Abstract
A significant impediment to the widespread use of noninvasive in vivo vascular imaging techniques is the current lack of suitable intravital imaging probes. We describe here a new strategy to use viral nanoparticles as a platform for the multivalent display of fluorescent dyes to image tissues deep inside living organisms. The bioavailable cowpea mosaic virus (CPMV) can be fluorescently labeled to high densities with no measurable quenching, resulting in exceptionally bright particles with in vivo dispersion properties that allow high-resolution intravital imaging of vascular endothelium for periods of at least 72 h. We show that CPMV nanoparticles can be used to visualize the vasculature and blood flow in living mouse and chick embryos to a depth of up to 500 μm. Furthermore, we show that the intravital visualization of human fibrosarcoma-mediated tumor angiogenesis using fluorescent CPMV provides a means to identify arterial and venous vessels and to monitor the neovascularization of the tumor microenvironment.
Intravital vascular imaging has the potential to be a powerful tool for the noninvasive detection and visualization of disease. The resolution of functionally significant changes in structure in the endothelium of microvasculature using fluorescence imaging in live animals has proven challenging, however, because of the inadequate tissue penetration of fluorescent signal1.
Current agents for fluorescence imaging of microvasculature include microspheres or nanospheres2, iron oxide particles3, liposomes4, dextrans5, lectins6, antibodies7 and, more recently, quantum dots8. Although many of these particles have specific strengths, issues related to toxicity, stability, bioavailability, cost or chemical flexibility have yet to be overcome. Inorganic synthetic particles tend to aggregate under physiological conditions and can be toxic upon exposure to ultraviolet light9. Multivalency with respect to fluorochrome is crucial for achieving the requisite sensitivity for adequate tissue penetration. Thus, a multivalent, biologically compatible platform for the development of fluorescent and magnetic resonance imaging agents is still much needed for both clinical and research applications.
As a biological imaging agent, plant viruses possess a number of distinct advantages over other particles. CPMV is a small plant virus that is composed of 60 identical copies of an asymmetric protein unit assembled around a bipartite single-stranded RNA genome10. As such, the most compelling advantage of a virus-based platform for imaging is the multivalency of chemically addressable sites on the capsid surface. This multivalency, together with its biological compatibility and inert nature, makes CPMV an ideal in vivo imaging agent11–16.
Here, we assess the utility of fluorescent CPMV particles for intravital imaging by injecting and visualizing the vasculature of mouse embryos and shell-free chick embryos. Furthermore, we conducted vascular mapping studies to visualize the extent of angiogenesis induced by HT1080 tumor onplants on the chick chorioallantoic membrane (CAM).
RESULTS
High-density conjugation of fluorescent dyes to CPMV
We isolated wild-type CPMV particles and conjugated them with fluorescent dyes using N-hydroxysuccinimide (NHS) ester chemistry. The atomic structure of CPMV describes a defined structural environment suitable for chemical conjugation of dyes to the inside and outside surface of the viral particle. Each asymmetric protein unit possesses five accessible lysine residues that are positioned on the exterior of the assembled virus particle, and their reactivity with chemical reagents such as activated NHS esters provides a total of 200 addressable sites per 31 nm virion17,18 (Fig. 1a,b). We used the fluorophores Alexa Fluor 555 (A555), Alexa Fluor 488 (A488) and fluorescein. We achieved an average labeling of 70 dyes per virus particle for A488 and 120 dyes per virus particle for A555. We analyzed the CPMV-A555 conjugate by size-exclusion chromatography and the fluorescence peak coeluted precisely with the virus particles (Fig. 1c). When we coated particles with larger molecules such as 3,400 Da polyethylene glycol (PEG), we achieved an average labeling of 65 dyes per particle. Conjugation of PEG molecules larger than 3,400 Da resulted in aggregation of the particles. Both the large and small subunits were conjugated to either fluorescent dye (Fig. 1d) or PEG and dye (Fig. 1d).
Figure 1.

