Abstract
The identification of the hepatitis C virus (HCV) strain JFH-1 enabled the successful development of infectious cell culture systems. Although this strain replicates efficiently and produces infectious virus in cell culture, the replication capacity and pathogenesis in vivo are still undefined. To assess the in vivo phenotype of the JFH-1 virus, cell culture-generated JFH-1 virus (JFH-1cc) and patient serum from which JFH-1 was isolated were inoculated into chimpanzees. Both animals became HCV RNA-positive 3 days after inoculation, but showed low-level viremia and no evidence of hepatitis. HCV viremia persisted 8 and 34 weeks in JFH-1cc and patient serum-infected chimpanzees, respectively. Immunological analysis revealed that HCV-specific immune responses were similarly induced in both animals. Sequencing of HCV at various time points of infection revealed more substitutions in the patient serum-inoculated chimpanzee and the higher level of sequence variations seemed to be associated with a prolonged infection in this animal. A common mutation G838R in the NS2 region emerged early in both chimpanzees. This mutation enhances viral assembly leading to an increase in viral production in transfected or infected cells.
Conclusion
our study shows that the HCV JFH-1 strain causes attenuated infection and low pathogenicity in chimpanzees, and is capable of adapting in vivo with a unique mutation conferring enhanced replicative phenotype.
Keywords: NS2, in vivo phenotype, virus assembly, T cell response
Introduction
Hepatitis C virus (HCV) infects about 170 million people worldwide and a major causative agent of chronic liver diseases including cirrhosis and hepatocellular carcinoma (1, 2). However, the underlying biological mechanisms of pathogenesis and persistence are still not well understood. No vaccine protecting against HCV infection is currently available (3). Therapy for HCV-related chronic hepatitis remains problematic, with limited efficacy, high cost and substantial adverse effects (1, 4, 5). Understanding the biology of this virus and the development of new therapies has been hampered by a lack of appropriate model systems for replication and infection of this virus.
Recent progress with a unique HCV genotype 2a strain JFH-1, isolated from a case of fulminant hepatitis in Japan, has led to the development of a robust HCV infectious cell culture system (6 - 9). This JFH-1 strain can replicate efficiently, produce the infectious viral particles, and show robust infection in vitro. However, in our previous report, the inoculation of cell culture-generated JFH-1 virus (JFH-1cc) induced only transient and attenuated infection in a chimpanzee (8). The observed low virulence of this strain in vivo was unexpected but consistent with the inverse relationship between in vivo and in vitro properties of cell culture adaptive mutations in the HCV replicon system (10).
In this study, we performed an extensive analysis of the in vivo replication and pathogenicity of the JFH-1 strain by inoculating chimpanzees with JFH-1cc and patient serum from which the JFH-1 strain was isolated. Furthermore we analyze viral sequences during the infection to identify mutations that might represent in vivo adaptive mutations with unique phenotype.
Materials and Methods
Cell culture
Huh7 derivative cell lines Huh7.5 and Huh7.5.1 were provided by Charles Rice (Rockefeller Univ, NY, NY) and Francis Chisari (Scripps Research Institute, La Jolla, CA), respectively (7, 9). The Huh7 derivative clone Huh7-25 that lacks CD81 expression was reported previously (11).
Inocula
The production of JFH-1cc has been reported previously (12). Briefly, the full-length JFH-1 RNA was synthesized by in vitro transcription with linearized pJFH-1 plasmid and MEGAscript kit (Ambion, Austin, TX)(8). Ten μg of full-length JFH-1 RNA was transfected into 3.0 × 106 Huh7 cells by electroporation and the culture medium with JFH-1cc was harvested 5 days after transfection. The culture medium was passed through a 0.45 μm filter unit. The case of fulminant hepatitis C from which the JFH-1 strain was isolated has been reported previously (6). An aliquot of acute phase serum (point A as indicated in the reference 6) was used in this study. To determine the HCV RNA titers in these inocula, total RNA was extracted from 140 μL of these samples by QIAamp Viral RNA Kit (QIAGEN, Valencia, CA) and copy numbers of HCV RNA were determined by real-time quantitative RT-PCR as described previously (13).
Infection study in chimpanzees
Housing, maintenance, and care of the chimpanzees used in this study conformed to the requirement for the humane use of animals in scientific research as defined by the Institutional Animal Care and Use Committee of the Centers for Disease Control and Prevention. Chimpanzee 10273 (CH10273, female, age 5, 20 kg) was inoculated intravenously with 100 μL of serum (9.6 × 106 copies) from the fulminant hepatitis patient mixed with 400 μL of DMEM culture medium. Chimpanzee 10274 (CH10274, female, age 5, 22 kg) was inoculated intravenously with 500 μL of DMEM culture medium containing JFH-1cc (1.4 × 107 copies). Serum and liver biopsy samples of these animals were obtained at baseline and weekly after inoculation.
Measurement of HCV RNA, anti-HCV and ALT
HCV RNA in chimpanzees was quantitatively measured by nested RT-PCR with a sensitivity of detection of approximately 50 IU/mL (COBAS Amplicor; Roche Molecular Systems, Pleasanton, CA) and was quantified using Amplicor Monitor (Roche Molecular Systems). Serum samples were tested for anti-HCV (ORTHO version 3.0 ELISA test system, Ortho-Clinical Diagnostics, Raritan, NJ). Serum alanine aminotransferase (ALT) values in chimpanzee’s sera were established using a commercially available assay kit in accordance with the manufacturer’s instructions (Drew Scientific, Dallas, TX). Cutoff values representing 95% confidence limit for the upper level of normal ALT activity were calculated individually for each chimpanzee using 10 pre-inoculation enzyme values obtained over a period of 4-6 weeks, and were 73 U/L in CH10274 and 76 U/L in CH10273.
