Summary
Saccharomyces cerevisiae is the simplest among the major eukaryotic model organisms for aging and diseases. Longevity in the chronological life span paradigm is measured as the mean and maximum survival period of populations of non-dividing yeast. This paradigm has been used successfully to identify several life-regulatory genes and three evolutionary conserved pro-aging pathways. More recently, Schizosaccharomyces pombe has been shown to age chronologically in a manner that resembles that of S. cerevisiae and that depends on the activity of the homologues of two pro-aging proteins previously identified in the budding yeast. Both yeast show features of apoptotic death during chronological aging. Here, we review some fundamental aspects of the genetics of chronological aging and the overlap between yeast aging and apoptotic processes with particular emphasis on the identification of an aging/death program that favors the dedifferentiation and regrowth of a few better adapted mutants generated within populations of aging S. cerevisiae. We also describe the use of a genome-wide screening technique to gain further insights into the mechanisms of programmed death in populations of chronologically aging S. cerevisiae.
Chronological aging in budding and fission yeast
S. cerevisiae has long been used as model system to uncover the mechanisms that lead to cellular and organismal aging. Thanks to its asymmetric cell division, a method to monitor the replicative life span of this budding yeast was established fifty years ago [1] and widely employed starting from the nineties [2–4]. Under the replicative life span (RLS) paradigm, the limited reproductive potential of individual mother cells is quantified by counting the total number of daughter cells generated before cell division stops. Each yeast strain is characterized based on its mean and maximum replicative life span, estimated by measuring the number of total buds produced by at least forty individual mother cells. Alternatively, the life span of S. cerevisiae is measured by monitoring the mean and maximum survival times of populations of post-mitotic yeast cells. The latter paradigm is referred to as the chronological life span (CLS) of yeast and is analogous to the way life span is monitored in metazoans. The CLS paradigm was introduced in 1996 [5] and since then has become more widely studied due to several reasons that have been discussed elsewhere [6]. For the purpose of this review it is sufficient to point out that both yeast aging paradigms have proven important for the identification of numerous gerontogenes. Here, we focus on the CLS paradigm.
For chronological aging studies, yeast cells are grown in synthetic complete medium containing glucose (SDC) and then either maintained in this medium without replenishing any of the nutrients or washed and incubated in water. Their survival is monitored until over 99% of the population dies by counting the number of cells that are able to form colonies on YPD rich medium (CFUs) [6]. Cells grown and left in SDC medium enter a post-diauxic phase and maintain high metabolic rates for the majority of their life span [6, 7]. Incubation in water, which we also refer to as “extreme calorie restriction”, instead causes entry into a longer-lived but low metabolism stationary phase [6]. Originally recognized as a life-extending intervention in rodents by McVay et al. [8], restriction of the number of calories consumed is known to promote longevity in organisms ranging from yeast to mice, although the anti-aging molecular mechanisms activated by this intervention are still not fully understood [9, 10]. In yeast, calorie restriction extends both CLS and RLS [6, 11–13]. Under the CLS paradigm, calorie restriction is induced either by incubation in water as described above, or by growing the cells in synthetic medium containing glucose ranging from 0.05% to 0.5% instead of the standard 2% [14]. Similar life spans are obtained when yeast are subject to either of the two calorie restriction regimes suggesting that switching yeast to water after saturation has been reached in standard SDC medium or growing them in low glucose medium from the beginning activate a common anti-aging response.
The CLS paradigm has recently been utilized to measure longevity in S. pombe [15]. Fission yeast are grown in either synthetic complete medium or in minimal medium (EMM) until the cultures reach saturation. The non-dividing cultures are maintained in medium and the CFUs are monitored by plating adequate dilutions of cells onto YE rich medium [15].