Labeling of CPMV particles with fluorescent dyes. (a) Subunit organization of CPMV: domain A (blue) represents the small subunit, domains B (orange) and C (yellow) represent the two domains of the large subunit. The maximum (31 nm) and minimum (25 nm) particle diameters according to the refined crystal structure14 are indicated. (b) Surface model of CPMV particle showing predicted arrangement of conjugated fluorochromes. (c) Size-exclusion FPLC analysis of CPMV-A555 conjugate. (d) SDS-PAGE analysis of CPMV-A555 (A555) and CPMV-PEG-FITC (PF) conjugates. Mobility of unmodified large (L, 42 kDa) and small (S, 24 kDa) virus subunits are indicated. The marker (M) is Bio-Rad broad-range prestained standard. Both panels are the same gel, Coomassie Blue staining (left) and ultraviolet illumination (right) to detect conjugated fluorescent dye. Unmodified subunits are visible with Coomassie staining but not under ultraviolet light (marked S, L). In PF lane, L1, L2 and L3 correspond to the conjugation of 1, 2 and 3 PEG molecules to the large subunit. S1 and S2 represent conjugation of 1 or 2 PEG molecules to the small subunit. (e) Fluorescence quantification per molecule expressed as a ratio of a single fluorescein dextran molecule. Average labeling of CPMV particles is indicated. Fd, 10 kDa fluorescein dextran; A4d, 10 kDa Alexa Fluor 488 dextran; Rhl, rhodamine-labeled L. culinaris lectin; CPMV PF, CPMV PEG-FITC. ‘0,04 μm nanospheres’ are FluoSpheres carboxylate-modified orange fluorescent microspheres, 0.04 μm.
We measured the fluorescence output per particle of the conjugated virus using an in vitro fluorescence assay and compared it to that of 10 kDa fluorescein dextran, 10 kDa A488 dextran, rhodamine-labeled Lens culinaris lectin and 0.04 μm fluorescent nanospheres (Fig. 1e). The dye-labeled CPMV particles had a fluorescence intensity that was proportional to the number of attached dyes, indicating that fluorescence quenching caused by unfavorable dye-dye or dye–amino acid interactions was not occurring. The CPMV-A555 nanoparticles had a fluorescence intensity that was considerably brighter per particle than conventional vascular imaging agents such as dye-labeled dextrans and lectins, and approximately one-half that of 0.04 μm polystyrene nanospheres containing Alexa fluor dye.
Dye-labeled CPMV localizes to vascular endothelium
We assessed the brightness, biodistribution and stability of CPMV-A555 in the circulation in the adult mouse after tail injection. Fluorescent CPMV particles were detected and easily visualized in all tissues examined. CPMV associated preferentially with the lumen periphery of the vasculature (Fig. 2a) and distributed throughout both large and small vessels and into the smallest capillaries (Fig. 2a). This allowed resolution of vascular structures in various organs, including kidney, heart, placenta and liver (Fig. 2a) when compared to fluorescein dextran (Fig. 2a). No CPMV particles were detected outside of the vasculature up to 72 h after injection. Notably, fluorescein dextran was sensitive to fixation time, with fluorescence signal dropping precipitously after 2 h in fixative, whereas CPMV-A555 particles remained bright and localized for periods of fixation up to 30 d. Dye-labeled CPMV particles began to accumulate in the liver (Fig. 2a) and spleen after injection, and were gradually depleted from the circulation over 72 h. We observed no deleterious effects (thrombosis, malaise, death) in the mice after injection with dye-labeled CPMV.
Figure 2.

