Abstract
A coarse-grained model for molecular dynamics simulations is extended from lipids to proteins. In the framework of such models pioneered by Klein, atoms are described group-wise by beads, with the interactions between beads governed by effective potentials. The extension developed here is based on a coarse-grained lipid model previously developed by Marrink et al., though further versions will reconcile the approach taken with the systematic approach of Klein and other authors. Each amino acid of the protein is represented by two coarse-grained beads, one for the backbone (identical for all residues) and one for the side-chain (which differs depending on the residue type). The coarse-graining reduces system size about ten-fold and allows integration time steps of 25 to 50 fs. The model is applied to simulations of discoidal high-density lipoprotein particles, involving water, lipids, and two primarily helical proteins. These particles are an ideal test system for the extension of coarse-grained models. Our model proved reliable in maintaining the shape of pre-assembled particles and in accurately reproducing overall structural features of high-density lipoproteins. Microsecond simulations of lipoprotein assembly revealed formation of a protein-lipid complex in which two proteins are attached to either side of a discoidal lipid bilayer.
1 Introduction
High-density lipoproteins (HDL), sometimes called “good cholesterol”, are protein-lipid particles responsible for carrying cholesterol, lipids, and fatty acids from arteries to the liver for degradation. Low levels of HDL have been implicated in increasing the risk of atherosclerosis (hardening of the arteries) and coronary heart disease.1 The major protein component of HDL particles is apolipoprotein A-1 (apo A-1). Apo A-1 is secreted by the liver and intestines and exists in three forms: unassociated apo A-1 (lipid-free/poor), nascent discoidal particles, and mature spherical forms.2 Apo A-1 consists of 243 amino acid residues with two distinct domains, an N-terminal globular domain and a C-terminal lipid binding domain. The lipid binding domain consists of eight 22-mer and two 11-mer amphipathic α-helical repeats punctuated by the presence of prolines and glycines.3,4 Each HDL particle contains two apo A-1 strands, which due to the amphipathic nature of the helices interact with both lipids and water. The interaction between the two apo A-1 strands is proposed to involve a series of salt-bridges between oppositely charged residues.5,6,7,8
Several models exist for the structure of apo A-1 in nascent discoidal HDL particles including the double-belt,5,9 picket-fence,10,11 and helical-hairpin12,13 models. Initial infrared spectroscopy studies supported the picket-fence model.14 However, methodologically updated infrared experiments15 contradict these earlier studies, instead favoring a double-belt model. Additionally, fluorescence studies16 and the X-ray crystal structure of a lipid-free apo A-117 support the double-belt model. More recently, cross-linking mass spectrometry results ruled out the picket-fence model and strongly suggest a double-belt model.18
The metabolism of HDL proceeds via the reverse cholesterol transport pathway, which regulates the transformation of HDL particles from an unassociated lipid-free form to nascent discoidal particles to mature spherical form. Apo A-1 is synthesized primarily in the liver, where it forms nascent discoidal HDL particles. These particles circulate in the plasma and remove cholesterol and phospholipids from peripheral tissues. The nascent discoidal HDL particles swell in size upon the addition of cholesterol and lipids. The associated cholesterol is then esterified by lecithin cholesterol acyl transferase and the HDL particles undergo a structural change, going from discoidal to spherical in shape.19
Nanodiscs are protein-lipid particles which are engineered to mimic nascent discoidal HDL particles. They are produced in order to create a platform in which to embed and solubilize membrane proteins in a native lipid environment.20,21,22,23,24 Each nanodisc consists of two membrane scaffold proteins surrounding a lipid bilayer in a belt-like fashion (Figure 1). Membrane scaffold proteins are engineered based on the structure of the lipid binding domain of apo A-1. Nanodiscs have been well characterized25,26,27 and in addition to acting as platforms for studying membrane proteins, their well-defined size and composition provide an ideal system for studying the HDL lipid-binding domain.
Figure 1.
Schematic presentation of the double-belt model of the nanodisc formed by two amphipathic helical membrane scaffold proteins wrapped around a lipid bilayer.
Under a set of conditions in which the ratio of membrane scaffold proteins to lipids is precisely controlled and optimized, nanodiscs self-assemble into discoidal particles of discrete size and composition.28 The assembly process by which homogeneous nanodisc particles form is initiated by the removal of cholate from a starting mixture of cholate, scaffold protein, and lipids. Although the assembly mechanisms of native HDL particles and of nanodisc particles are different, the information gathered from studying the assembly of nanodiscs should shed light on HDL particle formation.
Due to the absence of a high-resolution structure for lipid-associated nascent HDL particles and nanodiscs, comparisons between models resulting from all-atom molecular dynamics (MD) simulations with low-resolution structural techniques, such as small-angle X-ray scattering (SAXS), proved valuable in verifying the structural properties of these protein-lipid particles.27 All-atom simulations of high-density lipoprotein particles and nanodiscs have already provided insight into the secondary structure of the proteins surrounding the lipid bilayer,8,27 and an anti-parallel alignment of the two protein strands that maximizes salt-bridging.5,6,7,8 The apo A-1 and scaffold proteins in these simulations assumed a double-belt orientation5,9 and were built using insights provided by the X-ray crystal structure of lipid-free apo A-1.17
All-atom MD simulations of nascent discoidal HDL particles and nanodiscs are limited to nanosecond time scales due to the large size of the simulated systems.27 In order to investigate assembly and disassembly of these protein-lipid particles, longer time scales are required. A coarse-grained (CG) MD approach in which much longer time scales can be simulated appears to be a promising alternative to all-atom MD simulations.
CG MD models reduce the overall system size compared with all-atom models by mapping clusters of atoms onto CG beads. Recent advances in CG modeling have resulted in CG lipid models which allow the study of lipid assembly on a micrometer length scale and on a millisecond time scale. These CG lipid models have been successfully used to model the assembly of liposomes, micelles, inverted hexagonal phases, and lipid bilayers. In addition to the reduction in system size, CG models use a combination of short-range electrostatics and minimal number of bead types to further improve computational efficiency.29,30,31
One of the first CG MD lipid models to successfully show assembly of lipids was developed by Klein and coworkers.29,32 The developed DMPC CG model was parameterized to mimic structural features resulting from all-atom simulations. Successful assembly of lamellar and inverted hexagonal phases from a random distribution demonstrated the ability of the model to investigate the dynamics of lipid assembly. Recent refinement and extension of the CG model has led to the description of transmembrane peptide induced lipid sorting,33 lipid perturbation due to hydrophobic mismatch surrounding a nanotube,34 insertion of a model pore into a lipid bilayer,35 and effects of anesthetics on model membranes.36 Models which use a similar coarse-graining method (i.e. a ten-fold reduction in system size as compared with all-atom models) have recently been developed by Marrink et al.30,37,38,39,40,41 and Stevens.31,42
Numerous other groups have also developed various CG lipid models using approaches that are less tightly coupled to conventional MD. Lipowsky and coworkers have used simple lattice based models of multi-component bilayers to analyze membrane phase behavior,43 and applied dissipative particle dynamics to successfully simulate tension-induced vesicle fusion.44 The use of a continuum model known as the elastic membrane model has allowed analysis of large scale properties such as swelling of vesicles due to osmotic pressure45 and responses of bilayers to strain,46 and even allowed for limited shifting between CG and all-atom representations by using the results of the continuum model as boundary conditions for all-atom MD.47 Although these more simplified CG models are computationally efficient and have provided a good deal of data on lipid bilayers, because we also seek to model protein systems (to which continuum models and soft-core dynamics are less applicable) we restrict ourselves to considering models in which traditional MD simulations are performed with reduced number of particles.