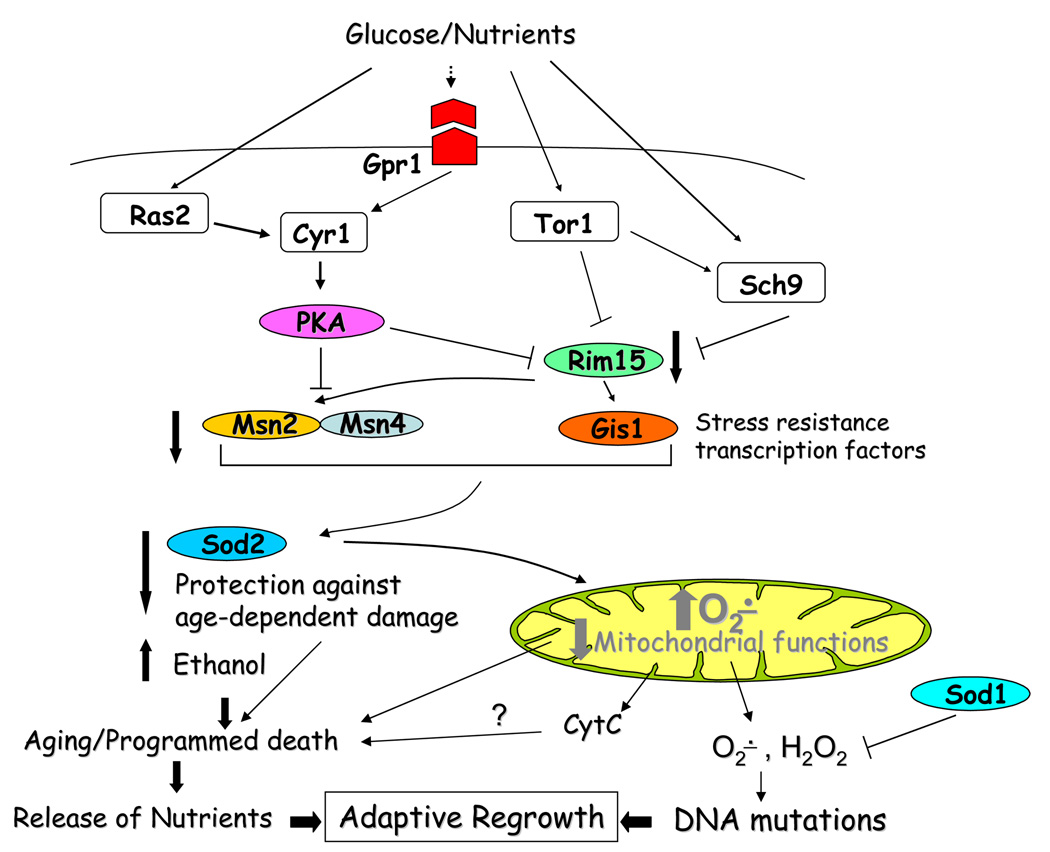
CLS studies in S. cerevisiae have led to the discovery of three life span-regulatory pathways, the Sch9, Tor, and Ras/PKA pathways [7, 16], which are conserved in part and regulate aging also in higher eukaryotes [9, 17–19] (FIG. 1). When glucose and other nutrients are present, the signaling cascades activated by these pathways promote cell growth and division. When nutrients are scarce, the reduction of the Sch9, Tor, and Ras/PKA signaling causes cell division arrest and the activation of mechanisms responsible for cellular protection. The deletion of the gene coding for the serine-threonine kinase Sch9, extends the yeast CLS by 3 fold and promotes stress resistance (FIG. 1)[16]. Sch9 shares a high degree of homology with Akt/PKB, SGK and S6K, whose inactivation promotes longevity extension in worms, flies, and mice [9, 20]. Similarly to Sch9, the serine/threonine activity of Tor1 is promoted by the presence of nutrients and its inactivation or that of other members of the Tor pathway leads to yeast CLS extension and promotes protection against stress [18, 21]. Tor1 is a component of the TORC1 complex, which has been recently shown to directly phosphorylate and promote the activation of Sch9 [22]. Genetic data indicate that Tor1 plays its pro-aging role partially by stimulation of Sch9 (P. Fabrizio, unpublished results) suggesting a certain degree of overlap between the Sch9 and Tor1 pathways. The TORC1 complex is widely conserved among species and in both C. elegans and Drosophila a diminished TORC1 activity slows down aging [23–25]. Ras/PKA, another CLS regulatory pathway identified in yeast, has recently been shown to play a conserved role in mice. In yeast, lack of Ras2 or a reduced activity of adenylyl cyclase (Cyr1), which is activated by the Ras proteins, causes life span extension and stress resistance [7] (FIG. 1). Similarly, mice lacking type 5 adanylyl cyclase (AC5) live 30% longer than wild type littermates [26]. Analogously to ras2 and cyr1 yeast mutant cells, fibroblasts and cardiac myocytes isolated from AC5 KO mice are resistant to oxidative stress. This resistance is, at least in part, mediated by mitochondrial superoxide dismutase, whose protein level is higher in AC5 KO cells as compared to wild type cells. Notably, the resistance at the myocite level translates into the resistance to age-induced cardiomyopathy in the mouse [26]. Together, these results confirm the association between longevity extension and ability to withstand stress. Importantly, this association has been observed in all the chronologically long-lived yeast and in the majority of the worm long-lived mutants identified so far. In some cases, long-lived flies and mice are also resistant to stress suggesting that increasing cellular protection against damage and possibly increasing repair and replacement may be conserved molecular strategies to delay aging in all species [17, 27].
Figure 1.