Fluorescent dye–conjugated CPMV particles enable visualization of vasculature intravitally and in fixed tissues. (a) Fluorescence images of tissue cryosections from kidney (asterisk indicates vessel lumen), heart, placenta and liver isolated from adult mice coinjected with CPMV-A555 (top panels) and fluorescein dextran (bottom panels). Scale bar, 50 μm. (b,c) Intravital imaging of CPMV-A555 perfused 11.5-d embryo with the yolk sac intact (b) and removed (c). White boxes indicate the regions magnified in (e) and (f). Scale bar, 1.1 mm. Images were captured approximately 1 h after injection. (d) Cryosection of an 11.5-d mouse embryo perfused with CPMV-A555. White box indicates the region magnified in (g). Scale bar, 1.1 mm. (e) Yolk sac vasculature, magnified. Scale bar, 25 μm. (f) Capillaries in the head region. Scale bar, 25 μm. (g) Intersomitic and placental vessels in embryo tissue section. Scale bar, 100 μm. (h–o) Comparison of intravital imaging with CPMV-A555 (h–k) and fluorescent nanospheres (e–o) in E11.5 mouse embryo. (h,l) Whole embryo. Scale bar, 1.1 mm. (i,m) Head region, arrows indicate areas of differential staining of anterior vasculature, brain vasculature. Scale bar, 770 μm. (j,n) CPMV staining allows increased resolution of intersomitic vessels. Scale bar, 540 μm. (k,o) Arrows indicate capillary and larger vessels of yolk sac membrane. Scale bar, 50 μm.
CPMV-A555 permits visualization of deep microvasculature
To evaluate the efficacy of CPMV-based intravital fluorescence imaging, we explanted mouse embryos between embryonic day (E)9.5 and E15.5 of development with the yolk sac and placenta intact and placed them in culture. When 10 μg of CPMV-A555 was injected into a small venule of the yolk sac, the particles circulated rapidly throughout the embryonic vasculature (Fig. 2b–g). Under the fluorescence microscope it was possible to visualize the yolk sac vasculature (Fig. 2b) to high resolution (Fig. 2e and Supplementary Video 1 online). Dye-labeled CPMV distributed evenly to large and small vessels and showed consistency of signal throughout the vasculature. After removal of the yolk sac, intravital imaging of the embryonic vasculature was possible (Fig. 2c,f). Larger vessels could be visualized with adequate resolution, in most cases at a depth of up to 500 μm. Microvasculature could be resolved well to a depth of 200–250 μm, or deeper with an unobstructed view. One hour after injection, we were able to clearly visualize the vascular endothelium and observe blood flow in real time at high frame rates (Supplementary Video 2 online). The signal intensity and biodistribution of dye-labeled CPMV was not altered substantially after paraformaldehyde fixation and cryosectioning (Fig. 2d,g).
The biodistribution of CPMV-based fluorescent nanoparticles was compared to that of similar-sized fluorescent nanospheres in E11.5 mouse embryos. Despite extensive efforts to disperse the nanospheres by vortexing and sonication, aggregates formed and tended to lodge in the capillaries and precapillary arterioles of the embryos after injection, resulting in the uneven distribution of particles (Fig. 2l–o) and reduced resolution of deeper vascular structures. No aggregates were observed using dye-labeled CPMV, resulting in high-resolution images at depths of up to 500 μm of capillaries and larger vessels in the head meningeal plexus, somites and yolk sac (Fig. 2i–k).
CPMV differentially labels embryonic arteries and veins
The CAM in the chick embryo is highly developed at the 10-d stage. Although identification of arterioles and venules by direction of blood flow is possible in the live chick embryo, unaided identification is difficult at high magnification in vivo and impossible in tissue sections. We injected chick embryos with 50 μg of CPMV-A555 and visualized them under the fluorescence microscope. CPMV particles distributed throughout the embryonic and extraembryonic vasculature within seconds, and provided sufficient signal to visualize the vasculature throughout the CAM to a depth of 500 μm (Fig. 3a and Supplementary Video 3 online). We observed considerable internalization of virus particles by vascular endothelial cells. Furthermore, uptake of CPMV in the vessels of the venous system occurred at a much higher rate than those of the arterial system (Fig. 3b,c), and this resulted in a markedly more intense labeling of the venous vasculature compared to the arterial vasculature within 1 min of injection. This behavior allowed for rapid identification of vascular origin both in live tissues at high magnification (Fig. 3a–c) and in fixed tissues (data not shown).