HCV sequencing
The total RNA was extracted from 280 μL of chimpanzee sera collected at appropriate time points by use of QIAamp viral RNA kit, and cDNA was synthesized by use of Superscript III (Invitrogen, Carlsbad, CA). The cDNAs were subsequently amplified with TaKaRa LA™ Taq DNA polymerase (Takara Mirus Bio, Madison, WI). Five separate fragments were amplified by nested-PCR covering the almost the entire open reading frame and a part of the 5′UTR of the JFH-1 strain as follows; nt 128 - 1829, nt 1763 - 4381, nt 4278 - 6316, nt 6172 - 7904 and nt 7670 - 9222. The sequence of each amplified fragment was determined directly. The fragment encompassing HVR-1 (nt 128 - 1829) was cloned into the pGEM-T easy vector (Promega, Madison, WI) by TA-cloning and 10 clones from each time point were sequenced.
T-cell Proliferation and IFN-γ ELISpot Assays
The cryopreserved PBMCs were used for immunological analysis. Standard T-cell proliferation assay was performed as described previously (14). Cells were stimulated with recombinant HCV genotype 2a core or NS5a protein (Fitzgerald Industries International, Concord, MA) and pulsed with 3H-thymidine (GE Healthcare BioSciences, Piscataway, NJ). T-cell stimulation was expressed as a stimulation index that was calculated as the ratio of average counts per minute of antigen-stimulated proliferation over average CPM of the medium background. A sample was considered positive when the average stimulation index was greater than 5. The numbers of antigen-specific IFN-γ producing cells were analyzed by ELISpot assay. PBMC were stimulated with recombinant protein antigens (HCV core and NS5a proteins) and HCV overlapping peptide pools (OLPs, 15mers overlapped by 10 aa) from core (38 peptides, aa 1-195) and NS3 (56 peptides, aa 1031-315)(Mimotopes, Raleigh, NC). The NS3 OLPs were divided into two sets. The number of spots was counted by using a computer-assisted AID ELISpot Reader System and AID software version 3.5 (Autoimmune Diagnostika GmbH, Strassberg, Germany). Antigen-specific spot forming units (SFU) were calculated by subtracting the average of background values (4 wells without antigen, typically less than 10 spots) from that of the antigen-stimulated sample. The sample was considered positive when the background-corrected SFU was greater than 10 and ≥ twice the mean SFU of the pre-infection samples in the same animal.
To specifically evaluate the T-cell response against the NS2 region containing the G838R mutation, two peptides of 18 amino acids (NS2-G: ITLFTLTPGYKTLLGQCL and NS2-R: ITLFTLTPRYKTLLGQCL were synthesized (Sigma-Genosys, The Woodlands, TX). PBMCs from both chimpanzees were stimulated with the WT and mutant peptides (2 μg/ml), and analyzed for IFN-γ production by IFN-γ ELISpot assays as described above.
Production of JFH-1 G838R mutant virus
The full genome JFH-1 construct with G838R mutation in NS2 region was generated by site directed mutagenesis. The replication-deficient clone of JFH1 generated by introducing a point mutation into the GDD motif of the NS5B to abolish the RNA dependent RNA polymerase activity was used as a negative control (JFH-1 GND)(8).
Quantification of HCV RNA and HCV core Ag
To determine the amount of HCV, total RNA was extracted with QIAamp Viral RNA Kit from 140 μL of culture medium, or with RNeasy mini kit (QIAGEN, Valencia, CA) from cell pellet. Copy numbers of HCV RNA were determined by real-time quantitative RT-PCR as described. HCV core antigen (Ag) in culture supernatant was quantified by highly sensitive enzyme immunoassay (Ortho HCV core antigen ELISA Kit, Ortho Clinical Diagnostics, Tokyo, Japan)(15). To determine intracellular HCV core Ag, the cell pellet was re-suspended with 100 μl of radioimmune precipitation assay buffer containing 1% sodium dodecyl sulfate, 0.5% NP40, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl and Complete Mini protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN) then sonicated 10 minutes and subjected to the Ortho HCV core antigen ELISA assay following centrifugation.
Titration of HCV infectivity
To assess the intracellular infectivity, cells were harvested by treatment with trypsin-EDTA and pelleted by centrifugation. Cell pellets were resuspended with 500 μL of DMEM with 10% FBS and lysed by four freeze-thaw cycles. The supernatant was collected after centrifugation and passage through a 0.45 μm filter. These cell lysates and culture supernatants were serially diluted 5-fold and inoculated into naïve Huh7.5.1 cells seeded at 1 × 104 cells / well in 96-well flat-bottom plates and assayed for focus-forming units (FFU) by anti-core immunofluorescence as described previously (16).
Statistical analysis
Data from repeated experiments were averaged and expressed as mean ± standard deviation. Statistical analysis was performed using the Mann-Whitney test. P values of less than 0.05 were considered statistically significant.