CG lipid models rely on the hydrophobic and hydrophilic properties of lipids to accurately describe the dynamics of assembly. HDL and nanodiscs are comprised of helical amphipathic proteins (apo A-1 or membrane scaffold proteins, respectively) which wrap around a lipid bilayer core. The simple amphipathic helical structure of these proteins appear to be ideal for the application of a protein-lipid CG model. Nanodiscs were chosen over native HDL particles as a test system for the CG model due to their discrete size and composition, and previous extensive characterization of their structure.25,26,27
Protein CG models were developed in the past, however, primarily for use in protein folding simulations. The earliest attempts at protein coarse-graining involved lattice models, in which the protein was split into beads with certain interaction rules, and then the beads were arranged on a 2D or 3D grid and the stability of different conformations scored based on interactions with neighboring beads.48,49 Recently, other approaches have been attempted to study protein folding dynamics, including modeling of the protein as an interacting network of points50 or mapping protein residues onto beads with interaction potentials chosen to reproduce a desired structure.51,52 Although these models have provided considerable insight into the protein folding process and show promise for reproducing some features of large scale protein motion, the development of a CG protein model capable of reproducing stable protein structures and large-scale motions during MD simulations has proven quite difficult due to the high complexity of proteins.
We describe here a CG model of lipoprotein assembly involving an extension of the Marrink et al.30 CG lipid-water model to include proteins. We also describe how this model has been implemented into the MD program NAMD.53 The Marrink CG lipid model was chosen for its ability to reproduce experimental properties of various lipid assemblies, including DPPC lipids, as well as for the ease of implementing the potential energy function into NAMD.53 Additionally, because the Marrink CG model30 is not as strictly derived as other CG models29,32 from existing forcefields or statistical mechanical properties, it was easier to apply and extend to non-lipid systems. The new protein-lipid CG model was developed to include a description for protein coarse-graining as well as for protein-lipid interactions. The developed model represents a first step in our attempt to provide a reliable protein-lipid CG model. The model described here needs to be improved further, in particular, by following the systematic CG approach of Klein29,32 that incorporates a link to atomic level forcefields and geometries, as well as a link to experimental and atomistic computer simulation data. We also intend to use an implicit solvent model for water in future versions of our CG approach.
In this paper we describe the application of the CG model to nanodiscs, their stability and self-assembly. In Section 2, we introduce the CG model applied in this work. Results of the CG MD simulations are presented and discussed in Section 3. Conclusions from the work are provided in Section 4.
2 Methods
To develop a CG model for scaffold proteins and water-lipid systems, several routes would be feasible. For the reasons stated above, we have chosen to extend the lipid-water model of Marrink et al.30 to include proteins. In the Marrink et al.30 CG model an all-atom system is converted to a CG representation by mapping clusters of atoms with similar physical properties onto one CG bead; these beads usually stand in for four heavy atoms plus their associated hydrogens. A bead is then assigned a class based on the properties of the group of atoms being converted to the CG representation. The bead classes are based on properties such as hydrophobicity, hydrogen bonding, or charge, and are used to determine the strength of nonbonded interactions between any two beads in the system. Standard bond lengths, angles, and force constants are used for virtually all bonded interactions in the system. To account for the lipids encountered in our studies, namely DPPC, DOPE, and DPC, and for water and ions, the Marrink et al.30 CG model is applied in a straightforward manner.
To develop a combined protein-lipid model, we began with previously existing parameters for lipid and water coarse-graining,30 and extended them to describe proteins. In the framework of the resulting model, groups of atoms are substituted by CG beads as described below. The CG beads are considered as point masses and their dynamics described by Newton’s equations of motion. E3ective interactions between the beads are accounted for by using a potential of the form
| (1) |
where the index j denotes one of the four components of the system: protein, lipid, water and ions. The sum in eq 1 includes the terms describing the interactions between bonded CG beads: accounts for the bonds between the beads, corresponds to the forces used to sustain certain angles between sets of three bonded CG beads, and describes the potential of dihedral angles for quadruples of bonded CG beads. The term Vnonbond in eq 1 accounts for non-bonded interactions between all CG beads in the system. CG beads for water and ions do not have any bonds in the present model, such that the terms and , as well as and , actually vanish. Water and ions contribute only to the non-bonded interactions (term Vnonbond).
The term in eq 1, describing the bonds for both lipids and proteins, has the form
| (2) |
where j is the component (proteins or lipids) and the summation is over all bonds i; is the distance between the bonded beads for bond i, is the force constant, and is the equilibrium bond length.
The potential term for angles ( in eq 1) differs between lipids and proteins. The cosine-containing term
| (3) |
is used for lipids (as in the Marrink lipid-water model30), and the harmonic term
| (4) |
is used for proteins to represent the stiffness of the protein backbone. Here θk is the angle formed by any three successively bonded CG beads, and the summation is over all such triples, for proteins and lipids, respectively. Values and are force constants, and are equilibrium angles.
The potential Vdihedral in eq 1 is defined for any four successively bonded CG beads. It is used for backbone protein residues only, i.e., we set Vdihedral = 0 for any combination of four successively connected CG beads, except in the case when all four beads represent the protein backbone. The summation over all such protein backbone quadruples ℓ makes up the dihedral potential, expressed in the form
| (5) |
where Φℓ is the force constant, the integer n is the multiplicity, and δℓ is the phase shift; the variable here is the angle χℓbetween the plane formed by the first three beads in the quadruple ℓ and the plane formed by the last three beads.
To describe non-bonded interactions we use the van der Waals (vdW) potential for all CG beads and in addition for charged beads the Coulomb potential; the corresponding potential is
| (6) |
where the summation is performed over all pairs of CG beads m and n (all components comprising the system, i.e., proteins, lipids, water, and ions contribute to this sum). Constants εmn and σmn are the vdW parameters for the interaction between beads m and n, qm is the charge of the mth bead, rmn is the distance between beads m and n, and ε0 is the vacuum dielectric permittivity; a relative dielectric constant ε = 20 is assumed for all electrostatic interactions.
Parameters for the vdW potential in eq 6 are identical to those in the Marrink et al. lipid-water model.30 Each CG bead, depending on the properties of the group of atoms it represents (hydrophobic/hydrophilic, charged/uncharged, and hydrogen bonding), is assigned to one of four classes, polar (P), non-polar (N), apolar (C), and charged (Q), with the non-polar and charged classes further broken down into normal (0), hydrogen bond donor (d), hydrogen bond acceptor (a), and donor-acceptor (da) classes. The force constant εmn of vdW interactions between beads m and n is assigned to one of five levels (I to V) depending on the class the bead belongs to, as presented in Table 1. The values of the force constants for each of the levels are εmn = 5 kJ/mol for level I, εmn = 4.2 kJ/mol for II, εmn = 3.4 kJ/mol for III, εmn = 2.6 kJ/mol for IV, and εmn = 1.8 kJ/mol for V. The vdW radius is σmn = 4.7Å for any pair of CG beads.
TABLE 1.
