredoxin-1 using 15N NMR relaxation measurements. Biochemistry. 1997;36:5029–5044. doi: 10.1021/bi962181g. [DOI] [PubMed] [Google Scholar]
- 67.Shimotakahara S, Ríos CB, Laity JH, Zimmerman DE, Scheraga HA, Montelione GT. NMR structural analysis of an analog of an intermediate formed in the rate-determining step of one pathway in the oxidative folding of bovine pancreatic ribonuclease A: automated analysis of 1H, 13C, and 15N resonance assignments for wild-type and [C65S, C72S] mutant forms. Biochemistry. 1997;36:6915–6929. doi: 10.1021/bi963024k. [DOI] [PubMed] [Google Scholar]
- 68.Laity JH, Lester CC, Shimotakahara S, Zimmerman DE, Montelione GT, Scheraga HA. Structural characterization of an analog of the major rate-determining disulfide folding intermediate of bovine pancreatic ribonuclease A. Biochemistry. 1997;36:12683–12699. doi: 10.1021/bi970878b. [DOI] [PubMed] [Google Scholar]
- 69.Kowalski JM, Parekh RN, Wittrup KD. Secretion efficiency in Saccharomyces cerevisiae of bovine pancreatic trypsin inhibitor mutants lacking disulfide bonds is correlated with thermodynamic stability. Biochemistry. 1998;37:1264–1273. doi: 10.1021/bi9722397. [DOI] [PubMed] [Google Scholar]
- 70.Scheraga HA, Wedemeyer WJ, Welker E. Bovine pancreatic ribonuclease A: Oxidative and conformational folding studies. Method Enzym. 2001;341:189–221. doi: 10.1016/s0076-6879(01)41153-0. [DOI] [PubMed] [Google Scholar]
- 71.Wedemeyer WJ, Xu XB, Welker E, Scheraga HA. Conformational propensities of protein folding intermediates: Distribution of species in the 1S, 2S, and 3S ensembles of the [C40A,C95A] mutant of bovine pancreatic ribonuclease A. Biochemistry. 2002;41:1483–1491. doi: 10.1021/bi011893q. [DOI] [PubMed] [Google Scholar]
- 72.Merlino A, Mazzarella L, Carannante A, Di Fiore A, Di Donato A, Notomista E, Sica F. The importance of dynamic effects on the enzyme activity: X-ray structure and molecular dynamics of onconase mutants. J Biol Chem. 2005;280:17953–17960. doi: 10.1074/jbc.M501339200. [DOI] [PubMed] [Google Scholar]
- 73.Notomista E, Cafaro V, Fusiello R, Bracale A, D'Alessio G, Di Donato A. Effective expression and purification of recombinant onconase, an antitumor protein. FEBS Lett. 1999;463:211–215. doi: 10.1016/s0014-5793(99)01623-3. [DOI] [PubMed] [Google Scholar]
- 74.Xu GQ, Narayan M, Welker E, Scheraga HA. A novel method to determine thermal transition curves of disulfide-containing proteins and their structured folding intermediates. Biochem Biophys Res Comm. 2000;311:514–517. doi: 10.1016/j.bbrc.2003.10.039. [DOI] [PubMed] [Google Scholar]