Figure 3.
Intravital fluorescence imaging of chick CAM vasculature and subcellular localization of CPMV. (a) Fluorescence image looking down through surface of chick CAM showing multiple levels of vasculature, through capillary bed and larger vessels below to arterioles and venules. Scale bar, 100 μm. (b) CAM arteriole. Scale bar, 22 μm. (c) CAM venule (arrows in b and c denote blood flow direction). Scale bar, 22 μm. (d) Intravital image of large CAM vein, CPMV-A555 (orange), endothelial cell nuclei (blue) stained with Hoechst 33258. Box indicates area magnified in e. Scale bar, 16 μm. (e) CPMV particles are restricted to perinuclear compartments in the vascular endothelial cells. Compartments show uniform cell polarity. Scale bar, 5.5 μm. (f,g) CPMV-A555 remains restricted to endothelial cells and allows intravital staining of the vasculature over long periods of time. 72 h after the initial injection of CPMV-A555, venular staining remains roughly equivalent to the initial staining. Scale bar, 160 μm. (h) Transmission electron micrograph of chick embryo CAM injected with CPMV-A555 showing CPMV particles being actively internalized into an endothelial cell (black arrows indicate CPMV particles). ec, endothelial cell; n, nucleus. Asterisk indicates vessel lumen. Scale bar, 100 nm. (i) Endothelial cell with CPMV-filled vesicle. Arrows indicate CPMV particles among many. Scale bar, 64 nm. (j) Macrophage (wbc) at the luminal periphery with large CPMV-containing vesicles (arrows). rbc, red blood cell. Scale bar, 693 nm.
Fluorescent CPMV is internalized by vascular endothelial cells
To examine the localization and distribution of CPMV, we injected chick embryos with 50 μg of CPMV-A555 and then slowly perfused them with Hoechst 33258 to stain the endothelial cell nuclei. We found CPMV-A555 mainly in perinuclear endosomal compartments (Fig. 3d,e). Embryos visualized for 1 h and 72 h after injection with CPMV-A555 showed a similar biodistribution (Fig. 3f,g), indicating that the virus remains localized in endothelial cells over long periods of time. Arterial staining improved slightly over the 72-h period.
We analyzed CAM sections of CPMV-A555–injected chick embryos by transmission electron microscopy (TEM). CPMV particles were actively endocytosed by vascular endothelial cells (Fig. 3h). In addition, vesicles containing CPMV particles were also seen in the majority of the vascular endothelial cells examined (Fig. 3i). CPMV particles reside in perinuclear vesicles that are associated with lysosomes and the Golgi apparatus, as evidenced by colocalization with lysosomal-associated membrane protein 2 and β-COP markers (Supplementary Fig. 1 online). Within the CAM vasculature we also found macrophages that contained large phagocytic vesicles filled with virus particles (Fig. 3j). No CPMV particles were found outside the vascular endothelial layer in other cell types or in the interstitium.
Coating CPMV with PEG inhibits its internalization
Coatings such as PEG are known to minimize molecular interactions and thereby increase half-life in circulation of blood-borne agents19. In adhesion assays using human umbilical vein endothelial cells (HUVECs) or mouse embryonic fibroblasts (MEFs), the ability of cells to bind to plates coated with PEG-coated CPMV-fluorescein particles was inhibited compared to plates coated with native CPMV particles (Supplementary Fig. 1 online). Furthermore, internalization of CPMV by HUVECs in culture was completely inhibited by the PEG coating (Supplementary Fig. 1 online).