Results
Clinical, virological and immunological profiles of JFH-1 infected chimpanzees
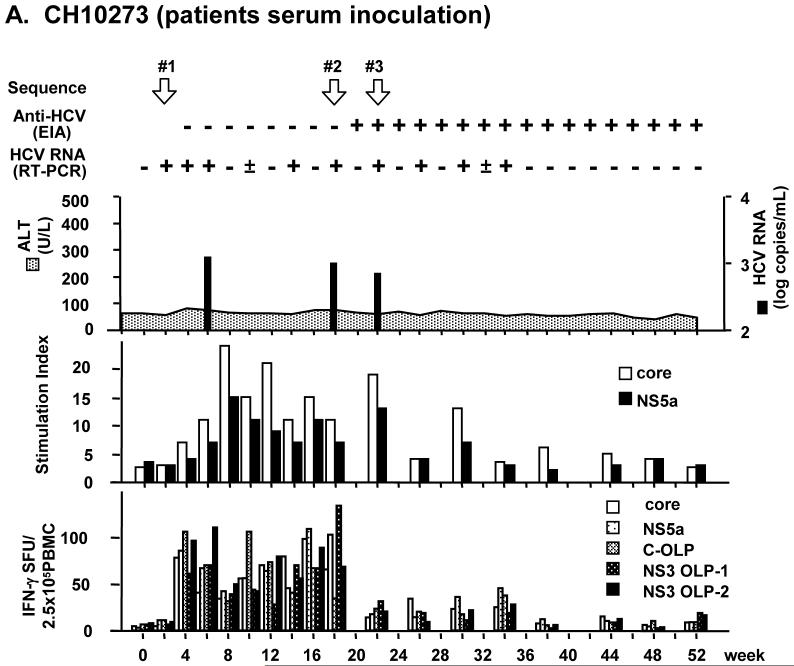
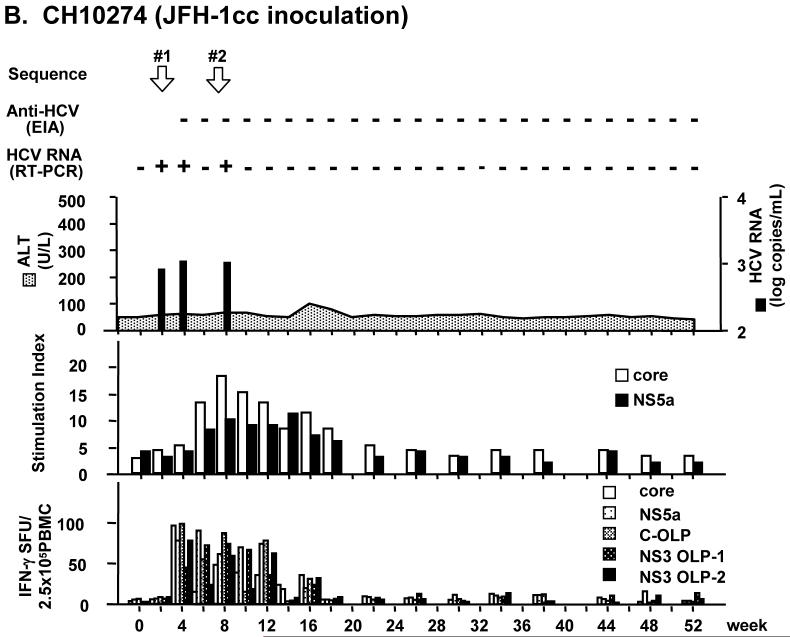
Chimpanzee 10273 (CH10273) was inoculated with patient serum containing 9.6 × 106 copies of HCV RNA. Chimpanzee 10274 (CH10274) was inoculated with 1.4 × 107 copies of JFH-1cc in culture medium. In both chimpanzees, HCV RNA became detectable in serum by RT-PCR 3 days after inoculation. Viremia was low with titers of about 103 copies/mL. Serum ALT levels were within normal limit and histological observation of liver biopsy showed no evidence of hepatitis (Fig. 1). In CH10273, HCV RNA in serum fluctuated but persisted for 34 weeks after inoculation, and anti-HCV was detected from 20 weeks after inoculation (Fig. 1A). In CH10274, serum HCV RNA disappeared at 9 weeks after inoculation and no anti-HCV seroconversion was observed (Fig. 1B).
Fig. 1.
Infection profiles and T cell immune responses in patient serum- and JFH-1cc-inoculated chimpanzees. (A) Chimpanzee CH10273 was inoculated with patient serum containing 9.6 × 106 copies of HCV. (B) Chimpanzee CH10274 was inoculated with JFH-1cc containing 1.4 × 107 copies of HCV. White arrows indicate the time points at which HCV sequences were determined. T cell proliferation assay results against HCV core and NS5a are shown as stimulation index (middle panel). IFN-γ responses against HCV core and NS5a proteins or OLPs of core and NS3 are shown as spot-forming units (SFU) per 2.5 × 105 cells (bottom panel).
Immunological analysis for T-cell proliferation and IFN-γ production revealed that HCV-specific immune responses were induced in both animals (Fig. 1). Their responses corresponded to the profiles of viremia, and remained at low levels after disappearance of viremia. The T-cell proliferative responses against the HCV core and NS5a proteins became positive 4 weeks after inoculation, and continued up to 30 and 18 weeks in CH10273 and CH10274, respectively. Likewise, the IFN-γ responses against HCV structural and nonstructural antigens were detected 4 weeks after inoculation and maintained 34 weeks and 16 weeks in CH10273 and CH10274, respectively (Fig. 1).
