Abstract
Bcor (BCL6 corepressor) is a widely expressed gene that is mutated in patients with X-linked Oculofaciocardiodental (OFCD) syndrome. BCOR regulates gene expression in association with a complex of proteins capable of epigenetic modification of chromatin. These include Polycomb group (PcG) proteins, Skp-Cullin-F-box (SCF) ubiquitin ligase components and a Jumonji C (Jmjc) domain containing histone demethylase. To model OFCD in mice and dissect the role of Bcor in development we have characterized two loss of function Bcor alleles. We find that Bcor loss of function results in a strong parent-of-origin effect, most likely indicating a requirement for Bcor in extraembryonic development. Using Bcor loss of function embryonic stem (ES) cells and in vitro differentiation assays, we demonstrate that Bcor plays a role in the regulation of gene expression very early in the differentiation of ES cells into ectoderm, mesoderm and downstream hematopoietic lineages. Normal expression of affected genes (Oct3/4, Nanog, Fgf5, Bmp4, Brachyury and Flk1) is restored upon re-expression of Bcor. Consistent with these ES cell results, chimeric animals generated with the same loss of function Bcor alleles show a low contribution to B and T cells and erythrocytes and have kinked and shortened tails, consistent with reduced Brachyury expression. Together these results suggest that Bcor plays a role in differentiation of multiple tissue lineages during early embryonic development.
Introduction
Mutations causing gene fusions, loss of function or misregulation of transcriptional corepressor proteins have been implicated both in genetically inherited diseases and cancer [1], [2]. BCOR (BCL6 corepressor) is a transcriptional corepressor that was originally identified by its ability to interact with the site specific transcriptional repressor BCL6 [3]. BCOR is found in an 800 kDa complex in which at least two proteins have chromatin modifying activity: the PcG transcriptional repressor protein, RNF2, is a histone H2A E3 ubiquitin ligase and FBXL10 (JHDM1B, KDM2B) is a Jmjc histone demethylase in addition to its presumed ubiquitin E3 ligase activity [4], [5]. BCL6 plays critical roles in specific immunological processes involving B and T cells, including germinal center formation and the generation and maintenance of memory T cells [6]–[10]. In addition, testicular germ cell apoptosis and defects in erythropoiesis have been reported in BCL6-deficient mice [11], [12]. Deregulated expression of BCL6, due to chromosomal translocations or point mutations, is associated with the formation of approximately one sixth of non-Hodgkin's lymphomas [13], [14]. In cell lines derived from such patients, BCOR is detected at a number of BCL6 target genes suggesting that BCOR is likely to play a role in mediating BCL6 driven lymphomagenesis. Since its original discovery, BCOR has been shown to directly interact with the transcriptional regulator AF9 (MLLT3), and to be in a complex with ENL (MLLT1). AF9 and ENL are known MLL (trithorax) fusion partners in acute leukemias [15]. AF9 itself is a regulator of Hox gene expression and skeletal development [16], [17].
In addition, BCOR has been shown to play multiple roles in the complex process of human development. Females who are heterozygous for X-linked Bcor mutations have the rare Oculofaciocardiodental (OFCD) syndrome, the primary subtype of OMIM #300166 microphthalmia, syndromic 2 (MCOPS2) [18]. Congenital disorders in patients suffering from OFCD include cataracts, microphthalmia, and cardiac, dental and digital anomalies [18], [19]. In the hematopoietic lineage of OFCD patients, Bcor clearly is critical as 90–100% of surviving white blood cells show inactivation of the X- chromosome carrying the mutant allele of Bcor [18]. Presumably, the selective disadvantage, caused by an active X-chromosome harboring a Bcor mutation, is less severe in some tissues leading to variable phenotypic effects in a mosaic fashion. Male OFCD patients do not exist and are presumed to die in early development. Almost all Bcor mutations in OFCD patients result in premature stop codons that are thought to cause nonsense-mediated decay of the mRNA [18]. The second form of MCOPS2, Lenz microphthalmia, results from a single missense mutation (p. P85L) in the fourth coding exon of Bcor and is inherited in an X-linked recessive pattern [18]. Patients with Lenz microphthalmia suffer from microphthalmia/anophthalmia, mental retardation, and skeletal and other anomalies [18], [20].
The severity and breadth of these two MCOPS2 syndromes illustrate the important roles Bcor plays during development. Currently two animal models have successfully recapitulated developmental abnormalities similar to those found in patients with OFCD and Lenz microphthalmia. RNAi knock down of Bcor in zebrafish (Danio rerio) results in colobomatous eye defects and perturbations in somite, skeletal and neural tube development [18]. Colobomas, microphthalmia and cardiac abnormalities were also seen in morpholino knock down of Bcor in frogs (Xenopus tropicalis). Additionally, laterality defects were observed in Bcor morpholino injected frogs, similar to those present in a subset of OFCD patients [21].
The expression of Bcor during mouse development correlates well with tissues and organs in adversely affected patients with OFCD or Lenz microphthalmia [22]. Three separate promoters differentially control the expression of Bcor through embryonic development and into adulthood. Section in situ analysis shows that Bcor mRNA is expressed strongly in extraembryonic tissues during gastrulation and embryonic turning, suggesting a significant role in placental formation. Bcor expression is upregulated in the embryo proper at embryonic day 9 in mice starting in the tail, limb buds and branchial arches. During the later fetal stages of mouse development, Bcor is expressed in multiple tissues including strong expression in lens and retina of the eye and the neural tube [22].
The abnormalities present in Bcor knockdown fish and frogs, combined with the severe clinical presentation of MCOPS2 syndromes in humans, clearly illustrate the importance of Bcor in development. The developmental requirement for Bcor, together with its potential role in BCL6-related lymphomagenesis, underscores the importance of generating and analyzing mutant alleles of Bcor. To this end, we have analyzed the effect of two independent Bcor loss of function alleles on early development in mice and in ES cell differentiation. We found that Bcor exhibits a parent-of-origin effect, in that only paternal transmission results in viable offspring. Since the paternal X chromosome is exclusively inactivated in the extraembryonic tissue, this strongly suggests Bcor is required for extraembryonic development. In addition, loss of Bcor results in compromised development during in vitro differentiation of embryonic stem cells.
Results
Generation and analysis of a conditional Bcor allele
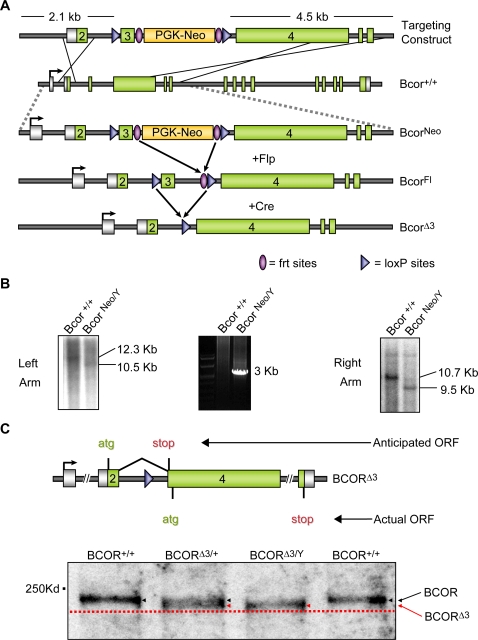
To investigate the role of the X-linked Bcor gene in development, we generated a conditional Bcor allele, BcorFl, in which exon 3 is flanked by loxP sites to allow its removal via expression of Cre recombinase (Figure 1A and targeting verification Figure 1B). We hypothesized that excision of exon 3 (Bcor Δ3) would result in a frame shift and a premature stop codon that should cause severe carboxy-terminal deletion of the BCOR protein and/or elimination of the mRNA by nonsense-mediated decay (Figure 1C). Based on human OFCD patients we expected that heterozygous female mice carrying this deletion in all tissues would recapitulate the phenotype of human OFCD patients and that hemizygous male mice would not be viable. Unexpectedly, breeding BcorFl/+ mice to ß-actin-Cre mice generated apparently normal female (Bcor Δ3/+) and male (Bcor Δ3/Y) mice. Western blot analysis of BCOR protein from the eyes of these mice and wild type controls revealed that an alternative start codon, 3′ to the engineered frame shift, was used for translation, generating BCOR protein with a predicted 71 amino acid amino-terminal deletion (Figure 1C). Thus, although this amino-terminal region is conserved, with 69% identity to the Xenopus protein and 90% identity to that of human, it is not essential.