PEG coating also completely inhibited the internalization by chick embryo endothelial cells in vivo (Supplementary Video 4 and Supplementary Fig. 2 online). Analysis of tissue sections of vasculature from these embryos with TEM indicated that CPMV-PEG-FITC particles were located exclusively in the vascular lumen (data not shown). Adult mice injected with CPMV-PEG-FITC showed greatly reduced uptake by the liver and spleen compared to those injected with CPMV-FITC (Supplementary Fig. 2 online). Thus, our results suggest that dye-labeled CPMV and CPMV-PEG will be useful to visualize the vascular endothelium, and to visualize the blood volume and blood flow, respectively.
CPMV is retained in the vascular endothelium over time
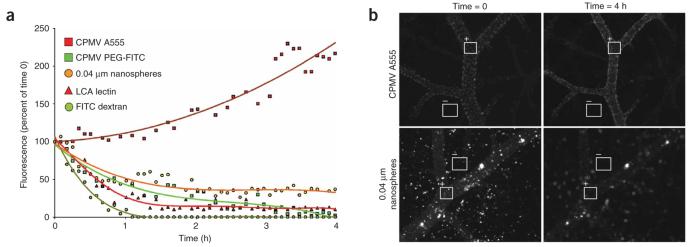
We next investigated the utility of dye-labeled CPMV for long-term intravital vascular imaging. We injected chick embryos with CPMV-A555, CPMV-PEG-FITC, 0.04 μm fluorescent nanospheres, rhodamine-labeled lectin (L. culinaris agglutinin) or fluorescein dextran (Fig. 4a,b). The vasculature in the CPMV-A555–injected embryos continued to increase in brightness over the course of the experiment and provided the best resolution of vascular structures (Fig. 4b). This increase in signal (Fig. 4a) probably resulted from continued internalization of circulating CPMV-A555 by endothelial cells. In contrast, the fluorescein dextran signal dropped off rapidly and showed nominal fluorescence after 1 h of incubation (Fig. 4a). In embryos injected with fluorescent lectin, the vascular staining dropped to approximately 20% of its initial intensity after 1.5 h and remained at this level over subsequent time points (Fig. 4a). The vasculature in embryos injected with fluorescent nanospheres declined in fluorescence over 2 h and then leveled off at 35–40% of the initial signal (Fig. 4a,b). We surmise that fluorescent nanospheres may be adequate for extended periods of intravital imaging. The deposition of aggregates throughout the vasculature, however, resulted in poor resolution of vascular structures (Fig. 4b). Fluorescence from CPMV-PEG-FITC declined slowly but consistently over a 4-h period (Fig. 4a), possibly resulting in part from their sensitivity to photobleaching in comparison to CPMV-A555.
Figure 4.
Comparison of intravital vascular staining intensity over time in the chick embryo. (a) Comparative analysis of changes in vascular staining intensity over time in the chick embryo using CPMV-A555, CPMV-PEG-FITC, 0.04 μm nanospheres, LCA lectin and FITC dextran. Reagent amounts were adjusted to equivalent total fluorescence. A single frame was captured every 5 min for a 4-h period. Regions of interest (ROI) within the vasculature were sampled at each time point for average fluorescence intensity, background values were subtracted, and the resultant values were graphed against initial fluorescence intensity. (b) Representative images captured immediately after injection (time = 0) and 4 h after injection (time = 4 h) showing sampled ROI during intravital retained fluorescence assay. Fluorescence average of negative ROI (−) was subtracted from positive ROI (+).
CPMV permits long-term vascular mapping of tumors
Intravital vascular mapping provides a new opportunity to identify areas of the vascular bed that are involved in neovascularization. Chick embryos bearing highly vascularized HT1080 tumor onplants on the CAM (Fig. 5a)20 were injected with CPMV-A555 and visualized by epifluorescence microscopy. Dye-labeled CPMV circulated freely through the tumor vasculature, labeling vessels both entering and exiting the tumor (Fig. 5b) as well as the capillary network within the tumor mass (Fig. 5c).
Figure 5.