Interaction matrix (adapted from Marrink et al. 30).
| P | N | C | Q | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | d | a | da | 0 | d | a | da | ||||
| P | I | IV | III | III | II | V | I | I | I | I | |
| N | 0 | IV | III | III | III | III | III | III | III | III | III |
| d | III | III | II | II | II | IV | III | III | II | II | |
| a | III | III | II | II | II | IV | III | II | III | II | |
| da | II | III | II | II | I | V | III | II | II | I | |
|
| |||||||||||
| C | V | III | IV | IV | V | III | V | V | V | V | |
|
| |||||||||||
| Q | 0 | I | III | III | III | III | V | III | III | III | II |
| d | I | III | III | II | II | V | III | III | II | I | |
| a | I | III | II | III | II | V | III | II | III | I | |
| da | I | III | II | II | I | V | II | I | I | I | |
2.1 Lipid-water model
The representation of water, ions and lipids by the CG beads in our model has been left the same as in Marrink et al.30 This choice ensures compatibility with earlier work and will have to be refined in later versions of our model. Clusters of heavy atoms as well as the associated hydrogen atoms are mapped onto one CG bead. Each bead has a mass of 72 amu. For water, four molecules are mapped onto one polar (class P) CG bead. Each ion, together with its hydration shell, is represented by one charged (class Q) CG bead. Functional groups of atoms (such as choline) are substituted by one CG bead for lipids and cholate (see Marrink 30,54 for a description of the coarse-graining of lipids and cholate).
A uniform set of bond and angle parameters is used for all bonded interactions (eqs 2, 3) between neighboring beads in lipids, with bond force constants of 1250 kJ mol−1 nm2, bond lengths of 4.7 Å, angle force constants of 25 kJ mol−1, and equilibrium angles of 180° except for the glycerol backbone (in DPPC lipids) which has an equilibrium angle of 120°.30 The CG beads representing charged groups or ions are given a charge of ±0.7|e|, where e is the charge of an electron.30
Before extending the CG model to proteins, we checked that the lipid-water model reproduced the results reported by Marrink et al.30 For this purpose, we simulated the assembly of identical systems of lipids in solution as in Marrink et al.30 MD of CG systems was performed using NAMD 2.5,53 modified to allow the use of either the cosine-based angle potential (3) or the harmonic angle potential (4), instead of using GROMACS.55 In agreement with Mar-rink et al.,30 we observed (results not shown) the aggregation of DPPC lipids into bilayers, and the formation of the inverted hexagonal phase from a random mixture of DOPE lipids. Another example, the assembly of DPC lipids into micelles, is presented in Figure 2. The time scales of these processes also agreed with that reported by Marrink et al.30
Figure 2.
Self-aggregation of DPC lipids into micelles in a CG representation. The DPC head groups are shown in red and the tails in blue. The initial random mixture of lipids (left) aggregates into a set of micelles as the lipids arrange to protect their hydrophobic tails from interaction with water. The configuration of formed micelles after 100 ns of evolution is shown on the right.
Marrink’s original model was designed for physiological or room temperature systems.30 Modeled water was reported to melt at 290 K and spontaneously freeze below 240 K. Nevertheless, caution needs to be exercised when applying the present model even at room temperature, since interactions between the water beads are accounted for by a pure Lennard-Jones 12-6 potential, which narrows the liquid range of the CG solvent (i.e., the range of temperatures at which the solvent is in a liquid state might be substantially narrower than the 100 K for real water). For instance, the CG water may eventually begin to freeze in a substantially longer simulation, even at temperatures where it should be liquid. However, even if such freezing occurs, the timescale for such a phenomenon is likely much larger than the microsecond timescale presently accessible by our CG model. In our simulations, performed over a timescale of microseconds and a temperature range of 300–353 K (see Section 3), no signs of freezing were observed.
2.2 Protein model
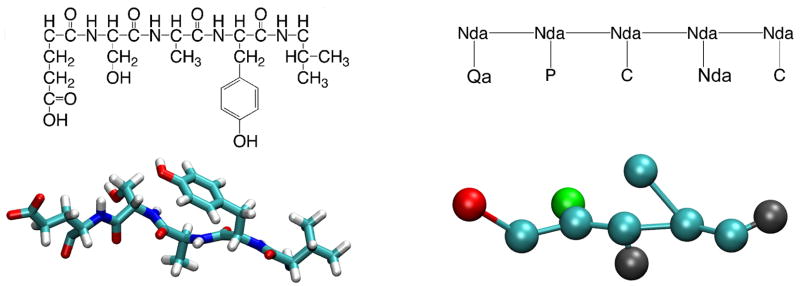
For coarse-graining of proteins each amino acid residue is mapped onto two CG beads, a class Nda bead for the backbone and a variable class bead for the side-chain. All twenty natural amino-acids are considered (glycine is represented without a side chain bead), and therefore we use 20 different types of CG beads for the description of proteins. Prolines are described through a particular choice of the angular (eq 4) and dihedral potentials (eq 5) as specified further below. Whenever an all-atom protein structure is mapped into a CG structure, beads are placed at the center of mass of the group of atoms they represent. Each CG protein bead has a mass equal to the mass of the atoms it represents. The conversion of an isolated stretch of the oligopeptide Glu-Ser-Ala-Tyr-Val, from the all-atom to the CG representation is illustrated in Figure 3.
Figure 3.
Structure of the oligopeptide sequence Glu-Ser-Ala-Tyr-Val in an all-atom representation (left) and the equivalent CG version (right). The CG representation is shown with class N beads in cyan, class C in grey, class Q in blue, and class P in green.
To describe the non-bonded interactions of protein CG beads, each bead is assigned to one of the interaction classes introduced above (see above and Table 1). A listing of all protein beads and their CG forcefield classes is provided in Table 2 (backbone beads are class Nda). The beads representing the charged side-chains are given a charge of ±0.7|e|.
TABLE 2.
CG bead class and distances between backbone and side-chain beads for each protein residue used in our simulations. Types follow the convention of Marrink et al.,30 (see also Table 1). Each bead may be polar (P), non-polar (N), apolar (C), or charged (Q); non-polar or charged beads may further be hydrogen bond donors (d) and/or acceptors (a). The distance between backbone beads was kept at 3.5 Å for all protein residues.
| Residue | Bead Class | Bond Length (Å) |
|---|---|---|
| Alanine | C | 2.0 |
| Arginine | Qd | 4.1 |
| Asparagine | Nda | 2.8 |
| Aspartate | Qa | 3.0 |
| Cysteine | P | 2.7 |
| Glutamine | Nda | 4.0 |
| Glutamate | Qa | 4.0 |
| Glycine | (none) | N/A |
| Histidine | P | 4.7 |
| Isoleucine | C | 2.7 |
| Leucine | C | 3.5 |
| Lysine | Qd | 4.2 |
| Methionine | C | 3.8 |
| Phenylalanine | C | 4.1 |
| Proline | C | 2.5 |
| Serine | P | 2.5 |
| Threonine | P | 2.7 |
| Tryptophan | C | 4.5 |
| Tyrosine | Nda | 4.6 |
| Valine | C | 2.7 |
The bond lengths for proteins ( in eq 2) depend only on the type of the protein beads comprising the pair i. To obtain the values of , we converted the satellite tobacco mosaic virus monomer structure (PDB code 1A34)56 into its CG representation and assigned the average distances between the beads of each pair of types to . Since this structure contains no histidines, we used the structure of ubiquitin (PDB code 1UBQ) to get the distance for the pair of beads representing the backbone and histidine side-chain. The force constant for any pair of protein CG beads was chosen to be the same as that used for bonds in the lipid model, 1250 kJ mol−1 nm−2. The lengths of bonds between the bead of each type and the backbone bead are presented in Table 2 (for the main chain beads we use a bond length of 3.5 Å to adjacent main chain beads).