Figure 1. Generating Bcor Δ 3/+ and Bcor Δ 3/Y mice.
(A) Diagram of targeting construct and recombination strategy. The Bcor+ allele was targeted by homologous recombination to generate BcorNeo embryonic stem cells. This allele contains loxP sites flanking the third exon, and a Neo resistance cassette flanked by frt sites, internal to the loxP sites immediately downstream of exon 3. Expression of Flp and/or Cre site specific recombinases in vivo or in vitro can be used to excise the intervening sequence between their respective recognition sites, frt and loxP, generating BcorFl and Bcor Δ3 alleles. PGK-Neo (phosphoglycerate kinase promoter–driven neomycin resistance gene cassette) (B) Southern blot and PCR analyses of targeted and wild type ES cell clones. Genomic DNA from individual ES cell clones was digested with the restriction enzymes, BamH1 (left arm) and BglII (right arm). The 5′ external probe (Left Arm) hybridizes to 10.7-kb (Bcor+/Y) and 9.5-kb (BcorNeo/Y) BamH1 fragments. Given the weak left arm signal, confirmatory PCR was performed on genomic DNA from wild type and targeted ES cells using a forward primer 5′ to the left arm and a reverse primer in the neomycin coding sequence. The 3′ external probe (Right Arm) hybridizes to 12.3-kb (Bcor+/Y) and 10.5-kb (BcorNeo/Y) BglII fragments. (C) The Bcor Δ3 allele encodes an N-terminally truncated version of BCOR. The anticipated and actual translation open reading frames for the Bcor Δ3 allele are diagrammed (top). Western blot analysis on the eyes of Bcor+/+, Bcor Δ3/+ and Bcor Δ3/Y mice (bottom) reveals the presence of an N-terminally truncated version of BCOR (red arrows) in both the Bcor Δ3/+ and Bcor Δ3/Y lanes, migrating at a slightly lower molecular weight in comparison to wild type BCOR (black arrows).
Generation and analysis of BcorNeo and BcorGt alleles in mice
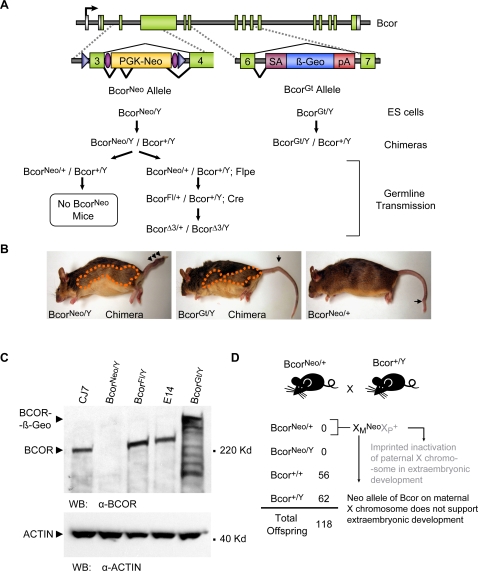
Although the BcorFl mutation did not provide the expected null allele, during the initial targeting of the Bcor locus we fortuitously created a loss of function Bcor allele (BcorNeo) that provided insights into Bcor function both in mice and in ES cell differentiation. We included a Neomycin (Neo) selection cassette in the Bcor targeting vector. Often the Neo cassette is excised during mRNA splicing to generate a normal transcript. However, in some cases a proportion of mRNAs are spliced into and then out of Neo, and the insertion of Neo sequences can result in a hypomorphic or null allele [23], [24]. Phenotypic effects of such aberrant splicing in chimeric animals would be most readily observed for X-linked genes like Bcor since XY ES cells are routinely used for gene targeting. Upon generation of chimeric mice with our BcorNeo/Y ES cells (Figure 2A) we observed two prominent phenotypes suggesting that the Neo cassette was indeed interfering with Bcor expression and function (Figure 2B). First, 3 out of 10 BcorNeo chimeras had kinked and sometimes shortened tails, suggesting a possible neural tube defect [25]. Second, 10 out of 10 chimeras exhibited a consistent bias in coat color contribution of the 129 (Agouti) ES cells to the sides of the bodies and legs of the mice. Since Agouti locus expressing cells in the skin are derived from the ectodermal lineage, this indicates a selective bias against BcorNeo/Y cells to contribute to certain ectodermal lineages. The same tail and coat color phenotype was also observed in 3 out of 3 chimeric mice that we generated from the Bay Genomics ES cell line, XE541 (BcorGt) (Figure 2B). The XE541 ES cell line harbors an insertional gene trap mutation (ß-Geo) in the 6th intron of mouse Bcor, which is predicted to result in an N-terminal BCOR-ß-Geo fusion protein (Figure 2A).
Figure 2. Transmission of the BcorNeo and BcorGt alleles in mice.
(A) Diagram of BcorNeo and BcorGt allele transmission. Location of each genomic alteration is shown, respective to the Bcor locus (top). The splicing pattern of each allele is indicated with the more predominant splicing pattern drawn below each allele in a darker line. See Supplementary Figure 1 for detailed analysis. The mating schemes for BcorNeo and BcorGt mouse lines are depicted below the genomic alterations. (B) BcorNeo and BcorGt chimeric mice and BcorNeo/+ mice display similar phenotypes. Coat color contribution bias (orange dashed outline) and malformed kinked tail phenotype (black arrows) were present in both BcorNeo and BcorGt chimeric mouse lines. BcorNeo/+ offspring of BcorNeo chimeric fathers also displayed the malformed kinked tail phenotype. (C) Western blot analysis of CJ7 (BcorNeo/Y parent), BcorNeo/Y, BcorFl/Y (rescued line), E14 (BcorGt/Y parent) and BcorGt/Y ES cell lines. The top panel was probed with an anti-BCOR antibody and anti-ACTIN antibody was used as a loading control on the bottom panel. BCOR protein is undetectable in BcorNeo/Y ES cells. BcorFl/Y rescued ES cells show wild type levels of BCOR. BcorGt/Y ES cells show a large BCOR-ß-Geo fusion protein with multiple breakdown products. (D) Parent-of-origin effect of BcorNeo allele. Matings between BcorNeo/+ females (offspring of BcorNeo male chimeras) and wild type males do not produce any offspring carrying the BcorNeo allele. In mice, the paternal X chromosome is selectively inactivated in extraembryonic tissue whereas in the embryo proper X inactivation occurs randomly (grey text indicates paternally imprinted X inactivation). The female lethality observed likely results from paternally imprinted X-inactivation of the wild type Bcor allele in extraembryonic tissue.
Molecular analysis of BcorNeo and BcorGt alleles
To establish the molecular nature of the expression defects of the BcorNeo and BcorGt alleles in ES cells, we examined Bcor mRNA splicing patterns by RT-PCR and proteins levels by western blot analysis. The BcorNeo allele ES cells had a mixture of transcripts spliced to exclude Neo (Figure 2A, left upper splicing pattern and Figure S1A), which encode intact BCOR, together with transcripts spliced to incorporate the Neo cassette (Figure 2A, left lower splicing pattern and Figure S1A). Sequencing of RT-PCR products demonstrated that the two regions of the Phosphoglycerate Kinase-Neomycin (Pgk-Neo) cassette were recognized as exons by the splicing machinery (Figure S1B). The first exon was 86 nucleotides from the Pgk promoter and the second utilized a splice acceptor site within the 5′ UTR of the Neo gene cassette and a donor site within the 3′UTR. This latter exon was identical to that used by other previously reported targeted genes [24]. The result of this splicing is the introduction of a premature in frame stop codon derived from the Pgk promoter sequence. Splicing into Neo predominates, as full length BCOR protein levels are not detectable by western blot analysis in the BcorNeo allele ES cells as compared to the parental ES cell line CJ7 (Figure 2C). We conclude that the BcorNeo allele is a severe loss of function allele but because we did detect some correctly spliced transcript it may not be a null allele.