Programmed aging and adaptive regrowth in S. cerevisiae. Glucose and nutrients activate the Sch9, Tor1, and Ras/PKA pathways, which in turn promote the down-regulation of Msn2/4/Gis1-dependent stress resistance systems and the production of superoxide. High levels of superoxide damage the mitochondria causing aging and apoptosis. Additionally, superoxide causes DNA damage and increases DNA mutations. Cell death leads to the release of nutrients that are utilized by “regrowth” mutants to reenter the cell cycle. Cytochrome C released from the mitochondria may function to signal apoptotic death. Ethanol, accumulated during exponential growth, is also involved in the activation of programmed aging.
The Sch9, Tor1, and Ras/PKA pathways have been investigated thoroughly and some of the principal mediators of these life regulatory pathways have been uncovered. Among these are the stress resistance transcription factors Msn2/Msn4, whose nuclear localization is inhibited by the PKA activity and the protein kinase Rim15 [16], whose cytosolic retention is promoted by both a PKA-dependent phosphorylation and Sch9 activation and blocks the activation of Gis1, another stress-dependent transcription factor [28] (FIG. 1). Lack of Rim15 causes the total reversion of the longevity extension phenotype observed in the three long-lived mutants indicating that the pro-aging pathways controlled by Sch9, Tor, and Ras2 converge on Rim15 [21]. However, although Msn2 and Msn4, whose activation is controlled by Rim15, are required for the longevity extension associated with a reduced Ras/PKA pathway activity [7], they do not seem to play a crucial role in the regulation the life span of the sch9 mutants [16]. Furthermore, the double mutant lacking both Ras2 and Sch9 lives significantly longer that either one of the single mutants suggesting that: 1) the Sch9 and Ras/PKA pathways differentially modulate the activation of signaling proteins downstream of Rim15, 2) either pathway may function also through the activation of yet to be identified pathway-specific mediators [21]. The Tor pathway functions through Rim15 to control longevity in part by modulating Sch9 (see above).
The use of S. pombe for chronological aging studies has only recently begun. S. pombe and S. cerevisiae are phyologenetically very distantly related and neither of them seems to be closer to humans than the other, although the physiology of S. pombe is considered more similar to that of higher eukaryotes [29, 30]. Thus, the two yeast can complement each other as aging models and both contribute to the identification of important longevity mediators relevant to aging of higher eukaryotes.
To date a pioneering study has shown that, in analogy with S. cerevisiae, the activation of the S. pombe homologues of PKA and Sch9 negatively affects the CLS of the fission yeast. However, of the two Sch9 homologues present in S. pombe only one, Sck2, controls aging [15]. Interestingly, in fission yeast the deletion of the only gene coding for the PKA catalytic subunit is not lethal and extends the CLS [15]. By contrast, in budding yeast the activity of at least one of the three redundant PKA catalytic subunits (Tpk1-3) is required for cell viability and the deletion of none of the Tpk coding genes has been directly associated with longevity extension. These differences between S. pombe and S. cerevisiae are not surprising in light of the distant phylogenetic relationship between these two species [31].