HCV sequence analysis
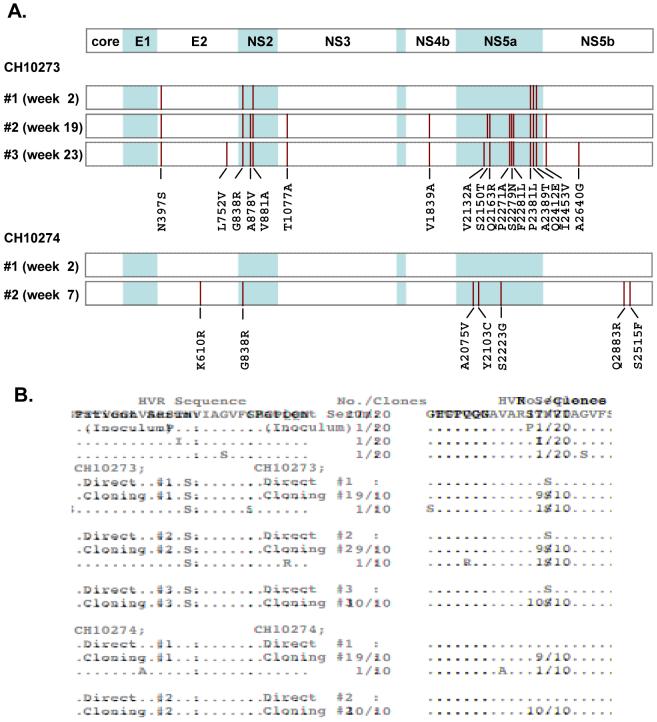
To investigate the difference and evolution of infected viruses, HCV sequences in both chimpanzees were determined directly at multiple time points as indicated in Fig. 1. In CH10273, HCV sequences were determined with sera collected at weeks 2, 19, and 23. Nineteen synonymous and 6 non-synonymous mutations were already observed at week 2, and the number of mutations increased gradually with time (Table 1). On the other hand, CH10274, showed no mutation at the earliest time point of infection (week 2), but subsequently developed 4 synonymous and 7 non-synonymous mutations at week 7 (Table 1). The mutated amino acids in the JFH-1 genome were distributed in E2, NS2, NS5a and NS5b regions (Fig. 2A). Among these mutations, only one mutation, G838R in NS2, was identified as a common mutation between the two chimpanzees. To assess the complexity of the quasispecies, the amplified fragment encompassing hypervariable region (HVR) -1 was cloned and 10 clones in each time point were sequenced. In both animals, HVR populations of isolated HCV indicated similarly low complexity of heterogeneity (Fig. 2B). HCV clones isolated from CH10273 contained one HVR-1 mutation N397S at the earliest time point of infection, and this mutation could not be found in clones of the inoculum (Fig. 2B). To exclude the possibility of PCR artifact, sequences were confirmed by independent analyses. To ensure that the common NS2 mutant was not present as a minor species at the earliest time point of CH10274 (week 2), cloning (15 clones) and sequencing was performed and showed wild-type sequence.
Table 1.
Evolution of JFH-1 in chimpanzees
| Synonymous mutationsA | Non-synonymous mutationsA | Total | |
|---|---|---|---|
| CH10273 | |||
| #1 (week 2) | 19 | 6 | 25 |
| #2 (week 19) | 33 | 15 | 48 |
| #3 (week 23) | 35 | 17 | 52 |
| CH10274 | |||
| #1 (week 2) | 0 | 0 | 0 |
| #2 (week 7) | 4 | 7 | 11 |
Compared to the consensus JFH-1 sequences.
Fig. 2.
HCV sequence analyses. (A) Distribution of amino acid substitutions in patient serum- (CH10273) and JFH-1cc- (CH10274) inoculated chimpanzees. Positions of amino acid substitutions are indicated as vertical bars, and the mutated amino acids are shown at the bottom of each panel. The amino acid numbers correspond to the JFH-1 sequence. (B) HVR-1 populations in patient serum (inoculum) and chimpanzees. HVR-1 sequence in patient serum has been reported previously (6). HVR-1 sequences determined by direct sequencing (Direct) or cloning and sequencing (Cloning)(10 clones at each time point) in each animal are shown. Investigated time points (#1, 2 and 3) are indicated in Fig. 1. Identical amino acids are indicated as dots.
Effect of the NS2 mutation on HCV life cycle
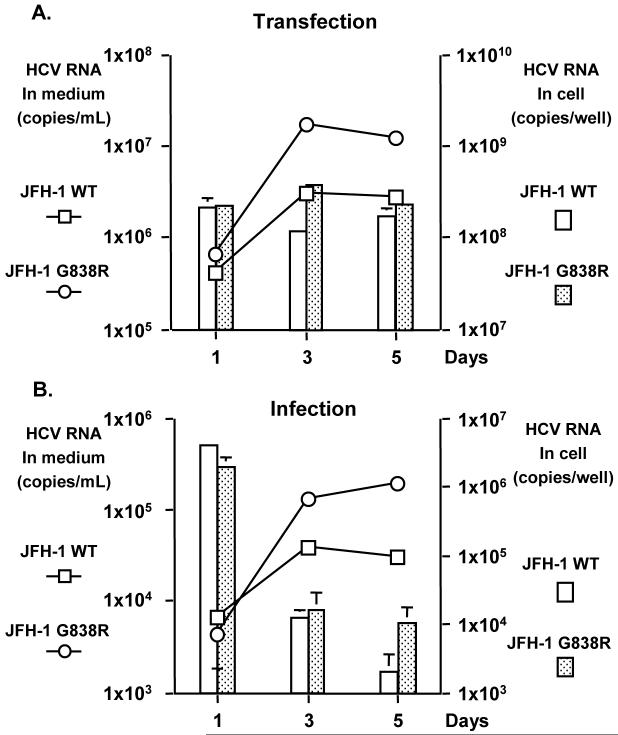
To assess whether this NS2 mutation could be a result of CTL escape, which has been described in acutely HCV-infected chimpanzees (17), we tested the T cell response of PBMC from various time points during the infection against 18-mer peptides encompassing this region (both the WT and mutant sequences were tested). No T cell response could be detected against either the WT or mutant peptides throughout the infection, therefore making CTL escape mutation highly unlikely. To assess the phenotype of the observed common mutation, G838R in the NS2 region, JFH-1 construct with this mutation was generated (JFH-1 G838R). Viral replication and production of the JFH-1 G838R mutant was compared to that of the wild type JFH-1 (JFH-1 WT) by transfecting the in vitro transcribed full-length genome RNA into Huh7.5.1 cells. HCV RNA levels in culture media of JFH-1 WT and JFH-1 G838R transfected cells were 2.96 × 106 ± 1.63 × 105 and 1.69 × 107 ± 3.61 × 105 copies/ml on day 3, and 2.67 × 106 ± 3.69 × 105 and 1.14 × 107 ± 2.23 × 105 copies/ml on day 5, respectively (P < 0.05)(Fig. 3A). In JFH-1 WT and JFH-1 G838R transfected cells, intracellular HCV RNA levels were 1.14 × 108 ± 1.36 × 107 and 3.66 × 108 ± 1.20 × 107 copies/well on day 3, and 1.67 × 108 ± 3.94 × 107 and 2.23 × 108 ± 1.90 × 107 copies/well on day 5, respectively (P < 0.05) (Fig. 3A). Thus, JFH-1 G838R could produce HCV RNA about 5-fold higher than the JFH-1 WT in culture media and transfected cells (days 3 and 5, P < 0.05).