The transcripts from the BcorG t allele are a mixture of those that splice over the gene trap cassette (Figure 2A, right upper splicing panel and Figure S1B) which encode intact BCOR and those that splice into the gene trap ß-Geo cassette, truncating the gene (Figure 2A, right, lower splicing pattern and Figure S1B). By western blot analysis the large BCOR-ß-Geo fusion protein and multiple breakdown products are observed (Figure 2C). Although a faint band corresponding to full length BCOR is present this may represent a co-migrating BCOR-ß-Geo degradation product. The BCOR-ß-Geo fusion protein lacks the region required for interaction with the PcG proteins [4] and therefore is predicted to lack transcriptional repression activity. Although the fusion protein appears to be unstable, the total amount of protein is significantly higher than the level of intact BCOR found in the parental ES cell line E14 (Figure 2C). Consistent with this we found, using quantitative-RT-PCR, that the Bcor-ß-Geo fusion transcript level is 10-fold higher than the level of intact Bcor transcript found in the parental ES cell line. This suggests that intact BCOR negatively autoregulates its own transcription while the BCOR-ß-Geo fusion is unable to function in this capacity. In summary, both the BcorNeo and BcorGt alleles represent severe loss of function mutations and result in kinked and shortened tails and a coat color bias in chimeric animals.
Parent-of-origin effect of BcorNeo allele
To further characterize the effect of these two loss of function mutations in mice we bred the male chimeras to wild type females to obtain germline transmission of the alleles. This was only successful with the BcorNeo allele and we were able to produce BcorNeo/+ females. (Because Bcor is X-linked, BcorNeo/Y males cannot be generated from male chimeras.) Germline transmission of the BcorGt allele did not occur and there are at least two possible explanations for this. First, the BcorGt allele may not be compatible with germline development. Second, since only three BcorGt chimeric mice were obtained and bred, it is possible that upon generation of more chimeras germline transmission would occur. We bred the female BcorNeo/+ mice extensively but, in contrast to the chimeric males, they never produced any BcorNeo/+ offspring (Figure 2A and D). (BcorNeo/Y males, based on the lack of male OFCD patients, are predicted to die during embryonic development.) However, we were able to breed the female BcorNeo/+ mice to FLPe recombinase expressing mice to remove Neo, and got viable BcorFl/+ and BcorFl/Y progeny (Figure 2A). In mice, X chromosome inactivation differs between the embryo and the extraembryonic tissue. The embryo undergoes random X-inactivation, in contrast to the extraembryonic tissue where the paternally-derived X chromosome is inactivated [26]. The female lethality observed, arising when the mutant X-linked allele comes from the mother, likely results from paternally-imprinted X-inactivation of the wild type Bcor allele in extra-embryonic tissue of the female offspring (Figure 2D and see discussion). Thus, our results are consistent with a requirement for Bcor in extraembryonic tissue where Bcor is normally highly expressed [22]. The practical consequence of this parent-of-origin effect was that we could not generate extensive numbers of BcorNeo/+ mice and thus phenotypic analysis was limited to the BcorNeo and BcorGt allele-containing chimeras and the remaining six aging female BcorNeo/+ mice that were generated from the two germline-transmitting male chimeras we obtained.
Effect of BcorNeo allele on eye development
In OFCD congenital cataracts have been described in all patients. We examined the eyes of the remaining BcorNeo/+ mice when they ranged in age from 7 to 22 months. Using a slit lamp, the mice were examined for lens opacification, which is indicative of catararacts. Eight out of twelve eyes (67%) in the six BcorNeo/+ mice displayed lens opacification. Although we do not know the age of onset, two of BcorNeo/+ mice that were 7 months of age had bilateral lens opacification. We did not have aged matched C57Bl/6∶129 mixed background control animals. However, the frequency of cataracts in the BcorNeo/+ mice was substantially higher than that reported for either C57Bl/6 or 129 inbred stains (C57Bl/6 is 25% at 14 months [27] and 129 0% at 13–17 months, as detected by slit lamp analysis [28]). Over seventy percent of OFCD patients have microphthalmia. By visual inspection we only observed one BcorNeo allele-containing chimera with obvious microphthalmia. However, we did not perform postmortem globe measurements so we cannot rule out the possibility that additional mice had more subtle microphthalmia.
Analysis of BcorNeo/Y and BcorGt/Y cell contribution to hematopoietic development
In OFCD patients 90–100% of surviving white blood cells show inactivation of the X chromosome carrying the mutant allele of Bcor, indicating that cells expressing the mutant allele of Bcor are at a selective disadvantage [18]. Because the majority of white blood cells are neutrophils and T cells (and to a lesser extent B cells) this strongly suggests that Bcor plays an important role in the development of these hematopoietic lineages.
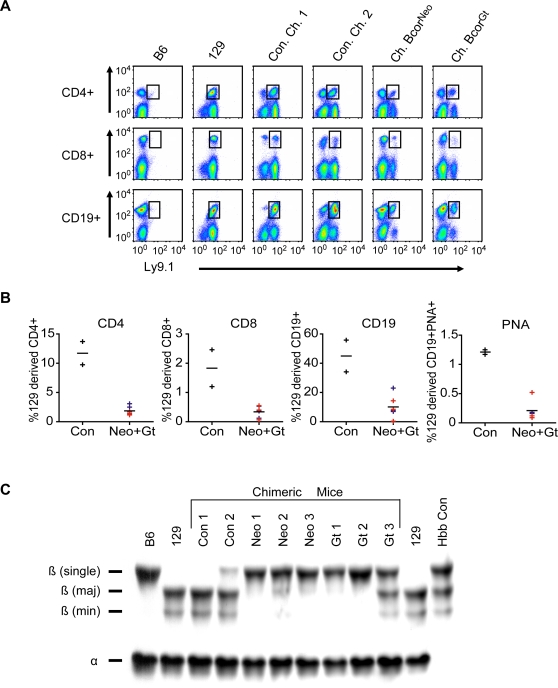
We hypothesized that the loss of function mutations in the BcorNeo/Y and BcorGt/Y ES cell would result in reduced or blocked contribution to hematopoietic lineages in our chimeric mice. The contributions of BcorNeo/Y and BcorGt/Y 129 strain ES cells to splenic B and T cells of 129/B6 chimeric animals were analyzed by fluorescence activated cell sorting (FACS). We used the 129 strain-specific B and T cell surface antigen Ly9.1 [29], [30], the commonly used T-cell markers CD4 and CD8, and the B-cell marker CD19. We found that Ly9.1 positive cells can contribute to CD4, CD8 and C19 positive hematopoietic populations in BcorNeo/Y and BcorGt/Y chimeric mice (Figure 3A and B). The 129 contribution to each hematopoietic cell type tested was reduced relative to two control chimeras in three BcorNeo/Y and three BcorGt/Y chimeric mice. CD4+, CD8+ and CD19+ were reduced compared to control chimeras on average by 87%, 86% and 85%, respectively. Thus, the BcorNeo/Y and BcorGt/Y ES cells can contribute to the B and T lineages but they appear to be at a selective disadvantage, as in OFCD patients, suggesting that Bcor is important for murine B and T cell development.
Figure 3. Analyses of BcorNeo/Y and BcorGt/Y cell contribution to hematopoietic development in chimeric mice.