Protein beads have equilibrium angles ( in eq 4) of 92° along the main chain, 134° for angles including a side chain bead, and 180° for main chain angles centered on a proline (based on the structure resulting from all-atom simulations of nanodiscs).27 The force constants for the angle term described by eq 4 were chosen to be = 12.5 kJ mol−1 rad−2 for all angles. The dihedral energy term described by eq 5 is used to maintain a helical secondary structure through a dihedral force constant Φℓ of 1.21 kJ mol−1, a multiplicity n of 1, and a phase shift δℓ of 130° for all non-proline backbone dihedrals and δℓof 180° for backbone angles involving proline.
2.3 Simulation methods
Simulations were performed using NAMD 2.5,53 modified as explained in Section 2.1. Non-bonded interactions were cut o3 at 12 Å, with shifting throughout the interaction range for electrostatic interactions and beginning at 9 Å for vdW interactions to implement a smooth cut-off. Simulations were performed using a 25 fs timestep for protein containing systems and a 30 to 50 fs timesteps for pure lipid-water systems. Pair lists were updated at least once per 20 steps, with a 16 Å pair list cut-o3. In all cases we performed Langevin dynamics with a damping coefficient of 5 ps−1. A constant pressure of 1 atm was maintained with a Nosé-Hoover Langevin piston,57 using a piston period of 1000 fs and a decay time of 500 fs.
The necessary calculations were carried out mainly on a cluster of 48 Athlon XP 2600+ processors. To illustrate the computational efficiency of the CG approach on increasing numbers of processors, data on how simulation time per step scales with the number of CPUs used are shown in Table 3. The performance data were obtained for a 33,573 bead system run with 50 fs timesteps on the SGI Altix cluster at the National Center for Supercomputing Applications.
TABLE 3.
Performance of NAMD simulations using the CG description. All runs were performed with 50 fs time steps on an SGI Altix cluster using the specified number of nodes, a 33,573 bead system consisting of a cholate/DPPC micelle solvated with water and neutralized with sodium ions. Ideal scaling corresponds to constant values in the right column.
| CPU | S/step | CPU ×s/step |
|---|---|---|
| 2 | 0.190 | 0.38 |
| 4 | 0.0953 | 0.38 |
| 8 | 0.0476 | 0.38 |
| 16 | 0.0249 | 0.40 |
| 24 | 0.0174 | 0.42 |
| 36 | 0.0121 | 0.44 |
| 48 | 0.00934 | 0.45 |
| 64 | 0.00774 | 0.50 |
| 128 | 0.00490 | 0.63 |
2.4 Conditions for nanodisc simulations
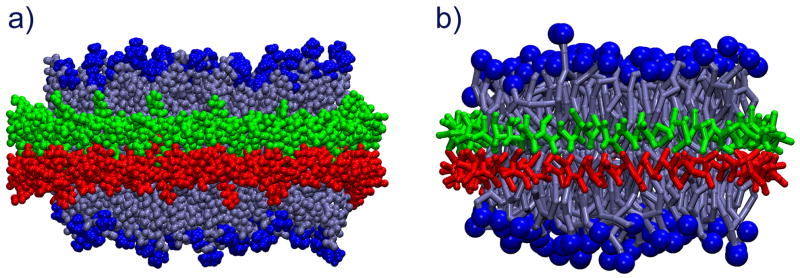
In our study, nanodiscs served as a test system for the CG model and as a subject for investigation into the self-aggregation of lipoproteins. Nanodiscs contain two anti-parallel amphipathic helical scaffold proteins, each with 178 amino acid residues, wrapped in a belt-like fashion around a lipid bilayer containing 160 DPPC lipids (Figure 1). The nanodiscs were simulated using the CG method described above, in which each amino acid residue was represented by two CG beads (except glycine that was represented by only a backbone bead) and each DPPC lipid by 12 CG beads. The full atom and CG model of a nanodisc are compared in Figure 4.
Figure 4.
Nanodisc shown in (a) all-atom and in (b) CG description. Coarse-graining reduces the number of particles by a factor of 10. Nanodiscs are shown in licorice representation with the two scaffold proteins in green and red and the lipids in light blue with their head groups highlighted as dark blue balls.
2.4.1 Nanodisc stability simulations
In order to test the stability of preformed nanodiscs in the CG representation, a CG nanodisc was solvated in a water box using the Solvate plug-in of VMD58 modified for use with CG water beads; sodium CG beads were added to neutralize the charge. The overall system had dimensions 127Å×128Å×83Å, and consisted of 11,444 CG beads. The system was minimized to eliminate steric clashes and equilibrated for 60 ns at 323 K. The nanodisc (DPPC lipids, scaffold proteins, and sodium ions) was then re-solvated into a larger water box with an overall size of 198 Å×207Å×177Å and 200 Na+ as well as 200 Cl− ions were added resulting in a 62,583 bead system. This re-solvated nanodisc system was then simulated for 62.5 ns at 300 K, for 100 ns at 323 K, and for 285 ns at 353 K.
2.4.2 Nanodisc self-assembly simulations
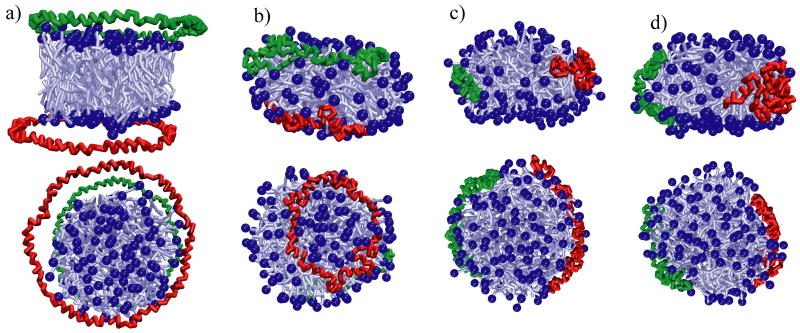
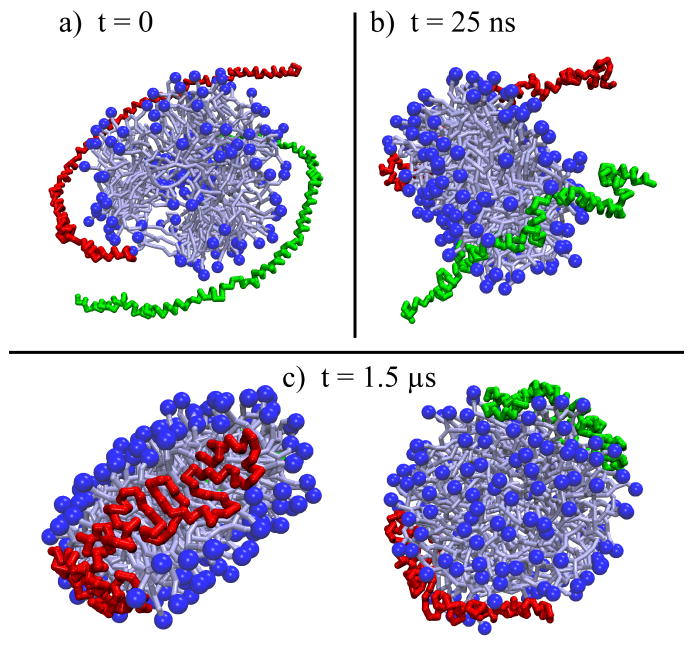
Nanodiscs self-assemble upon the removal of cholate from a mixture of cholate, scaffold proteins, and lipids, forming homogeneous discoidal particles of uniform size and shape.28 To test the ability of the model to reproduce nanodisc assembly, we performed a series of simulations. The first self-assembly simulation, referred to as SimND, started from a system in which the scaffold proteins were left in a circular shape, but shifted from their equilibrium position (Figure 4) on the nanodisc. Taking the z-axis as perpendicular to the plane of the nanodisc, the initial configuration of the proteins was the following: one scaffold protein was shifted 25 Å down the z-axis and 10 Å along the x-axis, and the other was shifted 25 Å up the z-axis and 10 Å along the y-axis (Figure 6a). The system was solvated in a water box (with ions added for neutrality) of dimensions 158 Å×158Å×115Å, resulting in a system of 24,046 beads, and simulated at a temperature of 323 K for 1 μs.