Fig. 3.
Comparison of viral replication between JFH-1 WT and JFH-1 G838R in Huh 7.5.1 cells. At various time points, HCV RNA was measured in culture media and cells by transfecting the same amount of in vitro transcribed full genome RNA (A) and by infecting the same FFU of JFH-1cc at a MOI of 0.003 (B). Means of triplicate samples + standard deviations are shown.
To confirm this observation, an infection study was also conducted with cell culture generated viruses. After transfection of JFH1 WT and JFH-1 G838R genome RNA, viruses in culture media were harvested and FFU of these viruses were titrated. Same titer of JFH1 WT or JFH-1 G838R viruses was inoculated into naïve Huh7.5.1 cells (9 × 102 FFU, MOI = 0.003). After infection, HCV RNA titer in culture medium and infected cells was determined. Consistent with the transfection study, HCV RNA levels in culture media of JFH-1 G838R virus infected cells were 3 to 6-fold higher than those of JFH-1 WT virus (days 3 and 5, P < 0.05; Fig. 3B). Intracellular HCV RNA level on day 5 also appeared to be higher (5-fold) in JFH-1 G838R-infected cells (P < 0.05). Based on these data, JFH-1 G838R replicates more efficiently than the WT.
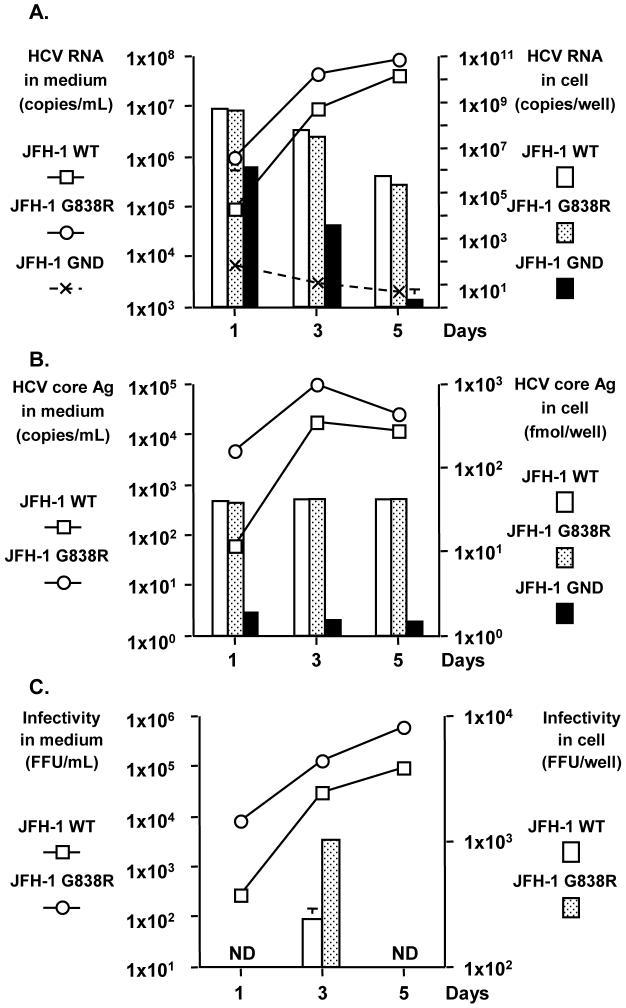
To further investigate the mechanism of this enhanced replication, we reasoned that this mutation could affect any of the viral RNA synthesis, assembly or secretion steps. To distinguish among these possibilities, we used Huh7-25 cells, a Huh7 cells derived cell line lacking of CD81 expression (11). This cell line cannot be re-infected by HCV, but can support and produce infectious HCV upon transfection with the HCV genome, therefore allowing us to address the above question without the confounding effect of re-infection. HCV RNA levels of JFH-1 G838R transfected cells in culture media were 8-fold higher on day 1 and 3-fold higher on day 3 compared with those of JFH-1 WT transfected cells (Fig. 4A, P < 0.05). On day 5, HCV RNA level was still higher in JFH-1 G838R-transfected cells but the difference was less. The HCV RNA levels of the replication deficient clone, JFH-1 GND, transfected cells were substantially lower than both NS2 mutant- and WT-transfected cells (Fig. 4A). Similarly, HCV core Ag in culture media showed a significant difference between JFH-1 WT- and JFH-1 G838R-transfected cells (days 1, 3 and 5, P < 0.05)(Fig. 4B). HCV core Ag of JFH-1 GND transfected cells was under the detection limit. In contrast to culture media data, intracellular HCV RNA and core Ag levels in JFH-1 G838R-transfected cells were similar or slightly lower than those of JFH-1 WT transfected cells. Therefore, the G838R mutation does not appear to affect RNA replication and probably enhances either the assembly or secretion step.