(A) The BcorNeo/Y and BcorGt/Y 129 strain derived ES cells can contribute to the B and T lineages. White blood cells from the spleens of BcorNeo/Y and BcorGt/Y and appropriate controls were stained with antibodies to the 129 strain specific surface antigen Ly9.1 and the T cell markers CD4 and CD8 and B cell marker CD19. Cells were analyzed by flow cytometry and results are displayed as density plots with black boxes gating T or B cell populations of 129 origin. B6 and 129 strain wild type mice control for Ly9.1 specificity. CJ7 and E14 129 mouse strain derived ES cells were used to generate control chimera 1 (Con. Ch. 1) and control chimera 2 (Con. Ch. 2). Flow cytometry analyses were performed on chimeric control mice and on three mice for each mutant chimeric line, of which an example scatter plot is displayed for each. (B) Compared to chimeric controls, BcorNeo/Y and BcorGt/Y 129 strain derived ES cell contribution to B and T cells is reduced. Graphs of scatter plot data show 129 strain contributions to each hematopoietic lineage. Black bars indicate the average contribution of control chimeric mice 1 and 2 (Con, black +) and the combined average of three BcorNeo (Neo, red +) and three BcorGt (Gt, blue +) mice. Contribution to germinal center B cells is also reduced compared to controls as indicated by the final plot showing the PNA+ (peanut agglutinin; germinal center marker) percentage of CD19+ 129 derived B cells. P values were ≤0.05 for all hematopoietic cell populations tested (CD4; 0.004, CD8; 0.034, CD19; 0.023 and PNA; 0.001). (C) BcorNeo/Y and BcorGt/Y ES cells can contribute to adult erythrocytes in chimeric mice but contribution is reduced in comparison to controls. Coomassie stained gel of hemolysates from erythrocytes of mice described above in panels A and B. BcorNeo and BcorGt chimeric mice (Neo 1, 2 and 3 and Gt 1, 2 and 3) primarily express B6 derived ß -single globin where as chimeric control mice 1 and 2 primarily express 129 derived ß -major and ß -minor globin. The hemoglobin control lane (Hbb Con) was prepared from samples that contain ß -single, ß -major and ß -minor globin.
Because BCOR is a known corepressor of BCL6, an important regulator of germinal center formation in B-cell maturation, we hypothesized that BcorNeo/Y and BcorGt/Y cell contribution to germinal center formation might be blocked or reduced in chimeras. To test this, we looked for the presence of PNA, a germinal center marker, in the CD19+ and Ly9.1+ cell population in cells harvested from the spleens of both mutant chimeric lines. Similar to the B and T cell analysis, we observed an 87% reduction compared to control chimeras in BcorNeo/Y and BcorGt/Y contribution to CD19+ PNA+ cells suggesting a compromised germinal center reaction (Figure 3B). To assess the ability of BcorGt/Y and BcorNeo/Y ES cells to contribute to adult erythrocyte lineages in chimeras, a hemoglobin analysis was performed, relying on genotype differences between host and ES-derived cells. Erythrocytes derived from the 129 genetic background express ß-major and ß-minor globin, while B6 erythrocytes express ß-single globin [31], [32]. We found that BcorNeo/Y and BcorGt/Y ES cells can contribute to mature red blood cells but the contribution is severely reduced compared to the controls (Figure 3C).
Analysis of in vitro differentiation potential of BcorNeo/Y and BcorGt/Y ES cells
The absence of OFCD males indicates an early and essential role for Bcor in human development. The parent-of-origin effect strongly suggests that Bcor plays an essential role in extraembryonic tissue but Bcor may play additional roles earlier in development. We used our two loss of function ES cell lines in in vitro differentiation assays to test whether Bcor plays a role in ES cell differentiation. ES cells cultured in suspension in the absence of leukemia inhibitory factor (LIF) undergo primary differentiation after 2 to 4 days to form simple embryoid bodies (EBs) [33], [34]. These contain an outer endoderm layer surrounding an inner cell mass. Around day 4, differentiation of columnar epithelium with a basal lamina and formation of a central cavity occurs. Such cystic EBs bear similarities to the egg cylinder-stage embryos [35]–[37]. By 6 days of differentiation, EBs are comparable to early organogenesis-stage embryos (E7.5) [38]. EB cells can be dispersed and induced with appropriate growth conditions to undergo secondary differentiation into particular cell lineages [34], [39].
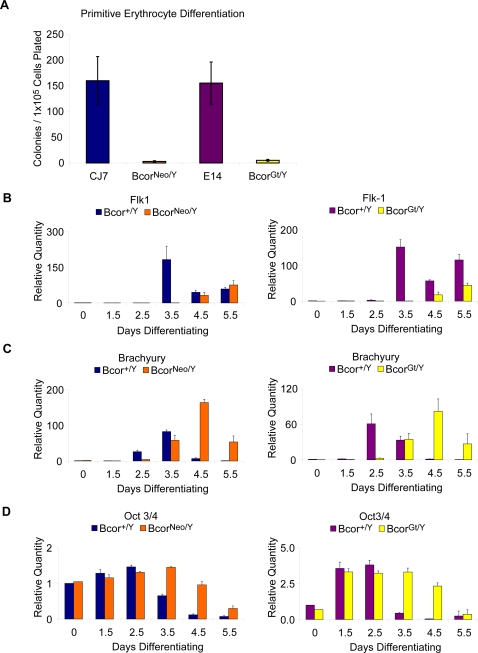
Both the severe skewing of X-inactivation seen in OFCD patients and the limited contribution of Bcor mutant ES cells to hematopoietic lineages in our chimeric animals indicate that Bcor plays a role in hematopoiesis. Primitive erythrocytes are the earliest hematopoietic cell types to form in vivo [34], [40]. Therefore, to determine whether Bcor plays an early role in hematopoiesis, we first examined the ability of dispersed embryoid bodies from our BcorNeo/Y and BcorGt/Y ES cell lines to differentiate into primitive erythrocyte (EryP) colonies in the presence of appropriate factors [41], [42]. We found EryP colony formation with CJ7 derived BcorNeo/Y and E14 derived BcorGt/Y cell lines is reduced relative to the matched parental ES cell lines (98% and 97% respectively, Figure 4A).
Figure 4. In vitro differentiation of BcorNeo/Y and BcorGt/Y ES cell lines.
(A) BcorNeo/Y and BcorGt/Y ES cell lines display a reduced ability to form primitive erythrocyte colonies in comparison to control CJ7 and E14 parental lines. ES cell were differentiated into embryoid bodies for 4.5 days, dispersed and replated in methylcellulose primitive erythrocyte colony forming assays. Error bars represent the standard error of the mean for each ES cell line differentiated (CJ7 and BcorNeo/Y; n = 3, E14 and BcorGt/Y; n = 5). (B–D) qRT-PCR analyses of gene expression were performed on embryoid body differentiations (0–5.5 days) of BcorNeo/Y and BcorGt/Y ES cell lines and respective parental control lines CJ7 and E14 (Bcor+/Y). Differentiations were performed in triplicate excluding the day 0 time point. Error bars represent the standard deviation within each time point of the samples analyzed. (B) The expression of hematopoietic lineage marker, Flk1, is delayed approximately 1 day in BcorNeo/Y and BcorGt/Y differentiating embryoid bodies. (C) The expression of mesoderm and early hematopoietic lineage marker Brachyury is similarly delayed 1–2 days in BcorNeo/Y and BcorGt/Y differentiating embryoid bodies. (D) The repression of pluripotency marker, Oct3/4, is delayed 1–2 days in BcorNeo/Y and BcorGt/Y differentiating embryoid bodies.
We next used quantitative real-time polymerase chain reaction (qRT-PCR) to examine expression of Flk1 (KDR), an early marker of hematopoietic differentiation potential that is required for primitive and definitive hematopoiesis [43], [44]. RNA samples were collected from ES cells that were allowed to differentiate into embryoid bodies for 0 to 5.5 days. We found that the induction of Flk1 expression in BcorNeo/Y and BcorGt/Y cell lines is delayed by one day relative to the parental cell lines and in the time course of the experiment Flk1 expression only reaches ∼40% of the peak expression seen in the parental cell lines (Figure 4B). Moving back in developmental time, we examined expression of Brachyury (T), a marker for the entire mesodermal lineage which in combination with Flk1 marks the hemangioblast cell population [43]. We saw a delay in expression of about 1–2 days but expression does eventually peak at levels comparable to the parental ES cell lines (Figure 4C). Finally, knowing that Bcor is expressed in ES cells, we examined the expression of Oct3/4 (Pou5f1), a marker of ES cell pluripotency [45], [46], and found a one to two day delay in repression of Oct3/4 (Figure 4D).