Apoptosis in chronologically aging yeast
Features typical of apoptotic death, including chromatin fragmentation and condensation and exposure of phosphatidylserine on the external layer of the cellular membrane, have been originally described in a S. cerevisiae cdc48 mutant and in wild type cells treated with hydrogen peroxide [32, 33]. Overexpression of mammalian pro- or anti-apoptotic Bax or Bcl2 in yeast causes or prevents an apoptosis-like death, respectively [34–36]. Furthermore, homologues of metazoan caspases and of the apoptotic inducing factor (AIF) have been identified on the yeast genome and shown to be implicated in hydrogen peroxide-induced apoptosis [37, 38]. Besides oxidative stress, other pro-apoptotic stimuli are known in yeast. These include acetic acid, the yeast mating factor alpha, the plant antibiotic osmotin, osmotic stress, decreased actin dynamics, an chronological aging [39–45]. Chronological aging-dependent apoptosis is discussed hereafter.
I. Programmed death and adaptive regrowth in S. cerevisiae: evidence for an altruistic aging program
Wild type yeast aging chronologically in medium show features of apoptotic death such as nuclear condensation/fragmentation, phosphatidylserine exposure, and caspase activation [44, 45]. Reactive oxygen species formation is enhanced in agreement with a central role for superoxide in the activation of yeast apoptosis during aging (see below) (FIG. 1) [44]. Almeida et al. have recently provided evidence for the production of nitric oxide (NO) in aging yeast. According to these authors, NO contributes to superoxide production and its removal by treatment with oxyhaemoglobin, a NO scavenger, extends the CLS of wild type cells [46]. A few genetic interventions have been described that delay chronological aging and the appearance of the apoptotic features associated to it. Among these are the disruption of the yeast caspase YCA1 gene, AIF homologue (AIF1), and NDE1 (coding for the yeast homologue of the AIF-homologous mitochondrion associated inducer of death, AMID) and the overexpression of the stress-dependent transcription factor Yap1 [37, 45, 47], although the most effective so far identified are the deletion of either RAS2, TOR1, or SCH9, which prolong the mean CLS by up to 3-fold [7, 16].
A non genetic intervention that promotes the life span extension of wild type yeast is calorie restriction (see previous section). Under extreme calorie restriction, yeast double their longevity indicating that they have the potential to modulate the length of their life span according to the environment. This is a crucial point in that incubation in medium after cell division has ceased, causes a sort of accelerated aging in cells that in a different environment would live much longer (see below). When wild type cells incubated in medium are compared to long-lived sch9Δ mutants, besides the apoptotic markers, a striking difference is observed in terms of activation of stress response. sch9Δ mutants are less susceptible to oxidative stress than wild type cells. Their resistance to oxidants depends largely on the expression of SOD2 and the activity of the corresponding protein (mitochondrial superoxide dismutase), which are both higher in the mutants [7, 44]. A similar phenotype has been observed in calorie restricted wild type cells (M. Wei and V. D Longo, unpublished results). Taken together, these results are consistent with the activation of an “aging program” that blocks cell protection and accelerates the death of the wild type yeast incubated in medium (FIG. 1). The existence of such a program may seem unlikely in a unicellular organism. However, several lines of evidence have demonstrated that, in the context of a population of millions of yeast, cellular “suicide” represents a survival strategy for the group [44, 48]. In fact, in approximately 50% of the wild type cultures used for chronological aging studies, after the majority of the population is dead, cellular “regrowth” is observed (CFUs increase by up to 100 folds). The percent of regrowth raises to 80% in mutants lacking SOD1, which codes for the cytosolic superoxide dismutase, suggesting a direct correlation between intracellular superoxide and frequency of regrowth. The regrowth phenotype, normally referred to as “adaptive regrowth”, has been characterized extensively [44]. Its principal features are: 1) Dependency on DNA mutations that accumulate during aging. 2) Requirement for the nutrients released by the dead cells present in the cultures. Both features are closely dependent on superoxide, which, on the one hand, promotes accelerated cell death and consequently nutrients accumulation and, on the other hand, causes DNA damage that facilitates the appearance of genetic mutants with the ability to reentry the cell cycle in conditions that normally do not promote growth (FIG. 1) [44]. Superoxide’s central role in causing DNA mutations is confirmed by a study showing that yeast mutants lacking SOD1 are among the top mutator strains with a mutation frequency (measured by monitoring the mutations in the gene coding for the arginine transporter CAN1) approximately six fold higher than that of wild type cells [49]. Importantly, the long-lived mutants, which are extremely protected against superoxide damage (including DNA damage), do not show adaptive regrowth, in agreement with a scenario of “escape” from an “altruistic” aging program that kills a vast majority of individuals in the culture in order for the few adapted mutants to survive (FIG. 1).[44]. The theoretical implications of the existence of an “altruistic aging program” will be discussed in the next subsection.