Fig. 4.
Comparison of viral replication among JFH-1 WT, JFH-1 G838R and JFH-1 GND in Huh 7-25 cells. At various time points, HCV production was assessed in culture media and cells by transfecting the same amount of in vitro transcribed full genome RNA. HCV RNA titer (A), HCV core Ag level (B), and infectivity titers (C) are shown. The data are expressed as means of triplicate samples + standard deviations. ND; not done.
To distinguish between these two possible effects, we determined the infectivity titer of intracellular viral particles in transfected cells as reported previously (18). On day 3 after transfection, the intracellular infectivity titer in JFH-1 G838R transfected cells was about 4-fold higher than that in JFH-1 WT transfected cells (P < 0.05, Fig. 4C and Table 2). Moreover, specific intracellular infectivity of JFH-1 G838R-transfected cells was about 8-fold higher than that in JFH-1 WT-transfected cells (P < 0.05, Table 2). Specific infectivity in culture medium was determined as the ratio of infectious virus (FFU) over HCV RNA copies. Specific infectivity of the JFH-1 G838R viruses was not significantly different from that of JFH-1 WT (Table 2). Finally the rate of secretion was determined by the ratio of extracellular FFU over the intracellular FFU (Table 2), and no difference was observed between JFH-1 WT and G838R-transfected cells. Based on these data, the G838R mutation in JFH-1 enhances the assembly step of HCV.
Table 2.
Specific infectivity and virus secretion in Huh7-25 cells
| Clone | Intracellular HCV RNA (copies/well) | AIntracellular infectivity (FFU/well) | ASpecific intracellular infectivity (FFU/copies) | AExtracellular HCV RNA (copies/well) | AExtracellular infectivity (FFU/well) | Specific exracellular infectivity (FFU/copies) | Infectious virus secretion (extra/intra) |
|---|---|---|---|---|---|---|---|
| JFH-1 WT | 4.40 × 107 ± 1.58 × 107 | 2.27 × 102 ± 5.17 × 101 | 1.09 × 10-5 ± 2.58 × 10-6 | 1.83 × 107 ± 1.95 × 106 | 6.17 × 103 ± 9.61 × 102 | 3.37 × 10-4 ± 1.38 × 10-3 | 7.20 ± 2.83 |
| JFH-1 G838R | 2.19 × 107 ± 1.11 × 106 | 9.89 × 102 ± 5.02 × 101 | 9.05 × 10-5 ± 2.76 × 10-6 | 5.14 × 107 ± 3.48 × 106 | 2.69 × 104 ± 6.96 × 103 | 5.33 × 10-4 ± 1.83 × 10-4 | 6.87 ± 2.07 |
The data are from day 3 after HCV RNA transfection of the Huh7-25 cells
P < 0.05 comparing JFH-1 WT and G838R
Discussion
Although HCV-associated fulminant hepatitis is rare, several cases have been reported (6, 19 - 25). The HCV JFH-1 strain was isolated from one of these cases, and its unique characteristic of robust replication in cell culture might be related to the etiology of fulminant hepatitis. Previously, HCV from a patient with fulminant liver failure has been shown to cause severe acute hepatitis with high viremia in a chimpanzee, although its molecular clone could not replicate in culture cells and did not induce severe hepatitis in the chimpanzee (26, 27). In our previous study, JFH-1cc induced a transient and attenuated infection in a chimpanzee (8). The infection profile was different from the typical course of HCV infection either with patient sera or infectious RNA molecules in chimpanzees (28 - 32). Because this observation was unexpected, we reasoned that the lower virulence of this strain in vivo might be related to the age of the chimpanzee. The chimpanzee used in the previous study was older (>25 years of age), and older chimpanzees typically do not develop significant disease upon HCV infection. Another possible cause was the characteristics of the viral inoculum. JFH-1cc inoculated in the chimpanzee was monotypic because it was generated in culture cells. The original JFH-1 virus replicating in the fulminant hepatitis patient existed as a mixture of various viral species and might induce a different outcome in vivo. Thus, to elucidate the pathogenesis and replication capacity of the original JFH-1 strain in vivo, the patient serum and the JFH-1cc were inoculated into juvenile chimpanzees (5 years old). However both chimpanzees showed attenuated infection with low-titer viremia, no ALT elevation and absence of histological hepatitis during the acute phase of infection. Therefore the manifestation of fulminant hepatitis of the original patient was likely a result of host factors, with the caveat that humans and chimpanzees might respond differently to HCV infection.