To confirm that these effects on ES cell differentiation are the result of the Neo insertion in Bcor, we removed the Neo cassette from the BcorNeo ES cell line. We generated three independent clonal BcorFl/Y ES cell lines via expression of Flpo recombinase [47] in BcorNeo ES cells. We found that these three BcorFl/Y “rescued” lines restore the timing of expression of various ES cell and differentiation markers to that of the parental wild type ES cells (Figure 5) demonstrating that the effects on differentiation were due to the interruption of Bcor by the Neo cassette. Included in the analysis were a second ES cell marker, Nanog, the primitive ectoderm marker, Fgf5, [48], [49] and the extraembryonic ectoderm marker, Bmp4 [35], [50], [51]. In the BcorNeo cells we observed a delay in the downregulation of the ES cell markers Oct3/4 and Nanog, whereas Fgf5 was still up regulated appropriately but its down regulation was delayed by two days. Subsequently, activation of Bmp4, Brachyury and Flk1 were all delayed by one to two days. For the genes that are normally expressed earlier in differentiation, the rescued lines restore both the timing and the level of gene expression to that of the parental cell line. However with Brachyury and Flk1, even though the normal timing of gene expression was restored, inexplicably the level was increased beyond the parental line. We also compared the proliferation rates and cell cycle profile of the parental CJ7 ES cell line with the BcorNeo/Y and the three rescued BcorFl/Y ES cell lines and detected no significant differences (data not shown).
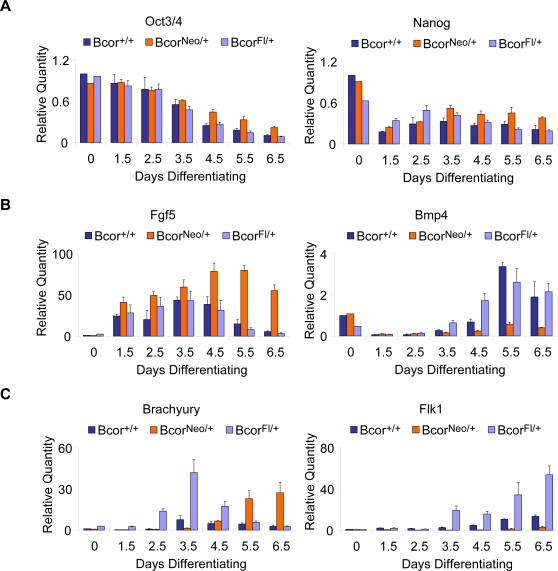
Figure 5. In vitro differentiation of BcorFl/Y “rescued” ES cell lines.
(A–C) BcorFl/Y “rescued” ES cell lines were created through removal of the Neo selection cassette via Flpo recombinase in vitro. BcorFl/Y ES cells were clonally selected, screened for Neo removal and assessed for rescue of BCOR protein expression by western blot (Figure 2C). qRT-PCR analyses of gene expression during embryoid body differentiation (0–6.5 days) of Bcor+/Y (CJ7, wild type parent line), BcorNeo/Y and three independent BcorFl/Y “rescued” ES cell lines. Differentiations were performed in triplicate excluding the day 0 time point. BcorFl/Y results are the average of 3 independent BcorFl/Y “rescued” lines each differentiated in triplicate. (A) The repression of pluripotency markers, Oct3/4 and Nanog, is restored in BcorFl/Y differentiating embryoid bodies in comparison to BcorNeo/Y. (B) Repression of the primitive ectoderm marker, Fgf5, and expression of extraembryonic ectoderm marker, Bmp4, is restored in BcorFl/Y differentiating embryoid bodies. (C) Timing of expression of the early mesoderm and hematopoietic lineage markers, Brachyury and Flk1, is restored in BcorFl/Y differentiating embryoid bodies; however expression levels are restored to a higher magnitude with respect to Bcor+/Y controls (CJ7, wild type parent line).
Together these ES cell differentiation and expression studies indicate that Bcor plays a role in the regulation of gene expression very early in ES cell differentiation and in the mesodermal and ectodermal lineages.
Discussion
In this study we have used a combination of mouse molecular genetics and ES cell differentiation to identify roles of Bcor in early development. These studies provide insights into the possible causes of male embryonic lethality in OFCD, the hematopoietic phenotype of female OFCD patients and the parent-of-origin effect in mice. We have two major findings, as discussed below.
First, we found that in mice the mutant BcorNeo allele exhibits a parent-of-origin effect, which together with the high level of Bcor expression in extraembryonic tissue [22] strongly suggest an essential requirement for Bcor in extraembryonic tissue. The X chromosome has been implicated in causing several malformations of the placenta [52] and our work suggests that Bcor is likely to be one of the genes responsible.
There are two alternative but less likely explanations for the parent-of-origin effect that we have considered. First, it is possible that Bcor is required very early in development (beginning as early as the 4 to 8 cell stage) when the whole embryo is subject paternal X chromosome inactivation. Once the inner cell mass has formed the cells of the embryo proper are subject to random X inactivation. Paternal X-inactivation is maintained and becomes more complete in the trophectoderm and primitive endoderm, which go on to form to the extraembryonic tissue. However, given the incomplete nature of the paternal imprinting during the 4 cell to blastocyst stage it is most likely that sufficient levels of wild type Bcor are transcribed to support development.
A second, albeit unlikely, alternative explanation for the parent-of-origin effect observed in the mouse is that Bcor is imprinted such that only the maternal allele is expressed in the embryo proper. However, if such imprinting [53]–[55] occurred in humans, the mother to daughter transmission seen in OFCD patients [56], [57] could not take place since the daughter would only express the mutant Bcor allele and would be equivalent to the inviable hemizygous males.
Our second major finding was that in ES cell differentiation studies Bcor is required for formation of primitive erythrocytes and proper expression of genes that regulate ES cell pluripotency and genes that drive ectodermal and mesodermal development. These defects together with the apparent limited contribution of BcorNeo/Y and BcorGt/Y cells to B and T cells and adult erythrocytes may provide an explanation for the severe skewing of X-inactivation seen in peripheral white blood cells of OFCD patients and indicate a critical early role for Bcor in helping to establish hematopoietic development. In data not included, we do not see significant changes in expression of the few endoderm specific genes we tested. Since the number of genes tested was not exhaustive, Bcor may be affecting the expression of other genes important for endoderm development that are yet unidentified.
One of the genes that we found to be misregulated in BcorNeo/Y and BcorGt/Y ES cell differentiation assays was Brachyury. Mice heterozygous for the Brachyury T mutation exhibit a variable short-tailed phenotype [58] while other mutations such as Brachyury T137 have a kinked or bent tail [59]. Thus mis-expression of Brachyury is a possible explanation for the kinked and shortened tails we observed in BcorNeo/Y and BcorGt/Y chimeras. The extraembryonic ectoderm marker, Bmp4, was also misregulated in BcorNeo/Y ES cell differentiation assays. Bmp4 knock out mice do not express Brachyury and display impaired mesoderm differentiation, including reduced extraembryonic mesoderm blood island formation and disorganized posterior structures, further implicating Bcor in mesoderm development [49].