Figure 6.
Self-assembly of nanodiscs with lipids initially in a bilayer from simulation SimND. For the nanodisc, the head groups are shown as dark-blue balls and the rest of the lipids as light-blue sticks; the scaffold proteins are drawn as green and red tubes. The side views of the system are shown in the upper row of snapshots, top-down views are shown in the bottom row. In (a) the initial configuration is shown: the disk of lipids is pre-assembled and the circular scaffold proteins are shifted 25 Å above and below the disk, as well as shifted asymmetrically o3 the z-axis by 10 Å. Other views present snapshots of the simulation at 300 ns (b), 500 ns (c) and 1 μs (d).
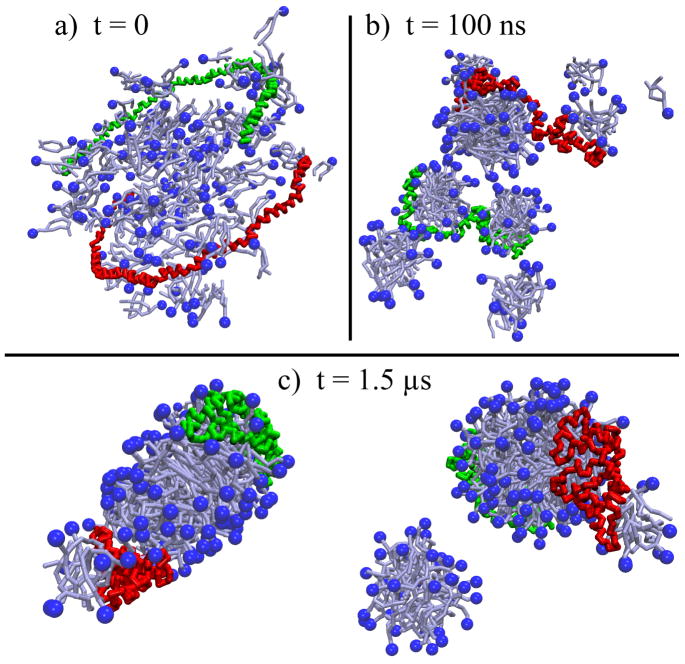
The second self-assembly simulation, referred to as SimRN, started with the two scaffold proteins in a half-circle shape and separated by 40 Å, and 160 DPPC lipids randomly scattered throughout the periodic cell (Figure 7a). The system was solvated in a water box of dimensions 180Å×181Å×137Å, and neutralized with ions, resulting in a system of 36,018 beads which was simulated at a temperature of 323 K for 1.5 μs.
Figure 7.
Self-assembly of nanodiscs with lipids initially randomly scattered from simulation SimRN. (a) Initially, the two membrane scaffold proteins (in green and red) are placed as half-circles separated by 40 Å with the lipids (shown in light-blue with the head groups highlighted in dark blue) randomly placed. (b) The lipids quickly form small micelles which then fuse into large micelles, some of which are associated with the scaffold proteins and others still disjointed. (c) At 1.5 μs, the membrane scaffold proteins are seen to have aggregated with the majority of the lipids and formed a discoidal structure (two views are shown, for clarity). The disjointed lipids are found in one large and one small micelle.
The third self-assembly simulation, referred to as SimMC, was performed in order to more accurately represent the actual assembly of a nanodisc particle. Initially, 160 DPPC lipids and 320 cholates were randomly scattered and simulated to form a lipid/cholate micelle. The system was solvated in a water box of dimensions 162Å×162Å×162Å and neutralized with ions resulting in a total size of 33,573 beads; the system was simulated for 150 ns at 323 K. Cholate, water, and ions were removed from the system leaving only the lipids, which were then used in the self-assembly simulation. Two scaffold proteins separated by 40 Å were placed as half-circles around the lipids in a pseudo-micelle (Figure 8a). The system was resolvated and neutralized with ions resulting in a system of 24,933 beads with dimensions 146Å×173Å×123Å; this system was simulated at a temperature of 323 K for 1.5 μs.
Figure 8.
Self-assembly of nanodiscs with lipids initially organized in a pseudo-micelle from simulation SimMC. The lipids (shown in light-blue with their head groups highlighted in blue) were initially equilibrated with cholate to form a micelle. The cholate was then removed leaving only the lipids in a pseudo-micelle structure. (a) Two membrane scaffold proteins (shown in green and red) are placed in half-circles around the pseudo lipid micelle. (b) The lipids quickly form a bilayer. (c) The proteins slowly aggregate with the lipid bilayer, forming a discoidal structure with the proteins aggregated on either side (top and side views are shown of the final structure, right and left respectively).
3 Results and Discussion
Using the model for coarse-graining protein-lipid systems described above, we performed a series of simulations on the intact nanodisc to asses the model’s stability, followed by a simulation of a disassembled nanodisc to test the ability of the system to self-assemble. We discuss the computational speed-up provided by our model, simulations on the stability of CG nanodiscs, and simulations SimND, SimRN, and SimMC of the final stages of nanodisc assembly.
3.1 Speed-up through the CG method
All-atom MD simulations must be performed with integration time steps of 1–2 fs, because the fastest atomic-level vibration in biopolymers is an oscillation (of hydrogen atoms covalently bound to heavier atoms) with a period of ~10 fs. For CG simulations a much longer integration time step of 25 fs can be used, rendering simulations of the CG model 25 times faster than equivalent all-atom simulations. In addition, CG relaxation processes are known to occur faster due to differences in the potential energy surface. For example, we calculated the self-diffusion coefficient of water from our CG simulations to be 4.0 × 10−5 cm2/s, which is two times the experimental value. Marrink et al.30 reported that during their simulations CG diffusion coefficients were 3–6 times larger than those of the equivalent all-atom systems.30 This implies that CG dynamics speed up relaxation processes 2–6 fold compared with all-atom MD. The combination of longer time steps and faster relaxation results in a computational speedup of 50–150 times from an all-atom to a CG description. The reduction in particle number in the CG approach implies another factor of 10 in speedup. For judgement of the actual speed of simulations for the same system in all-atom and CG representations one should analyze the results of actual calculations. In the present study, simulations were done on a cluster of 48 Athlon 2600+ processors. With a working size of approximately 30,000 particles for the CG system of the nanodisc in solution, the performance of simulations on this cluster reached 150 ns in a day. The same system (nanodisc in solution) in an all-atom representation including 300,000 particles achieved only 0.1 ns a day; thus the CG simulations are 1500 times faster.