Here we want to emphasize that programmed death/adaptive regrowth is only one of the strategies available to yeast populations to overcome periods of starvation. As mentioned above, calorie restriction promotes life span extension and high levels of stress resistance in agreement with the activation of a “survival program” at the level of individual yeast cells [44]. Notably, adaptive regrowth as survival strategy is observed also in budding yeast isolated from the natural environment ruling out the possibility that it is a phenotype associated only with “genetically modified” yeast. Moreover, when laboratory strains are grown in grape extract (a more “natural” medium for yeast), the survival curves are very similar to those obtained when SDC medium is used excluding the possibility of artifacts due to the use of synthetic medium [44].
Further analysis of yeast apoptosis during chronological aging has revealed that the pH of the SDC medium of the aging cultures affects cell survival. By day 3 the pH of the SDC medium of yeast cultures reaches 3–3.5. When it is adjusted to 6–7, the survival of the culture is significantly extended [44]. In mammals the release of cytochrome C, which triggers the caspase cascade activation in apoptotic cells, is preceded by mitochondrial alkalinization and cytosol acidification [50]. Cytochrome C release has also been observed in yeast treated with acetic acid, alpha factor or overexpressing mammalian Bax, although its function in apoptosis activation has not been established yet [36, 40, 51]. A possibility still unexplored is that the extracellular and consequentely intracellular acidification observed in yeast aging cultures triggers apoptosis via the release of cytochrome C.
Another mediator of yeast apoptosis is ethanol. This carbon source is normally accumulated during fermentative growth but, in contrast with the common knowledge of its utilization when glucose is exhausted (diauxic shift), in chronologically aging cultures of wild type cells it is not metabolized and functions to shorten life span. In fact, when ethanol is removed from the incubation medium by evaporation, a significant life span extension is observed [11]. Moreover, ethanol addition to cultures switched to water rapidly kills the yeast suggesting an active role of ethanol in the activation of a death program [11].
Recent work by Allen et al. has characterized yeast stationary phase cultures grown in rich medium by using an equilibrium density-gradient centrifugation method [52]. After ~24 hours from the inoculation, the authors were able to distinguish two fractions of cells. The density of the lower fraction (high-density) increased steadily until day 7. An extensive characterization of the two fractions revealed that: 1) the high-density cells were mostly unbudded and showed several features of quiescent cells while the low-density cells were both budded and unbudded and morphologically more similar to dividing cells (non-quiescent). 2) The low-density cells lost viability more rapidly than the others over time, were more sensitive to heat-shock and produced more ROS than the quiescent cells. 3) The non-quiescent cells showed higher levels of apoptotic markers (DNA fragmentation and phosphatidylserine exposure) than non-quiescent cells [52].