Polycomb group proteins (PcG) have been shown to play a central role in stem cell maintenance and lineage specification [60]. The BCOR complex contains several PcG proteins, including RNF2, an H2A ubiquitin ligase, and NSPC1, a BMI1 homolog [4]. RNF2 has been shown to be present at over 1,200 targets in ES cells [61]. At many genes, the PcG complex, PRC2, serves to recruit RNF2 containing complexes. However, about one quarter of ES cell RNF2 targets are not co-occupied with PRC2 components, suggesting RNF2 can be recruited via other means. We hypothesize that BCOR, via interaction with site specific transcription factors, can recruit RNF2 and other BCOR complex components to regulate expression of genes in ES cells and their differentiating progeny. Recently, NSPC1 was identified as a protein that when over-expressed can partially rescue the phenotypic properties of undifferentiated mouse ES cells under differentiation-inducing conditions [62]. Over-expression of NSPC1 may be sequestering or diluting the activity of the Bcor complex thereby preventing full transcriptional repression of target genes. This, together with our data, suggests that inappropriate expression of BCOR complex components affects ES cell differentiation.
Our studies suggest two possible causes of male embryonic lethality in OFCD. First, inappropriate regulation of key developmental genes in Bcor−/Y embryos may cause lethality before implantation. Second, male embryonic lethality may be due to improper development of extraembryonic derived tissue, potentially resulting in placental failure or incomplete chorioallantoic fusion. Conditional inactivation of Bcor in a spatial and temporal manner will be critical in future experiments in order to bypass the strict requirement for Bcor function in early development.
Materials and Methods
Animals
All experimental protocols involving mice described in this publication have been approved by the University of Minnesota Institutional Animal Care and Use Committee. The mice were not kept in specific pathogen free (SPF) rodent housing, thus they were exposed to multiple pathogens that are present at sub clinical levels in conventional rodent housing. Due to continual pathogen exposure, PNA+ CD19+ germinal center positive cell populations were already present in our mice thus, we did not specifically induce germinal center reactions through immunization.
Generation of Bcor mutant mice
A mouse Bcor cDNA fragment containing sequences from within exon 4 to within exon 7 was used to screen the RPCI-22 129S6/Sv EvTac Bac library (Stratagene) [63], and positive BAC clones were used to clone Bcor genomic sequence. The left and right homology arms were cloned into the backbone vector pDZ157 [64]. The final targeting construct is diagrammed in Figure 1A. The Bcor targeting vector was linearized with PmeI and electroporated into CJ7 ES cells (originally derived from the 129S1 strain). One homologous recombinant was identified from 768 G418-resistant colonies by Southern hybridization using a DNA probe from sequences upstream of the targeting vector to screen genomic DNA digested with BamH1. Homologous recombination was confirmed on both ends of the targeted region by Southern hybridization. Probes for Southern hybridization were generated from a BAC clone containing Bcor genomic sequence by PCR using primers VBp525/VBp526 (5′ probe, left arm) and VBp582/ VBp583 (3′ probe, right arm). The targeted ES cell clone containing the BcorNeo allele was injected into C57Bl/6 blastocysts to generate chimeras. Chimeric males were bred with C57Bl/6 females to generate heterozygotes carrying the BcorNeo allele. BcorNeo /+ females were bred with male β-actin-Flpe transgenic mice to delete the frted sequence and generate heterozygotes carrying the BcorFl allele. BcorFl/+ females were bred with male β-actin-Cre transgenic mice to delete the floxed sequence and generate heterozygous BcorΔ3/+ and hemizygous BcorΔ3/Y deletion mutants.
VBp525 5′-CTCTACTTGCTCAGTCTGCCTGCAATG-3′
VBp526 5′-AAGTCGACACATTTCCTTTGTTAGCAG-3′
VBp582 5′-GAGTTGTATCTCATAAATTGTGGTTG-3′
VBp583 5′-CTGTCATTCACTTTGAGCCTGGTGT-3′
The XE541 ES cell line was created by the BayGenomics genetrap consortium through insertional genetrap mutagenesis of the wild type E14 ES cell line (originally derived from the 129P2 strain). The genetrap mutation resides in the sixth intron of the Bcor gene and results in a loss of function allele we named BcorGt. XE541 ES cells harboring the BcorGt allele were injected into C57Bl/6 blastocysts by the University of Minnesota Mouse Genetics Laboratory to generate chimeric mice.
Genotyping
The Bcor+, BcorFl and Bcor Δ3 alleles were detected by polymerase chain reaction (PCR) from tail clip genomic DNA using primer set VBp573 /VBp981. The described PCR generates an 1122 base pair (bp) amplicon for the BcorFl, a 993 bp amplicon for the Bcor+ allele and a 332 bp amplicon for the Bcor Δ3. The BcorNeo allele was detected by separate PCR reaction using primer set, VBp1023/VBp981 and generates a 693 bp amplicon.
VBp573 5′-GCCTGAAGTAGCTGACATGTCTCTGAT-3′
VBp981 5′-AAAGCCCTAGGAACTACTTGGAGGC-3′
VBp1023 5′-CTATCGCCTTCTTGACGAGTTCTTC-3′
Western Blot Analyses
In both western blot analyses performed, mouse eyes or ES cells were incubated in lysis buffer (1× phosphate buffered saline (PBS), 10% glycerol, 0.5% Nonidet-P40, 2 mM dithiothreitol (DTT), 0.2 mM phenylmethanesulphonylfluoride (PMSF) and 1× Complete Protease Inhibitor (Roche)), sonicated, normalized by Bradford Assay and resolved on a NuPAGE 3–8% Tris-Acetate gel (Invitrogen). Proteins were transferred overnight at 4°C to a nitrocellulose membrane, blocked with non-fat dry milk and incubated with polyclonal anti-BCOR [4] and monoclonal anti-B-Actin antibodies (Abcam 8226).
Generation of “Rescued” BcorFl/+ Embryonic Stem Cells
To remove the Neo selection cassette and restore BCOR expression, BcorNeo /Y ES cells were transfected with a mouse codon-optimized FLPo site specific recombinase as described by Raymond and Soriano [47]. Five clonally selected rescued lines in which the Neo cassette had been deleted were identified by PCR screening of gDNA with primer sets VBp573 /VBp981 and VBp1023/VBp981.
Flow Cytometry and Hemoglobin Analyses
White blood cells were harvested into Hank's buffered saline solution (HBSS) at 4°C from the spleens of sacrificed BcorNeo, BcorGt and appropriate control mice. Cells were pelleted at 300×g at 4°C for this step and all subsequent centrifugation steps. Red blood cells were lysed and white blood cells were then pelleted and resuspended in a 1∶3 dilution of clone 2.4G2 hybridoma (ATCC HB-197) supernatant for 10 minutes at room temperature to block Fc receptors. Cells were then counted and 2×106 cells were respuspended in 200 uL of FACs buffer (PBS, 1% FCS, and 0.02% azide, pH 7.2) in a 96 well round bottom plate for antibody staining. Cells were stained with a 1∶200 dilution of appropriate primary antibody or lectin (Anti-Ly9.1-FITC (BD Pharmingen), Anti-CD19-PE (eBioscience), Anti-CD4-PerCP (BD Pharmingen), Anti-CD8-APC (eBioscience) and PNA-biotin (BD Pharmingen)) in FACs buffer for 20 minutes at room temperature and washed 1 time with 200 uL of FACs buffer. Cells incubated with PNA-biotin were then incubated with streptavidin-APC for 20 minutes at room temperature. All samples were then washed another 2 times with FACs buffer, fixed in 1% paraformaldehyde and stored at 4°C until flow cytometry was performed on the following day. Cell events were recorded on a flow cytometer (FACSCalibur; BD Biosciences) and analyzed with FlowJo software (Tree Star Inc.). Clarified hemolysates prepared from the erythrocytes of the same mice were resolved by Triton-acid-urea gel electrophoresis and visualized by Coomassie blue staining [32], [65]
ES Cell Culture and Differentiation
ES cells were maintained in 37°C 5% CO2 incubator on irradiated mouse embryonic fibroblasts (MEFs) in Knock Out Dulbecco's Modified Eagles Medium (KO-DMEM, Invitrogen) supplemented with (15% ES cell certified fetal bovine serum (ES-FBS, Hyclone), 0.1 mM non-essential amino acids (NEAA), 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol (BME), 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen) and 1000 U/mL ESGRO (Chemicon)). Protocols for differentiation of ES cells were essentially as described by Zhang et. al. [41], [42] with minor modifications. Briefly, ES cells were passaged one time onto 0.1% gelatinized tissue culture plates to remove MEFs. After 2 days, cells were trypsinized for 5 minutes to a single cell suspension, stopped with fetal bovine serum, counted and plated at low density (5–10×104 cells/mL depending on the experiment) in embryoid body (EB) differentiation media (Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 15% fetal bovine serum (Atlas Biologicals), 50 ug/mL Ascorbic Acid (Sigma), 2 mM L-glutamine, 4.5×10−4 M monothioglycerol (MTG) in biological triplicate for each sample on ultra low attachment tissue culture plates (Corning)). EBs were maintained at 37°C 5% CO2 for duration of indicated experiments. Primitive erythrocyte colony forming assays utilized EBs, as described above, incubated until day 4.5. EBs were then trypsinized to a single cells suspension, stopped with FBS, counted and replated at 5×103 cells/mL in triplicate in primitive erythrocyte differentiation media containing (IMDM, 1% methylcellulose (Sigma), 10% plasma derived serum (Animal Technologies Inc.), 12.5 ug/mL Ascorbic Acid (Sigma), 2 mM L-glutamine (Invitrogen), 200 ug/mL transferrin (Roche), 4.5×10−4 M MTG (Sigma), 2 U/mL Erythropoietin (Epogen, Roche) and 5% protein free hybridoma media –II (Invitrogen). On day 4 secondary differentiation plates were counted for the presence of primitive erythrocyte colonies based on morphology and color.