3.2 Stability of nanodiscs in the CG description
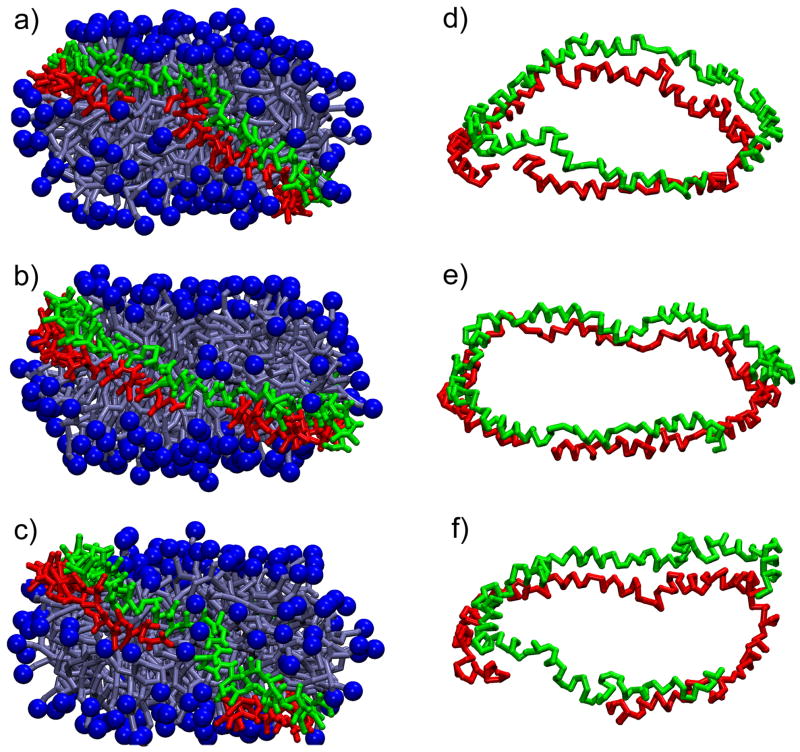
Previous all-atom MD simulations revealed that nanodiscs maintain their discoidal shape with minimal distortion for up to 7 ns.27 The CG model should reproduce this stability. A series of simulations was performed (at 300 K for 62.5 ns, at 323 K for 100 ns, and at 353 K for 285 ns) to test the ability of the CG model to maintain the nanodisc geometry. The simulations are presented in Figure 5. The nanodiscs, at all temperatures, indeed maintained their initial discoidal shape. Experimentally, nanodiscs are stable at temperatures of up to about 330 K, at which point irreversible aggregation of the nanodisc particles occurs, albeit on an hour time scale.26 Due to the short simulation times the nanodiscs were not expected to denature. However, even though full denaturation was not expected, the diameter of nanodiscs was expected to increase with rising temperature. The diameters of the nanodiscs, 9.5 nm at 300 K, 10.5 nm at 323 K, and 10.6 nm at 353 K (as measured from the CG simulations) did, in fact, increase with temperature and correlated well with experimental SAXS measurements (see Table 4).25,26 The gaps between the ends of membrane scaffold protein strands increase with temperature suggesting that perhaps disassembly of nanodiscs results from the loss of favorable salt-bridging (represented in our CG model through CG particle charges) between the two scaffold protein strands (Figure 5d, e, and f).
Figure 5.
CG nanodiscs consisting of a DPPC lipid bilayer (shown with the head groups highlighted as dark blue balls and the remainder of the lipid depicted in light blue using a licorice representation) and two membrane scaffold proteins (green and red licorice representation). Nanodiscs resulting from simulations at 300 K for 62.5 ns (a), at 323 K for 100 ns (b), and 353 K for 285 ns (c) are shown. The scaffold proteins (shown with their backbone only) at each temperature, 300 K (d), 323 K (e), and 353 K (f), maintained a helical content of about 70%.
TABLE 4.
Comparison of nanodisc diameter and lipid bilayer thickness resulting from CG simulations with values experimentally observed.25,26
| temperature | CG | experimental | |
|---|---|---|---|
| Nanodisc diameter | 300 K | 9.5 nm | 9.6 nm |
| 323 K | 10.5 nm | 10.5 nm | |
| 353 K | 10.6 nm | – | |
|
| |||
| bilayer thickness | 300 K | 5.5 nm | 5.7 nm |
| 323 K | 5.2 nm | 5.4 nm | |
| 353 K | 5.0 nm | – | |
The scaffold proteins are comprised of 11 and 22 residue helical segments punctuated by prolines and glycines, resulting in a primarily helical secondary structure.5,6,7,8 All-atom simulations showed that the proteins maintain a helical structure with kinking around the proline and glycine residues.27 The range of scaffold protein dihedral angles (χℓ in eq 5) from the initial, minimized structure was taken to represent the range indicating α-helical structure; using this criterion, the scaffold proteins in the CG nanodisc simulations remained primarily (~70%) helical; the non-helical part became unstructured (Figure 5d, e, and f)
The conformation and thickness of the lipid bilayer can also be used as a measure of the stability of the CG model. All simulations of the nanodiscs showed that the boundary lipids (the lipids closest to the membrane scaffold proteins) were perturbed in such a way as to minimize the hydrophobic mismatch at the protein-lipid interface which resulted in distortion of the lipid packing in this region (Figure 5a, b, and c). These observations are consistent both with experimental investigations25,26 and with all-atom simulations,27 which revealed a similar perturbation of boundary lipids. A CG nanodisc simulated at 300 K achieved a bilayer thickness of ~5.5 nm, whereas at higher temperature the bilayer was 0.2–0.5 nm thinner. This decrease in bilayer thickness with increased temperature was similarly observed in SAXS experiments25,26 studying the effect of temperature on nanodiscs (see Table 4).
3.3 Simulations of nanodisc self-assembly
The process by which nanodiscs self-assemble into homogeneous discoidal particles upon the removal of cholate from an initial mixture of cholate, lipid, and membrane scaffold proteins is unknown. The development of the presented CG model provides an opportunity to investigate the intermediate states of nanodisc assembly and disassembly.
Simulating the complete assembly process of nanodiscs would require beginning with a large number of cholate-lipid micelles in solution with added scaffold proteins, and then removing the cholate over a time scale of milliseconds to seconds. Such long times are beyond the realm of current coarse-graining simulation methods. With future refinements of the CG approach that may employ, for example, an implicit solvent model for water, the time scales of full assembly may become accessible. For the present purpose, however, we restricted our CG assembly simulations to investigating a hypothetical final step of nanodisc assembly (simulation SimND), starting with an initial random distribution of lipids (simulation SimRN), and assembly of nanodiscs with instantaneous removal of cholate (simulation SimMC).