Evaluation of tumor angiogenesis in an intravital CAM/HT1080 fibrosarcoma model. (a) Bright-field image of HT1080 tumor CAM onplant at 7 d. Opaque object is a nylon mesh grid used for quantification of angiogenesis. (b) Fluorescence image of tumor onplant after injection of embryo with 50 μg of CPMV-A555. (c) High-magnification (×20) image of tumor interior shown in b; tumor microvasculature is clearly visualized. (d,e) Visualization of HT1080 tumor angiogenesis using CPMV-A555. (d) Left, visualization of preexisting vasculature in the CAM immediately after HT1080 tumor cell injection using CPMV-A555. Middle, GFP-expressing HT1080 tumor bolus under the surface of CAM. Right, merge. Scale bar, 100 μm. (e) Left, visualization of preexisting CAM vasculature and neovasculature arising from tumor angiogenesis 24 h after tumor-cell injection. Middle, GFP-expressing HT1080 tumor bolus. The extensive migration over 24 h indicates a high level of tumor-cell viability. Right, merge. Scale bar, 100 μm. (f) Intravital vascular mapping of tumor angiogenesis using CPMV-A555 and CPMV-A488. Human fibrosarcoma HT1080 tumor cells were implanted within CAM. After 48 h of incubation, embryos were injected with 50 μg of CPMV-A555 (left panels). After a further 24-h incubation, embryos were injected with 50 μg of CPMV-A488 (right panels) and visualized. Arrows indicate a site of angiogenic sprouting and arrowheads indicate newly formed vessels.
We injected a small bolus of green fluorescent protein (GFP)-expressing HT1080 cells underneath the capillary bed within the chick embryo CAM. The preexisting vasculature surrounding the newly introduced tumor cells was visualized using CPMV-A555 (Fig. 5d). After 24 h, we injected additional CPMV-A555 to visualize the newly formed vasculature around the tumor cells. We observed substantial vascularization of the region occupied by the HT1080 tumor cells (Fig. 5e), indicating that CPMV will be useful for the intravital labeling of tumor neovasculature.
Taking advantage of the fact that dye-labeled CPMV is stably internalized by endothelial cells, we surmised that it could be used to label and distinguish endothelial cells arising at different points in time. To investigate this, a bolus of HT1080 was introduced into the CAM and incubated for 48 h. We then injected embryos with CPMV-A555 reagent to label the vasculature. After a further 24 h of incubation, we injected embryos with CPMV-A488 to label both the preexisting and newly formed vasculature (Fig. 5f). Endothelial cells that were formed in the 24 h between injections were labeled only by CPMV-A488, whereas those in existence before that time were labeled with both reagents. This technique allowed for the clear visualization of vessels entering and exiting the tumor, and entire regions of newly formed vasculature could be identified using this method (Fig. 5f). Thus, our data suggest that dye-labeled CPMV is a suitable reagent for the intravital vascular mapping of tumor vasculature at multiple time points.
DISCUSSION
Using basic crosslinking chemistry, we conjugated CPMV particles to a variety of commercially available fluorescent dyes to a density of up to 120 dyes per virus. Injected into adult mice, dye-labeled CPMV remained monodisperse and localized to the vascular endothelium, allowing resolution of macro- and microvasculature in vivo and in fixed tissues using fluorescence microscopy. Our studies indicate that dye-labeled CPMV reagents are superior to similar-sized fluorescent nanospheres for the resolution of microvasculature during intravital imaging. In the chick embryo, fluorescent CPMV particles differentially label the arterial and venous systems as a result of internalization of the particles by endothelial cells, allowing for rapid identification of vascular origin. Although this internalization can be blocked by coating the CPMV particles with PEG, internalized virus particles remain localized within endothelial cells for at least 72 h. This allows for long-term intravital imaging of either the endothelium or blood volume of microvasculature.