Gene Expression Analysis
Total RNA was extracted with TRIzol (Invitrogen), normalized by spectroscopy and reverse transcribed using either M-MLV or SuperScript III (Invitrogen) according to the manufacturer's protocol. Real-time quantitative PCR was performed using FastStart SYBR Green Master mix (Roche) on a Stratagene M×3000P (Stratagene). Gene expression was determined using the relative quantitation method [66] and Hprt expression was used to normalize all sample template concentrations. Denaturing curves were performed on all reactions to verify homogeneity of the amplified product. The following gene specific primers were used.
Hprt For 5′-AGCTACTGTAATGATCAGTCAACG-3′
Hprt Rev 5′-AGAGGTCCTTTTCACCAGCA-3′
Oct4 For 5′-GAAGCAGAAGAGGATCACCTTG-3′
Oct4 Rev 5′-TTCTTAAGGCTGAGCTGCAAG-3′
Nanog For 5′-CCTCAGCCTCCAGCAGATGC-3′
Nanog Rev 5′-CCGCTTGCACTTCATCCTTTG-3′
Brachyury For 5′-CTCACCAACAAGCTCAATGG-3′
Brachyury Rev 5′-GGTCTCGGGAAAGCAGTGGC-3′
Flk1 For 5′-CACCTGGCACTCTCCACCTTC-3′
Flk1 Rev 5′-GATTTCATCCCACTACCGAAAG-3′
Fgf5 For 5′-CAAAGTCAATGGCTCCCACGAAG-3′
Fgf5 Rev 5′-CTACAATCCCCTGAGACACAGCAAATA-3′
Bmp4 For 5′-CACTGTGAGGAGTTTCCATCACGAAG-3′
Bmp4 Rev 5′-GGATGCTGCTGAGGTTGAAGAGGA-3′
Statistical analysis
P values in Figure 3 were determined using a Student's t-test, using the natural log of the percent contribution values, assuming unequal variances.
Supporting Information
Analysis of splicing pattern in BcorNeo/Y and BcorGt/Y ES cells. (A) Clone 2B1 ES cells, containing the BcorNeo/Y allele, aberrantly splice two portions of the Pgk-Neo coding sequence into the Bcor transcript. S1–S4 shows the sequence surrounding the splice junctions as determined by sequencing of reverse transcription PCR products generated by primers A–D. Amplicon AB and CD were sequenced to determine splice junctions S1–S3 and S4, respectively. The predominant splice pattern found is indicated by bolded black splice connector lines. VBp1114 (A) ATGCTTTCTGCAACCCCTCTGTAT, VBpNeoRev (B) TCGGCAGGAGCAAGGTGAGAT, VBpNeoFor (C) CCGGTTCTTTTTGTCAAGACCG, VBp1116 (D) TTGTATCCCAGGCGGTGTTTTG. (B) Clone XE541 ES cells, containing the BcorGt/Y allele, predominantly splice from exon 6 of Bcor into the splice acceptor of the genetrap cassette. Reverse transcription PCR of XE541 ES cell total RNA using primers F and G, generates the expected amplicon of 529 base pairs. Wild type splicing can also be detected in XE541 ES cells as shown by reverse transcription PCR using primers E and H to generate the expected amplicon of 445 base pairs. The predominant splice pattern found is indicated by bolded black splice connector lines. VBp1616 (E) CGTGCAATGATGCGCTTCTC, VBp1074 (F) AGATTCCAGTCAGCTCAGCCGAGA, VBp1071 (G) ATTCAGGCTGCGCAACTGTTGGG, VBp1617 (H) CTTTGGAGATCCGTCTTCGCTT.
(0.97 MB DOC)
Acknowledgments
We thank members of the Zarkower and Bardwell labs for many helpful discussions; Anna Petryk and Lisa Schimmenti for help in initially characterizing our chimeric mice; Erick Mendenhall and Meri Firpo for insights into differentiating mouse embryonic stem cells; Laura Bursch, Troy Baldwin and Keli Hippen for help with immunological analyses; and Eric Russel and Zhennig He for help with hemoglobin analyses. We thank the University of Minnesota Mouse Genetics Laboratory for ES cell injection.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this work was provided by NCI (R01 CA071540), NIH (T32DE07288) and the University of Minnesota Cancer Center. The funding agencies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 2.Choi Y, Elagib KE, Goldfarb AN. AML-1-ETO-Mediated erythroid inhibition: new paradigms for differentiation blockade by a leukemic fusion protein. Crit Rev Eukaryot Gene Expr. 2005;15:207–216. doi: 10.1615/critreveukargeneexpr.v15.i3.30. [DOI] [PubMed] [Google Scholar]
- 3.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 4.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, et al. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichii H, Sakamoto A, Arima M, Hatano M, Kuroda Y, et al. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int Immunol. 2007;19:427–433. doi: 10.1093/intimm/dxm007. [DOI] [PubMed] [Google Scholar]
- 9.Jardin F, Ruminy P, Bastard C, Tilly H. The BCL6 proto-oncogene: a leading role during germinal center development and lymphomagenesis. Pathol Biol (Paris) 2007;55:73–83. doi: 10.1016/j.patbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 11.Asari S, Sakamoto A, Okada S, Ohkubo Y, Arima M, et al. Abnormal erythroid differentiation in neonatal bcl-6-deficient mice. Exp Hematol. 2005;33:26–34. doi: 10.1016/j.exphem.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Kojima S, Hatano M, Okada S, Fukuda T, Toyama Y, et al. Testicular germ cell apoptosis in Bcl6-deficient mice. Development. 2001;128:57–65. doi: 10.1242/dev.128.1.57. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Bereschenko O, Niu H, Klein U, Basso K, et al. Molecular pathogenesis of non-Hodgkin's lymphoma: the role of Bcl-6. Leuk Lymphoma. 2003;44:S5–12. doi: 10.1080/10428190310001621588. [DOI] [PubMed] [Google Scholar]
- 14.Staudt LM, Dent AL, Shaffer AL, Yu X. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int Rev Immunol. 1999;18:381–403. doi: 10.3109/08830189909088490. [DOI] [PubMed] [Google Scholar]
- 15.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan RS, de Erkenez AC, Hemenway CS. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene. 2003;22:3395–3406. doi: 10.1038/sj.onc.1206361. [DOI] [PubMed] [Google Scholar]
- 17.Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, et al. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 19.Hilton E, Black GC, Bardwell VJ. The BCL-6 corepressor (BCOR) and Oculofaciocardiodental syndrome. In: Epstein C, editor. Inborn Errors of Development: Oxford University Press; 2008. In press. [Google Scholar]
- 20.Ng D, Hadley DW, Tifft CJ, Biesecker LG. Genetic heterogeneity of syndromic X-linked recessive microphthalmia-anophthalmia: is Lenz microphthalmia a single disorder? Am J Med Genet. 2002;110:308–314. doi: 10.1002/ajmg.10484. [DOI] [PubMed] [Google Scholar]
- 21.Hilton EN, Manson FD, Urquhart JE, Johnston JJ, Slavotinek AM, et al. Left-sided embryonic expression of the BCL-6 corepressor, BCOR, is required for vertebrate laterality determination. Hum Mol Genet. 2007;16:1773–1782. doi: 10.1093/hmg/ddm125. [DOI] [PubMed] [Google Scholar]