In simulation SimND, a preformed nanodisc was used with an intact lipid bilayer (Figure 4b); the two scaffold proteins were left in their circular shape, but shifted horizontally and vertically away from the bilayer (Figure 6a). Initially, the scaffold proteins are too far from the lipids to have any interaction with them (Figure 6a), so they were seen to drift in the water bath until randomly coming close enough to the lipids to interact with them; the initial contact occurred within the first 10 ns. The early stages of contact appear to be driven primarily by hydrophilic interactions between charges on the scaffold proteins and the lipid head groups. After 100 ns both scaffold protein strands came to rest along the lipid head groups, and the lipid bilayer became distorted into a micelle-like structure with a few of the lipids shielding the remaining lipid tails from the water. The system remained in this conformation for almost 400 ns (see Figure 6b). Eventually the hydrophobic faces of the scaffold proteins became organized and started to interact with the lipids along the edges of the nanodisc. As this occurred for each of the two scaffold proteins separately, the proteins’ original ring shape was broken and the scaffold proteins began to orient themselves with their hydrophobic face toward the lipid and hydrophilic face toward water. In this process the proteins lost most of their previous contacts with the lipid head groups (Figure 6c). This process began around 100 ns into simulation SimND for one of the scaffold proteins and 220 ns for the other. After approximately 500 ns of simulation, the scaffold proteins reoriented and wrapped themselves around the exposed edge and the lipids returned to their original bilayer structure. The scaffold proteins wrapped themselves around most of the lipids and only a few lipids remained outside of the usual bilayer organization (Figure 6c). The structure of the proteins did not change significantly between 500 ns and 1 μs in simulation SimND (compare Figure 6c and d). At the end of 1 μs of simulation the lipids were arranged as a discoidal bilayer, and the proteins were attached to opposite edges of the lipid bilayer core (Figure 6d). The distances between the ends of the two protein strands fluctuated during the last 500 ns of simulation SimND from between 25 to 45 Å for one protein and 50 to 80Å for the other protein; the two protein strands did not interact with each other.
In simulation SimRN, the two scaffold proteins were placed 40 Å away from each other in a half-circle and lipids were randomly scattered around them (Figure 7a). Within 1 ns the lipids formed small micelles, with those initially close to the scaffold proteins already associated and interacting with the hydrophobic face of the proteins and those not close enough forming individual micelles. Over time the various small micelles diffused around, eventually colliding and fusing to form larger micelles (Figure 7b). At ~200 ns, the micelles and the membrane scaffold proteins already formed a nanodisc-like particle. From 200 ns to 525 ns, smaller micelles continued to fuse with larger micelles as well as fusing with and increasing the size of the nanodisc particle. By 525 ns, all but two micelles had fused with the nanodisc particle; by the end of the simulation (1.5 μs) the two separate micelles still had not fused with the nanodisc; at this point one contained 37 lipids and the other 7 lipids. (Figure 7c). The nanodisc particle had a main discoidal lipid bilayer portion consisting of 108 lipids to which the two membrane scaffold proteins were associated. One of the membrane scaffold proteins was also bent allowing for 8 lipids to aggregate, but turned outward from the main lipid bilayer (Figure 7c). The membrane scaffold proteins, although attached to the lipid bilayer were very disordered.
In simulation SimMC, the two scaffold proteins were placed as half-circles around a large pseudo-micelle (Figure 8a). This micelle represented a lipid-cholate micelle after the instantaneous removal of cholate. Within 25 ns the lipids formed a bilayer structure even though only a portion of the membrane scaffold proteins were associated with the lipids at this point (Figure 8b). From 25 ns to 175 ns the scaffold proteins slowly aggregated with the lipids, and by 175 ns both scaffold proteins had completely associated with the lipids. The scaffold proteins were attached to either side of the lipid bilayer and continued to rearrange themselves until the end of the simulation (1.5 μs), although the two scaffold proteins never came close enough to interact with each other (Figure 8c). The membrane scaffold proteins were disordered, but resembled loosely a picket-fence like orientation.
The results of simulations SimND, SimRN, and SimMC show that the lipids can easily form a discoidal bilayer structure; however, the scaffold proteins do not assume the expected “belt-like” conformation (Figure 1). Instead, once the two scaffold proteins attach to the lipid bilayer, they interact separately with the bilayer, with minimal interaction with each other; their arrangement resembles the picket-fence model,11 in which the two membrane scaffold proteins bind on opposite sides of the nanodisc with their helices directed up and down the z-axis covering the hydrophobic surface like fence posts. However, closer inspection reveals that the scaffold proteins in simulation SimND and SimMC are rather disordered with only a very faint resemblance of the picket-fence model and in SimRN the scaffold proteins are even more disordered. It is possible that with further simulation the two strands of membrane scaffold protein start interacting with each other, forming favorable salt-bridges with the other strand as opposed to themselves,5,6,7,8 and perhaps arranging themselves towards the generally accepted double-belt model.5 The picket-fence structure seen in simulations SimND, SimRN, and SimMC, could be only an intermediate in the assembly process, not the final state. It is, of course, also possible that this state is an artifact of the present CG model or of the initial conditions chosen for the simulations.
4 Conclusion
We have developed a protein-lipid CG model and employed it in describing nanodiscs and their aggregation on a microsecond time scale. The simulations suggest that pre-assembled double belt nanodiscs in the CG representation are stable over long periods of time, with diameter and thickness remaining close to experimental values (Figure 5). However, self-assembly simulations in which the lipids are initially in a bilayer (Figure 6), randomly placed (Figure 7), or in a pseudo-micelle (Figure 8) with added apo A-1 proteins lead to discoidal protein-lipid particles that are stable, but with the proteins forming a disordered fence instead of a double belt structure. Such an arrangement of proteins may correspond to an intermediate arrangement in HDL formation, that will eventually adhere to the more ordered double belt form. Alternatively, the disordered fence structure may represent a final state for discoidal lipoprotein particles. Further simulations covering longer timescales and employing improved CG forcefields should allow us to decide between these alternatives.
The results of our simulations show that the developed CG model, although simple, can capture the dynamics of large molecular aggregates with presently available computational resources. The clearest path to improving the present method is to incorporate more quantitative data into the model, an approach already employed for the development of other CG lipid models.29,32 To this end, we have begun working on the introduction of statistically developed potentials for the interaction between CG beads. Such potentials can be obtained from all-atom simulations and analysis of interactions of the groups that are assigned to single CG beads. These new potentials could lead to a more accurate CG protein-lipid model. As a further improvement, an implicit solvent model replacing water seems to be particularly well suited for a CG system, and might be easier to implement in the CG case than for all-atom descriptions because in CG models one has already dispensed with many of the fine details that are difficult to treat using implicit solvents. The use of an implicit solvent model combined with a CG description promises a further computational speed-up, which may permit one to capture assembly dynamics on the time scale of milliseconds.
One must keep in mind that while the CG approach can be used for large-scale descriptions of macromolecular complexes like lipoprotein complexes, it cannot account for the detailed behavior of individual residues. Presently, the CG approach seems suitable for studies of stability, self-assembly, and large mechanical motions and transformations of systems such as biological membranes, protein-lipid complexes, and mechanical nano-machines. Future improvements, including the use of derived potentials and of an implicit solvent model, may yield faster and more accurate results, although by their very nature CG models will never offer the same level of detail as atomistic simulations.
Acknowledgments
The authors would like to acknowledge the great inspiration they have received in this research through the published work of Michael Klein and through important personal encounters and discussions with him. The authors also acknowledge insight provided by Siewert Marrink in the field of CG modeling, and by Stephen Sligar and the Sligar group into the properties of nanodiscs. The authors thank Jim Phillips for his assistance in modifying NAMD for CG simulations. The work reported was supported by grants from the National Institutes of Health PHS-5-P41-RR05969 and 1 R01 GM067887. The authors gladly acknowledge supercomputer time provided by the Pittsburgh Supercomputer Center and the National Center for Supercomputing Applications via National Resources Allocation Committee grant MCA93S028. P.F. also acknowledges support from the NSF Graduate Research Fellowship Program.