It will be very important to study the dynamics of the two types of cells to establish whether their ratio changes over time and whether quiescent cells become non-quiescent once the apoptotic program is activated. The apoptotic features described above for the non-quiescent cells resemble closely those observed in wild type cells aging chronologically in medium containing ethanol suggesting the presence of non-quiescent cells in these cultures as well. Indeed budded cells have been identified in chronologically aging populations by Weinberger et al. These authors used flow cytometry to estimate the percent of G1/G2 cells in aging cultures and showed a tighter G1-arrest in the sch9 and ras2 long-lived mutants than in the wild type cell [53]. Consistently, the budding index of the wild type cells (budded cells are those that have passed the Start and exited the G1 phase) was higher than that of the mutants. The authors’ hypothesis is that improper G1 arrest represents a form of cellular stress (replicative stress) that leads to apoptosis and contributes to chronological aging [53]. However, the importance of this type of stress in determining yeast life span seems to be limited given that the fraction of budded cells is ~5% in the wild type and does not increase overtime. Importantly, flow cytometry analysis of wild type cultures revealed an “apoptotic” peak that becomes more evident during aging in agreement with the presence of the other apoptotic markers discussed above [53].
II. The programmed and altruistic aging theory
The existence of an “altruistic aging program” is certainly controversial even if the results reported above can be explained in light of it. The vast majority of the gerontologists supports theories of aging as non-adaptive and due to the stochastic accumulation of cellular damage caused by the decline of the force of natural selection and consequently the decline in cellular protection and maintenance during aging [48]. However, studies conducted in the last fifteen years have shown that longevity is genetically determined and depends on evolutionary conserved pathways (see previous section). In simple organisms such yeast and worms the down-regulation of these pathways is induced by starvation and promotes the activation of somatic maintenance to guarantee survival until nutrients become available. The discovery of these aging pathways has led to the “programmed longevity theory” according to which aging, rather than depending on stochastic damage, is caused by the genetically programmed inactivation or decline of a repair and maintenance system that can control cellular damage [48]. The “programmed and altruistic aging theory” described above for populations of aging yeast hypothesizes instead the existence of a genetic program activated in order to “sacrifice” the majority of the population to allow the survival of a few adapted organisms. According to this theory natural selection functions on the level of the group rather than on that of the individual. More specifically, when observed in yeast cultures originated from the same progenitor cell, which are isogenic except for the DNA mutations that accumulate over time, adaptive regrowth can be seen as a form of kin selection [48, 54]. Both group and kin selection have been very controversial theories. Either of them appears to apply well to cultures of aging yeast. The fact the pathways that regulate altruistic aging in yeast are conserved throughout evolution raises the possibility that aging and death are programmed in higher eukaryotes as well.
III. Chronological aging-dependent apotosis in S. pombe
As discussed in the previous section, S. pombe shares several similarities with S. cerevisiae in terms of chronological aging and its regulation. Although fewer studies have been performed on apoptosis in S. pombe, apoptotic markers have been detected in cells expressing mammalian Bax/Bak and in mutants lacking the enzymes responsible for the biosynthesis of triacylglycerol [55, 56]. Furthermore, a homologue of the budding yeast metacaspase Yca1 has been identified on the S. pombe genome [55]. With respect to chronological aging, fission yeast show activation of caspase activity, which is reduced in the long-lived pka1 Δ and sck2 Δ mutants [15]. Although still preliminary, these results and the conservation of the life regulatory pathways in the two yeast species suggest the possibility of the existence of an aging program and possibly of adaptive regrowth in fission yeast as well.