Similar to our previous study, the chimpanzee inoculated with monotypic JFH-1cc showed a short duration of infection and absence of serconversion. On the other hand, the chimpanzee inoculated with the patient serum showed a longer course of infection and developed anti-HCV antibodies. Immunological analysis with T-cell proliferation and IFN-γ ELISpot assays revealed that HCV-specific immune responses were similarly induced in both animals and abated with disappearance of viremia. Consistent with the longer viremia, the chimpanzee inoculated with the patient serum had a longer duration of detectable HCV immune response (Fig. 1). These differences could be explained by the sequence variations of the infecting HCV. In the chimpanzee inoculated with the patient serum, the infecting HCV showed a low sequence complexity, but exhibited some sequence diversity already at week 2. The infecting HCV had a sequence alteration in the HVR-1 (N397S), but this sequence alteration could not be found in any of the 20 clones of the inoculum (Fig. 2B) (6). In addition, the NS2 G838R mutation was also not detected by cloning (6 clones) and sequencing of the inoculum. Thus, this infecting HCV was probably selected from a minor species in the patient serum. It has been reported that minor clones in human serum were selected during HCV infection in chimpanzees (33). The selected clones were in the lighter fraction of the sucrose density gradient of the inoculum, which is devoid of immunoglobulins. Similar selection might have occurred in our study. The dominant clones in the inoculum might not be infectious due to binding to neutralizing antibodies. As a result, the infection-competent minor clone, selected during the infection, became the dominant species. Furthermore, this infecting minor clone could persist longer, although the characteristics of this clone and mechanisms for persistence are still unknown. HCV clones in CH10273 showed several other mutations at 2 weeks post-infection, and accumulated additional mutations in E2, NS2, NS3, NS4b, NS5a and NS5b regions over time (Fig. 2). Some of these regions contain known T-cell epitopes, although the MHC haplotype of this animal is unknown. In this chimpanzee, heterogeneity of the inoculating viruses might have contributed to the emergence of escape mutants from the host immune system resulting in a prolonged infection. Similar observations have been reported in acute HCV infection in chimpanzees and humans (34 - 36).
In HCV strains isolated from these two chimpanzees, 1 common mutation G838R in the NS2 region was identified. This mutation has not been reported among the adaptive mutations emerged in the JFH-1 virus passaged in cell culture (37 - 39). This mutation likely arose de novo because one of the chimpanzees was inoculated with a molecular clone, and the week 2 sample did not harbor this mutation. NS2 is a membrane-associated cysteine protease, composed of 3 transmembrane domains and a protease domain (40). Although NS2 region is dispensable for RNA replication, it is essential for production of infectious virus in cultured cells (41 - 43). Furthermore, the significance of this region has been shown in the establishment of replication and infection competent intergenotypic chimeric viruses (44, 45). The identified common mutation G838R was at the end of the first transmembrane domain (46), and mutations in the transmembrane domains have been shown to improve the yield of infectious virus production in several studies (45, 47). Thus, some advantage of this mutation in HCV replication and production could be expected. This mutation was shown to enhance HCV production in Huh7.5.1 cells. Detailed analysis with CD81-negative Huh7-25 cell demonstrated that viral assembly was affected by this mutation. Production of infectious virus in JFH-1 G838R-transfected cells was 8-fold higher than that in the JFH-1 WT-transfected cells. Thus, this mutation enhances the assembly of infectious virus particle in cultured cells, and as a result, increases infectious virus production in the culture medium. This mutation represents the first identified in vivo adapted mutation that is not immunologically mediated, and probably confers a replication advantage to the virus in vivo. This adaptive mutation, unlike the other adaptive mutations reported in vitro with poor infectivity in vivo, likely results from a highly biologically relevant event in the dynamic interaction between HCV and host. Finally it is possible that compensatory mutations in other regions of the virus may contribute to the overall biological adaptive response of the virus in vivo.
This study demonstrates that the HCV JFH-1 strain either generated in cell culture as a monotypic virus or obtained from patient serum is associated with attenuated infection in chimpanzees; however the virus can rapidly evolve with adaptive mutations to facilitate propagation of the virus in a susceptible host.
Acknowledgment
We are grateful to Charles Rice and Francis Chisari for providing the cell lines and the veterinary staff for technical assistance in the chimpanzee experiment.
Financial Support:
TK and TW were partially supported by a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science and from the Ministry of Health, Labor, and Welfare of Japan; and by the Research on Health Sciences Focusing on Drug Innovation from the Japan Health Sciences Foundation. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH, and Division of Viral Hepatitis, Center for Disease Control and Prevention.
Nonstandard abbreviations used
- HCV
hepatitis C virus
- JFH-1cc
cell culture generated JFH-1 virus
- ALT
alanine aminotransferase
- HVR
hyper variable region
- FFU
focus-forming unit
- Ag
antigen
- OLP
overlapping peptide pool
- SFU
spot forming unit.