References
- 1.Wang M, Briggs MR. Chem Rev. 2004;104:119–137. doi: 10.1021/cr020466v. [DOI] [PubMed] [Google Scholar]
- 2.Davidson WS, Silva RAGD. Curr Opin Lipidol. 2005;16:295–300. doi: 10.1097/01.mol.0000169349.38321.ad. [DOI] [PubMed] [Google Scholar]
- 3.Boguski MS, Freeman M, Elshourbagy NA, Taylor JM, Gordon JI. J Lipid Res. 1986;27:1011–1034. [PubMed] [Google Scholar]
- 4.Nolte RT, Atkinson D. Biophys J. 1992;63:1221–1239. doi: 10.1016/S0006-3495(92)81698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 6.Klon AE, Jones MK, Segrest JP, Harvey SC. Biophys J. 2000;79:1679–1685. doi: 10.1016/S0006-3495(00)76417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klon AE, Segrest JP, Harvey SC. Biochemistry. 2002;41:10895–10905. doi: 10.1021/bi020315m. [DOI] [PubMed] [Google Scholar]
- 8.Klon AE, Segrest JP, Harvey SC. J Mol Biol. 2002;324:703–721. doi: 10.1016/s0022-2836(02)01143-9. [DOI] [PubMed] [Google Scholar]
- 9.Segrest JP. Chem Phys Lipids. 1977;18:7–22. doi: 10.1016/0009-3084(77)90023-8. [DOI] [PubMed] [Google Scholar]
- 10.Jonas A, Kézdy KE, Wald JH. J Biol Chem. 1989;264:4818–4824. [PubMed] [Google Scholar]
- 11.Phillips JC, Wriggers W, Li Z, Jonas A, Schulten K. Biophys J. 1997;73:2337–2346. doi: 10.1016/S0006-3495(97)78264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brouillette CG, Anantharamaiah GM. Biochim Biophys Acta. 1995;1256:103–129. doi: 10.1016/0005-2760(95)00018-8. [DOI] [PubMed] [Google Scholar]
- 13.Rogers DP, Roberts LM, Lebowitz J, Engler JA, Brouillette CG. Biochemistry. 1998;37:945–955. doi: 10.1021/bi9713512. [DOI] [PubMed] [Google Scholar]
- 14.Wald JH, Coormaghtigh E, De Meutter J, Ruysschaert JM, Jonas A. J Biol Chem. 1990;265:20044–20050. [PubMed] [Google Scholar]
- 15.Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. J Biol Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 16.Panagotopulos SE, Horace EM, Maiorano JN, Davidson WS. J Biol Chem. 2001;276:42965–42970. doi: 10.1074/jbc.M106462200. [DOI] [PubMed] [Google Scholar]
- 17.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Proc Natl Acad Sci USA. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva RAGD, Hilliard GM, Li L, Segrest JP, Davidson WS. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Miller A, Rusch SL, Kendall DA. Biochemistry. 2004;43:13185–13192. doi: 10.1021/bi049485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayburt TH, Sligar SG. Proc Natl Acad Sci USA. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayburt TH, Sligar SG. Prot Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Biotechniques. 2003;35:556–560. 562–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 23.Duan H, Civjan NR, Sligar SG, Schuler MA. Arch Biochem Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Baas BJ, Denisov IG, Sligar SG. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 26.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Biophys J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayburt TH, Grinkova YV, Sligar SG. Nano Letters. 2002;2:853–6. [Google Scholar]
- 29.Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Moore PB, Klein ML. J Phys Chem B. 2001;105:9785–9792. [Google Scholar]
- 30.Marrink SJ, de Vries AH, Mark AE. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- 31.Stevens MJ. J Chem Phys. 2004;121:11942–11948. doi: 10.1063/1.1814058. [DOI] [PubMed] [Google Scholar]
- 32.Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML. J Phys Chem B. 2001;105:4464–4470. [Google Scholar]
- 33.Nielsen SO, Lopez CF, Srinivas G, Klein ML. J Phys: Condens Matter. 2004;16:R481–R512. [Google Scholar]
- 34.Nielsen SO, Ensing B, Ortiz V, Moore PB, Klein ML. Biophys J. 2005;88:3822–3828. doi: 10.1529/biophysj.104.057703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez CF, Nielsen SO, Ensing B, Moore PB, Klein ML. Biophys J. 2005;88:3083–3094. doi: 10.1529/biophysj.104.053769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickholz M, Saiz L, Klein ML. Biophys J. 2005;88:1524–1534. doi: 10.1529/biophysj.104.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faller R, Marrink SJ. Langmuir. 2004;20:7686–7693. doi: 10.1021/la0492759. [DOI] [PubMed] [Google Scholar]
- 38.Marrink SJ, Mark AE. J Am Chem Soc. 2003;125:15233–15242. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- 39.Marrink SJ, Mark AE. J Am Chem Soc. 2003;125:11144–11145. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 40.Marrink SJ, Risselada J, Mark AE. Chem Phys of Lipids. 2005;135(2):223–244. doi: 10.1016/j.chemphyslip.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 41.de Vries AH, Yefimov S, Mark AE, Marrink SJ. Proc Natl Acad Sci USA. 2005;102:5392–5396. doi: 10.1073/pnas.0408249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens MJ, Hoh JH, Woolf TB. Phys Rev Lett. 2003;91:188102. doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 43.Kumar PS, Gompper G, Lipowsky R. Phys Rev E. 1999;60:4610–4618. doi: 10.1103/physreve.60.4610. [DOI] [PubMed] [Google Scholar]
- 44.Shillcock JC, Lipowsky R. Nature Mater. 2005;4:225–228. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 45.Ayton GS, Smondyrev AM, Bardenhagen SG, McMurtry P, Voth GA. Biophys J. 2002;83:1026–1038. doi: 10.1016/S0006-3495(02)75228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayton GS, Voth GA. Biophys J. 2002;83:3357–3370. doi: 10.1016/S0006-3495(02)75336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang R, Ayton GS, Voth GA. J Chem Phys. 2005;122:244716–244728. doi: 10.1063/1.1931651. [DOI] [PubMed] [Google Scholar]
- 48.Cieplak M, Hoang TX. Phys Rev E. 1998;58:3589–3596. [Google Scholar]
- 49.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 50.Doruker P, Jernigan RL, Navizet I, Hernandez R. Int J Quantum Chem. 2002;90:822–837. [Google Scholar]
- 51.Das P, Matysiak S, Clementi C. Proc Natl Acad Sci USA. 2005;102(29):10141–10146. doi: 10.1073/pnas.0409471102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matysiak S, Clementi C. J Mol Biol. 2004;343(1):235–248. doi: 10.1016/j.jmb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marrink SJ. Proceedings of Falk Symposium. Vol. 139. Dordrecht: Kluwer Academic; 2004. pp. 98–105. [Google Scholar]
- 55.Lindahl E, Hess B, van der Spoel D. J Mol Mod. 2001;7(8):306–317. [Google Scholar]
- 56.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feller SE, Zhang YH, Pastor RW, Brooks BR. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]