- 22.Wamstad JA, Bardwell VJ. Characterization of Bcor expression in mouse development. Gene Expr Patterns. 2007;7:550–557. doi: 10.1016/j.modgep.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Moens C, Ivanyi E, Pawling J, Gertsenstein M, et al. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol. 1998;8:661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- 25.Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, et al. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proc Natl Acad Sci U S A. 2005;102:12843–12848. doi: 10.1073/pnas.0501963102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 27.Wolf NS, Li Y, Pendergrass W, Schmeider C, Turturro A. Normal mouse and rat strains as models for age-related cataract and the effect of caloric restriction on its development. Exp Eye Res. 2000;70:683–692. doi: 10.1006/exer.2000.0835. [DOI] [PubMed] [Google Scholar]
- 28.Newkirk KM, Chandler HL, Parent AE, Young DC, Colitz CM, et al. Ultraviolet radiation-induced corneal degeneration in 129 mice. Toxicol Pathol. 2007;35:819–826. doi: 10.1080/01926230701584197. [DOI] [PubMed] [Google Scholar]
- 29.Ledbetter JA, Goding JW, Tsu TT, Herzenberg LA. A new mouse lymphoid alloantigen (Lgp100) recognized by a monoclonal rat antibody. Immunogenetics. 1979;8:347–360. [Google Scholar]
- 30.Tovar V, de la Fuente MA, Pizcueta P, Bosch J, Engel P. Gene structure of the mouse leukocyte cell surface molecule Ly9. Immunogenetics. 2000;51:788–793. doi: 10.1007/s002510000209. [DOI] [PubMed] [Google Scholar]
- 31.Whitney JB, 3rd, Copland GT, Skow LC, Russell ES. Resolution of products of the duplicated hemoglobin alpha-chain loci by isoelectric focusing. Proc Natl Acad Sci U S A. 1979;76:867–871. doi: 10.1073/pnas.76.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rovera G, Magarian C, Borun TW. Resolution of hemoglobin subunits by electrophoresis in acid urea polyacrylamide gels containing Triton X-100. Anal Biochem. 1978;85:506–518. doi: 10.1016/0003-2697(78)90248-8. [DOI] [PubMed] [Google Scholar]
- 33.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 34.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 35.Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83:279–287. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 36.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc Natl Acad Sci U S A. 1975;72:1441–1445. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leahy A, Xiong JW, Kuhnert F, Stuhlmann H. Use of developmental marker genes to define temporal and spatial patterns of differentiation during embryoid body formation. J Exp Zool. 1999;284:67–81. doi: 10.1002/(sici)1097-010x(19990615)284:1<67::aid-jez10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 40.Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy M, Keller GM. Hematopoietic commitment of ES cells in culture. Methods Enzymol. 2003;365:39–59. doi: 10.1016/s0076-6879(03)65003-2. [DOI] [PubMed] [Google Scholar]
- 42.Zhang WJ, Chung YS, Eades B, Choi K. Gene targeting strategies for the isolation of hematopoietic and endothelial precursors from differentiated ES cells. Methods Enzymol. 2003;365:186–202. doi: 10.1016/s0076-6879(03)65013-5. [DOI] [PubMed] [Google Scholar]
- 43.Park C, Ma YD, Choi K. Evidence for the hemangioblast. Exp Hematol. 2005;33:965–970. doi: 10.1016/j.exphem.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Shalaby F, Ho J, Stanford WL, Fischer KD, Schuh AC, et al. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–990. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 45.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 46.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 47.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 50.Haub O, Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development. 1991;112:397–406. doi: 10.1242/dev.112.2.397. [DOI] [PubMed] [Google Scholar]
- 51.Hebert JM, Boyle M, Martin GR. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- 52.Hemberger M. The role of the X chromosome in mammalian extra embryonic development. Cytogenet Genome Res. 2002;99:210–217. doi: 10.1159/000071595. [DOI] [PubMed] [Google Scholar]
- 53.Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 54.Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 55.Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Hedera P, Gorski JL. Oculo-facio-cardio-dental syndrome: skewed X chromosome inactivation in mother and daughter suggest X-linked dominant Inheritance. Am J Med Genet A. 2003;123:261–266. doi: 10.1002/ajmg.a.20444. [DOI] [PubMed] [Google Scholar]
- 57.McGovern E, Al-Mudaffer M, McMahon C, Brosnahan D, Fleming P, et al. Oculo-facio-cardio-dental syndrome in a mother and daughter. Int J Oral Maxillofac Surg. 2006;35:1060–1062. doi: 10.1016/j.ijom.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Stott D, Kispert A, Herrmann BG. Rescue of the tail defect of Brachyury mice. Genes Dev. 1993;7:197–203. doi: 10.1101/gad.7.2.197. [DOI] [PubMed] [Google Scholar]
- 59.Abe K, Yamamura K, Suzuki M. Molecular and embryological characterization of a new transgene-induced null allele of mouse Brachyury locus. Mamm Genome. 2000;11:238–240. doi: 10.1007/s003350010044. [DOI] [PubMed] [Google Scholar]
- 60.Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 62.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 64.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russell JE, Liebhaber SA. Reversal of lethal alpha- and beta-thalassemias in mice by expression of human embryonic globins. Blood. 1998;92:3057–3063. [PubMed] [Google Scholar]
- 66.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of splicing pattern in BcorNeo/Y and BcorGt/Y ES cells. (A) Clone 2B1 ES cells, containing the BcorNeo/Y allele, aberrantly splice two portions of the Pgk-Neo coding sequence into the Bcor transcript. S1–S4 shows the sequence surrounding the splice junctions as determined by sequencing of reverse transcription PCR products generated by primers A–D. Amplicon AB and CD were sequenced to determine splice junctions S1–S3 and S4, respectively. The predominant splice pattern found is indicated by bolded black splice connector lines. VBp1114 (A) ATGCTTTCTGCAACCCCTCTGTAT, VBpNeoRev (B) TCGGCAGGAGCAAGGTGAGAT, VBpNeoFor (C) CCGGTTCTTTTTGTCAAGACCG, VBp1116 (D) TTGTATCCCAGGCGGTGTTTTG. (B) Clone XE541 ES cells, containing the BcorGt/Y allele, predominantly splice from exon 6 of Bcor into the splice acceptor of the genetrap cassette. Reverse transcription PCR of XE541 ES cell total RNA using primers F and G, generates the expected amplicon of 529 base pairs. Wild type splicing can also be detected in XE541 ES cells as shown by reverse transcription PCR using primers E and H to generate the expected amplicon of 445 base pairs. The predominant splice pattern found is indicated by bolded black splice connector lines. VBp1616 (E) CGTGCAATGATGCGCTTCTC, VBp1074 (F) AGATTCCAGTCAGCTCAGCCGAGA, VBp1071 (G) ATTCAGGCTGCGCAACTGTTGGG, VBp1617 (H) CTTTGGAGATCCGTCTTCGCTT.
(0.97 MB DOC)