References
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Feld JJ, Liang TJ. Hepatitis C -- identifying patients with progressive liver injury. Hepatology. 2006;43:S194–S206. doi: 10.1002/hep.21065. [DOI] [PubMed] [Google Scholar]
- 3.Shiina M, Rehermann B. Hepatitis C vaccines: Inducing and challenging memory T cells. Hepatology. 2006;43:1395–1398. doi: 10.1002/hep.21210. [DOI] [PubMed] [Google Scholar]
- 4.De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 5.Di Bisceglie AM, Hoofnagle JH. Optimal therapy of hepatitis C. Hepatology. 2002;36:S121–S127. doi: 10.1053/jhep.2002.36228. [DOI] [PubMed] [Google Scholar]
- 6.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, et al. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 7.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 10.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. Cell culture and infection system for hepatitis C virus. Nat Protoc. 2006;1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi T, Katsume A, Tanaka T, Abe A, Inoue K, Tsukiyama-Kohara K, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116:636–642. doi: 10.1016/s0016-5085(99)70185-x. [DOI] [PubMed] [Google Scholar]
- 14.Elmowalid GA, Qiao M, Jeong SH, Borg BB, Baumert TF, Sapp RK, et al. Immunization with hepatitis C virus-like particles results in control of hepatitis C virus infection in chimpanzees. Proc Natl Acad Sci U S A. 2007;104:8427–8432. doi: 10.1073/pnas.0702162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoyagi K, Ohue C, Iida K, Kimura T, Tanaka E, Kiyosawa K, et al. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato T, Matsumura T, Heller T, Saito S, Sapp RK, Murthy K, et al. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–895. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 18.Gastaminza P, Kapadia SB, Chisari FV. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol. 2006;80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright TL, Hsu H, Donegan E, Feinstone S, Greenberg H, Read A, et al. Hepatitis C virus not found in fulminant non-A, non-B hepatitis. Ann Intern Med. 1991;115:111–112. doi: 10.7326/0003-4819-115-2-111. [DOI] [PubMed] [Google Scholar]
- 20.Liang TJ, Jeffers L, Reddy RK, Silva MO, Cheinquer H, Findor A, et al. Fulminant or subfulminant non-A, non-B viral hepatitis: the role of hepatitis C and E viruses. Gastroenterology. 1993;104:556–562. doi: 10.1016/0016-5085(93)90426-d. [DOI] [PubMed] [Google Scholar]
- 21.Yoshiba M, Dehara K, Inoue K, Okamoto H, Mayumi M. Contribution of hepatitis C virus to non-A, non-B fulminant hepatitis in Japan. Hepatology. 1994;19:829–835. [PubMed] [Google Scholar]
- 22.Villamil FG, Hu KQ, Yu CH, Lee CH, Rojter SE, Podesta LG, et al. Detection of hepatitis C virus with RNA polymerase chain reaction in fulminant hepatic failure. Hepatology. 1995;22:1379–1386. [PubMed] [Google Scholar]
- 23.Vento S, Cainelli F, Mirandola F, Cosco L, Di Perri G, Solbiati M, et al. Fulminant hepatitis on withdrawal of chemotherapy in carriers of hepatitis C virus. Lancet. 1996;347:92–93. doi: 10.1016/s0140-6736(96)90212-3. [DOI] [PubMed] [Google Scholar]
- 24.Farci P, Alter HJ, Shimoda A, Govindarajan S, Cheung LC, Melpolder JC, et al. Hepatitis C virus-associated fulminant hepatic failure. N Engl J Med. 1996;335:631–634. doi: 10.1056/NEJM199608293350904. [DOI] [PubMed] [Google Scholar]
- 25.Fukai K, Yokosuka O, Fujiwara K, Tagawa M, Imazeki F, Saisho H, et al. Etiologic considerations of fulminant non-A, non-B viral hepatitis in Japan: analyses by nucleic acid amplification method. J Infect Dis. 1998;178:325–333. doi: 10.1086/515619. [DOI] [PubMed] [Google Scholar]
- 26.Farci P, Munoz SJ, Shimoda A, Govindarajan S, Wong DC, Coiana A, et al. Experimental transmission of hepatitis C virus-associated fulminant hepatitis to a chimpanzee. J Infect Dis. 1999;179:1007–1011. doi: 10.1086/314653. [DOI] [PubMed] [Google Scholar]
- 27.Sakai A, Takikawa S, Thimme R, Meunier JC, Spangenberg HC, Govindarajan S, et al. In vivo study of the HC-TN strain of hepatitis C virus recovered from a patient with fulminant hepatitis: RNA transcripts of a molecular clone (pHC-TN) are infectious in chimpanzees but not in Huh7.5 cells. J Virol. 2007;81:7208–7219. doi: 10.1128/JVI.01774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 29.Hong Z, Beaudet-Miller M, Lanford RE, Guerra B, Wright-Minogue J, Skelton A, et al. Generation of transmissible hepatitis C virions from a molecular clone in chimpanzees. Virology. 1999;256:36–44. doi: 10.1006/viro.1999.9603. [DOI] [PubMed] [Google Scholar]
- 30.Beard MR, Abell G, Honda M, Carroll A, Gartland M, Clarke B, et al. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 31.Thomson M, Nascimbeni M, Gonzales S, Murthy KK, Rehermann B, Liang TJ. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology. 2001;121:1226–1233. doi: 10.1053/gast.2001.28669. [DOI] [PubMed] [Google Scholar]
- 32.Major ME, Mihalik K, Puig M, Rehermann B, Nascimbeni M, Rice CM, et al. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J Virol. 2002;76:6586–6595. doi: 10.1128/JVI.76.13.6586-6595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojima M, Osuga T, Tsuda F, Tanaka T, Okamoto H. Influence of antibodies to the hypervariable region of E2/NS1 glycoprotein on the selective replication of hepatitis C virus in chimpanzees. Virology. 1994;204:665–672. doi: 10.1006/viro.1994.1582. [DOI] [PubMed] [Google Scholar]
- 34.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 35.Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–895. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 36.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, et al. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgrange D, Pillez A, Castelain S, Cocquerel L, Rouille Y, Dubuisson J, et al. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J Gen Virol. 2007;88:2495–2503. doi: 10.1099/vir.0.82872-0. [DOI] [PubMed] [Google Scholar]
- 38.Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J Virol. 2007;81:13168–13179. doi: 10.1128/JVI.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell RS, Meunier JC, Takikawa S, Faulk K, Engle RE, Bukh J, et al. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc Natl Acad Sci U S A. 2008;105:4370–4375. doi: 10.1073/pnas.0800422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature. 2006;442:831–835. doi: 10.1038/nature04975. [DOI] [PubMed] [Google Scholar]
- 41.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 42.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi M, Ma Y, Yates J, Lemon SM. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J Virol. 2007;81:629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaga AK, Ou JH. Membrane topology of the hepatitis C virus NS2 protein. J Biol Chem. 2002;277:33228–33234. doi: 10.1074/jbc.M202304200. [DOI] [PubMed] [Google Scholar]
- 47.Murray CL, Jones CT, Tassello J, Rice CM. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J Virol. 2007;81:10220–10231. doi: 10.1128/JVI.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]