Abstract
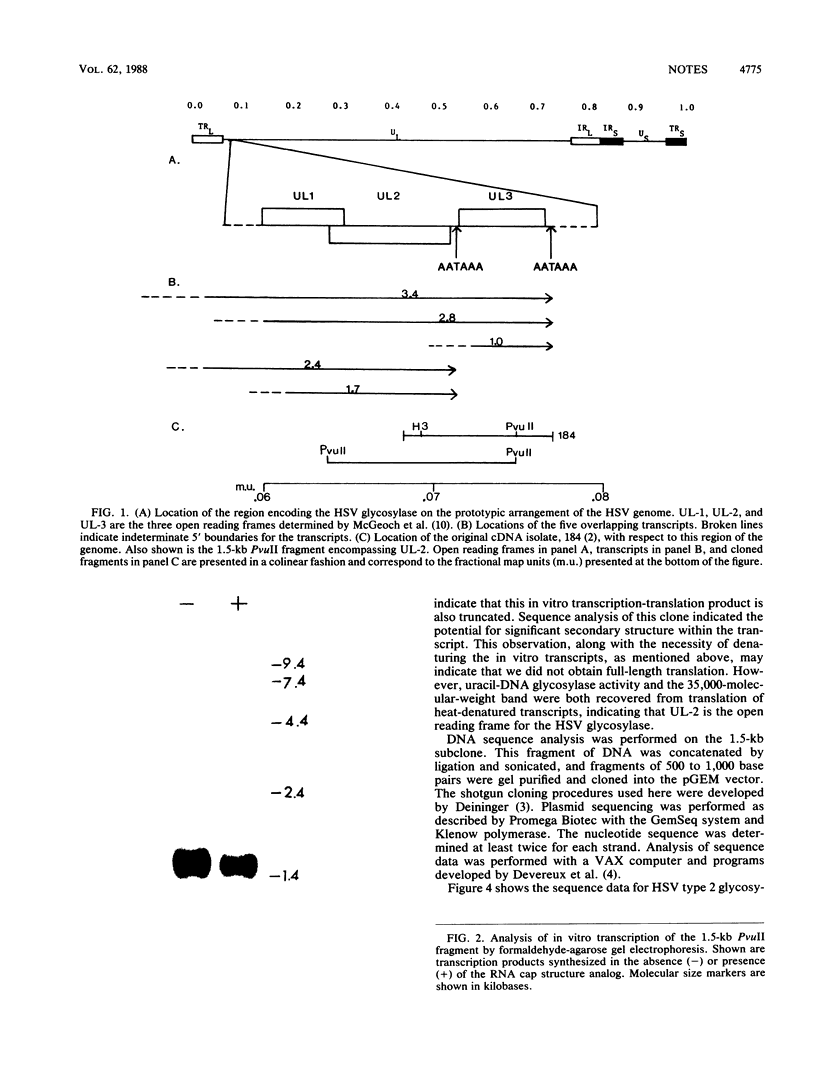
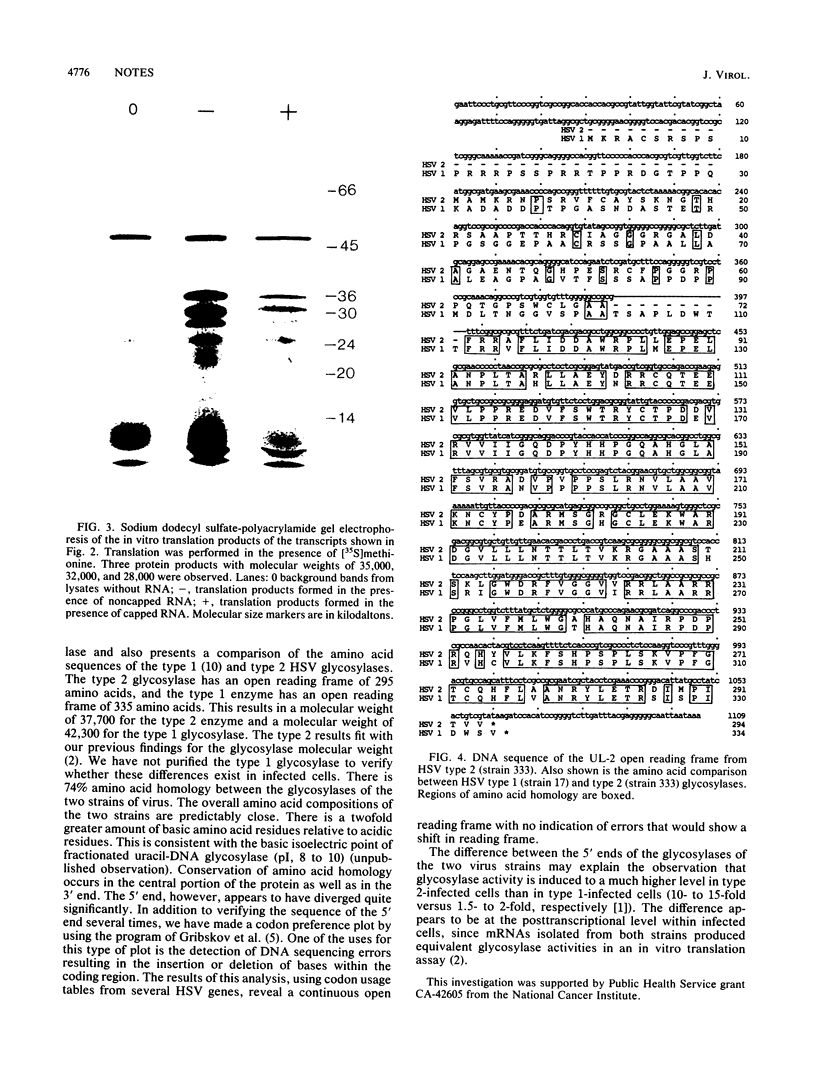
We have recently isolated a herpes simplex virus (HSV) type 2 (strain 333)-specific cDNA that encodes uracil-DNA glycosylase. This cDNA lies between 0.065 and 0.08 map units on the HSV genome. Within this region there are five overlapping transcripts which encompass three open reading frames. We have determined that the second open reading frame, UL-2, codes for glycosylase. In vitro transcription of the UL-2 region and subsequent translation yielded uracil-DNA glycosylase activity. Sequence analysis of the UL-2 open reading frame indicated a coding capacity of 295 amino acids. Comparison to the HSV type 1 (strain 17) sequence indicated that there is 74% amino acid homology between the two strains, with most of the conservation occurring in the middle and the 3' end. The 5' end, however, has diverged considerably.
Full text
PDF

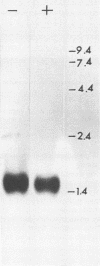
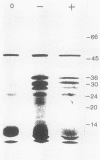
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Caradonna S. J., Cheng Y. C. Induction of uracil-DNA glycosylase and dUTP nucleotidohydrolase activity in herpes simplex virus-infected human cells. J Biol Chem. 1981 Oct 10;256(19):9834–9837. [PubMed] [Google Scholar]
- Caradonna S., Worrad D., Lirette R. Isolation of a herpes simplex virus cDNA encoding the DNA repair enzyme uracil-DNA glycosylase. J Virol. 1987 Oct;61(10):3040–3047. doi: 10.1128/jvi.61.10.3040-3047.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]