Screening of the yeast knock-out collection to identify novel aging/apoptosis-related genes
Novel S. cerevisiae genomics approaches, which have recently become available, facilitate the identification of additional genes and mechanisms implicated in the regulation of chronological aging and yeast apoptosis. One of these relies on the use of the yeast gene-deletion collection (YKO collection) [57]. This collection covers almost all the yeast open reading frames (ORFs) and is constructed in a way that each deletion represents a complete loss of function of the gene and is uniquely tagged with two 20-nucleotide sequences [58]. The abundance of specific deletion mutants in a pool can be identified quantitatively by amplifying the tag sequences and hybridizing them to a high-density oligonucleotide arrays (tag microarrays) [57]. The YKO collection represents a powerful tool for yeast geneticists in that it overcomes the main limitation of the standard mutagenesis methods used for mutant screening, that of underrepresenting the yeast genome. The diploid homozygous deletion collection was used by Powers et al. to develop the first high-throughput assay for CLS, which led to the identification of the Tor pathway as a central regulator of CLS [18] (see above). Powers and colleagues relied on an optical density-based system to monitor the survival of cultures of individual mutants in 96-wells plates weekly. Their method may be complemented by measuring the chronological survival of a pool of gene-deletion mutants and monitoring the representation of each different deletion strain over time by tag microarray analysis. By determining the relative abundance of each strain every 48 hours, it will be possible to generate accurate survival curves for each deletion mutant and identify further genes whose expression is either required for long-term survival (the corresponding deletion mutants disappear early in the experiment) or reduces the life span (the corresponding deletion mutants are found in the pool after the death of the majority of the population). It will be also possible to identify deletions that cause adaptive regrowth directly, although this may be a more challenging task. In fact, as we have shown for the sod1 deletion mutant, in several cases the regrowth mutation may appear as a consequence of a primary deletion mutation that affects DNA stability.
Using this approach, we expect to find several deletions that can interfere with the apoptotic program and that should enhance our understanding of its mediation and purpose. We also predict that most deletions will either have no effect or a negative effect on life span and only a handful will decrease apoptosis and extend longevity. Considering how conserved the regulation of aging is in all species, the chances that these few genes will be relevant to aging of higher eukaryotes are very good. Furthermore, both life-span extending- and shortening mutations might provide hints to further characterize the apoptotic mechanisms that contribute to aging and death.
Concluding remarks
The field of yeast apoptosis is expanding and so is our knowledge of the similarities and differences between yeast and metazoan apoptosis. The use of yeast is also contributing to further understand the molecular mechanisms of mammalian apoptosis and its relationship with aging and age-related damage.
Here we have discussed the link between apoptosis and yeast chronological aging. Our current knowledge suggests that an apoptotic program has evolved in microorganisms as a survival strategy beneficial to the group. This program is dependent on cellular pathways like the Sch9, Tor1, and Ras/PKA pathways and its activation reduces cell protection and maintenance and raises the level of superoxide production, which in turn contributes to cell damage and death (FIG.1). Superoxide also elevates DNA damage and mutation frequency in the aging cultures, facilitating the appearance of “regrowth mutants” that guarantee the survival of the group. Notably, these dedifferentiating/regrowth mutants are reminiscent of cancer cells that escape apoptosis and grow under conditions that are normally not permissive for growth. Aging and apoptosis are intrinsically related in yeast and the mechanisms that cause them have just begun to be elucidated. The use of high throughput screenings available to the yeast community, such as that described in the final section of this review, will accelerate this process and, given the conservation of the life-regulatory pathways in higher eukaryotes, will provide important information to understand the fundamental biology of aging in other species and investigate the controversial hypothesis that an “aging program” might be conserved in higher eukaryotes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mortimer RK. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy BK, Austriaco NRJ, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jazwinski SM. The genetics of aging in the yeast Saccharomyces cerevisiae. Genetica. 1993;91:35–51. doi: 10.1007/BF01435986. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 5.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 6.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 7.Fabrizio P, Liou LL, Moy VN, Diaspro A, SelverstoneValentine J, Gralla EB, Longo VD. SOD2 Functions Downstream of Sch9 to Extend Longevity in Yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCay CM. Effect of restricted feedig upopn aging and chronic diseases in rats and dogs. American Journal of Public Health. 1947;37:521–528. [PMC free article] [PubMed] [Google Scholar]
- 9.Longo VD, Finch CE. Evolutionary Medicine: from Dwarf Model Systems to Healthy Centenarians. Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- 10.Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 12.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DL, Jr., McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 15.Roux AE, Quissac A, Chartrand P, Ferbeyre G, Rokeach LA. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- 16.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 17.Longo VD, Fabrizio P. Regulation of longevity and stress resistance: a molecular strategy conserved from yeast to humans? Cell Mol Life Sci. 2002;59:903–908. doi: 10.1007/s00018-002-8477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. doi: 10.1016/s0092-8674(01)00306-3. [DOI] [PubMed] [Google Scholar]
- 21.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 24.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 25.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, Vatner DE, O'Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 27.Longo VD. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol. Aging. 1999;20:479–486. doi: 10.1016/s0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- 28.Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsburg SL. The best yeast? Trends Genet. 1999;15:340–344. doi: 10.1016/s0168-9525(99)01798-9. [DOI] [PubMed] [Google Scholar]
- 30.Toone WM, Jones N. Stress-activated signalling pathways in yeast. Genes Cells. 1998;3:485–498. doi: 10.1046/j.1365-2443.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 31.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 32.Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longo VD, Ellerby LM, Bredesen DE, Valentine JS, Gralla EB. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J. Cell Biol. 1997;137:1581–1588. doi: 10.1083/jcb.137.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65. doi: 10.1016/s0014-5793(98)01227-7. [DOI] [PubMed] [Google Scholar]
- 36.Jin C, Reed JC. Yeast and apoptosis. Nat Rev Mol Cell Biol. 2002;3:453–459. doi: 10.1038/nrm832. [DOI] [PubMed] [Google Scholar]
- 37.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 38.Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F. An AIF orthologue regulates apoptosis in yeast. J Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludovico P, Sousa MJ, Silva MT, Leao C, Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- 40.Severin FF, Hyman AA. Pheromone induces programmed cell death in S. cerevisiae. Curr Biol. 2002;12:R233–R235. doi: 10.1016/s0960-9822(02)00776-5. [DOI] [PubMed] [Google Scholar]
- 41.Narasimhan ML, Damsz B, Coca MA, Ibeas JI, Yun DJ, Pardo JM, Hasegawa PM, Bressan RA. A plant defense response effector induces microbial apoptosis. Mol Cell. 2001;8:921–930. doi: 10.1016/s1097-2765(01)00365-3. [DOI] [PubMed] [Google Scholar]
- 42.Silva RD, Sotoca R, Johansson B, Ludovico P, Sansonetty F, Silva MT, Peinado JM, Corte-Real M. Hyperosmotic stress induces metacaspase- and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol Microbiol. 2005;58:824–834. doi: 10.1111/j.1365-2958.2005.04868.x. [DOI] [PubMed] [Google Scholar]
- 43.Gourlay CW, Ayscough KR. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:6487–6501. doi: 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almeida B, Buttner S, Ohlmeier S, Silva A, Mesquita A, Sampaio-Marques B, Osorio NS, Kollau A, Mayer B, Leao C, Laranjinha J, Rodrigues F, Madeo F, Ludovico P. NO-mediated apoptosis in yeast. J Cell Sci. 2007;120:3279–3288. doi: 10.1242/jcs.010926. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Sun L, Liang Q, Wang J, Mo W, Zhou B. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol Biol Cell. 2006;17:1802–1811. doi: 10.1091/mbc.E05-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longo VD, Mitteldorf J, Skulachev VP. Programmed and altruistic ageing. Nat Rev Genet. 2005;6:866–872. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- 49.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 51.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberger M, Feng L, Paul A, Smith DL, Jr., Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS ONE. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skulachev VP. Programmed death in yeast as adaptation? FEBS Lett. 2002;528:23–26. doi: 10.1016/s0014-5793(02)03319-7. [DOI] [PubMed] [Google Scholar]
- 55.Low CP, Liew LP, Pervaiz S, Yang H. Apoptosis and lipoapoptosis in the fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 2005;5:1199–1206. doi: 10.1016/j.femsyr.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Chieu HK, Low CP, Zhang S, Heng CK, Yang H. Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J Biol Chem. 2003;278:47145–47155. doi: 10.1074/jbc.M306998200. [DOI] [PubMed] [Google Scholar]
- 57.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 58.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]