The biology and chemistry of CPMV have been studied extensively12,14,17,21, and the virus has a number of key qualities that make it particularly well suited to intravital imaging. CPMV particles can be produced in large quantities in plants22. They are also very stable and can withstand a variety of solvents and extremes of temperature and pH while maintaining structural integrity23. Although plant viruses themselves are noninfectious in animals15,16, CPMV is disseminated systemically after oral or intravenous administration24. CPMV is an inert substance already present in the human food chain, and preliminary studies indicate lack of systemic toxicity in mice at doses of up to 100 mg/kg (P. Singh & M.M., unpublished data).
The internalization of CPMV by vascular endothelial cells is advantageous for intravital imaging. The preferential labeling of the venous vessels provides a convenient means to identify vascular origin and directionality within the tissue. Furthermore, because the CPMV particles ‘arrest’ in endothelial cells for extended periods, they are particularly valuable for the mapping of vascular endothelium and quantification of endothelial cellularity during long-term intravital imaging. Several explanations for this differential staining are possible. First, the impact of flow velocity and shear stress in the different classes of vessels could influence uptake. Furthermore, uptake may either be the result of nonspecific internalization based on particle size or, alternatively, involve a specific molecular interaction with cell-surface proteins.
Although much is known about the molecular components responsible for regulating angiogenesis, the visualization of the vascularization and specifically physical changes in the vasculature have been limited to endpoint analysis such as histological microscopy of harvested tissue. Vascular mapping would provide a means to determine which vascular beds function as a portal for metastatic cells to enter the circulation. Our studies show that vascular mapping that uses the well-established chick angiogenesis model and dye-labeled CPMV as a vascular labeling reagent are straightforward for long-term intravital studies.
Our results suggest that CPMV nanoparticles will be well suited to the visualization of rare molecular targets, because of their high signal per molecule and favorable biocompatibility. The use of near-infrared fluorochromes and multiphoton confocal imaging will enhance the quality of imaging at greater tissue depths in vivo25,26. Furthermore, the multivalent display of vascular targeting peptides or proteins on the surface of CPMV would probably enhance their binding or targeting ability13. Viral nanoparticles are not limited to fluorescent labeling, and their multivalent properties may be exploited to display a wide variety of molecular tags, including, but not limited to, radioactive isotopes, magnetic resonance imaging contrast agents, enzymatic moieties or a combination thereof13. Because the viral genome is contained on two molecules of RNA that retain host infectivity without encapsidation, it can be manipulated at a genetic level to introduce desired mutations22. Bioengineered virus particles can be produced in large numbers, and the incorporation of new peptide sequences12 opens the door for future targeted molecular bioimaging studies.
METHODS
Propagation of CPMV in plants
The primary leaves of cowpea seedlings were mechanically inoculated with 10 μg each of cDNA plasmids encoding RNA1 (pCP1) and RNA2 (pCP2)27. We extracted the initial virus inoculum from infected cowpea leaves with 0.1 M potassium phosphate, pH 7.0 (phosphate buffer), 7 d after infection. Typically, 50 plants were infected with the plant extract, and the symptomatic leaves were harvested after 3 weeks. Virus was purified using standard techniques as previously described17.
Conjugation of CPMV with fluorescent dyes
We dissolved Alexa Fluor 555 carboxylic acid, succinimidyl ester (Molecular Probes) in DMSO and introduced it at a ratio of 50/1 mol/mol into a solution of virus (2 mg/ml) so that the final solvent mixture was composed of 80% buffer and 20% DMSO. In addition, we prepared CPMV particles using Alexa Fluor 488 NHS (Molecular Probes), fluorescein (FITC)-NHS (Molecular Probes) and fluorescein-NHS with a 3,400 Da PEG spacer (Nektar Pharmaceuticals). After incubation at 24 °C for 24 h, we performed initial separation of virus from unconjugated dye by ultracentrifugation at 42,000 r.p.m. over 3 ml of a 30% sucrose cushion. We then resuspended the pellet in 0.1 M potassium phosphate and loaded it on a 10–40% sucrose gradient for ultracentrifugation at 28,000 r.p.m. for 3 h. We further pelleted the resulting virus by ultracentrifugation at 42,000 r.p.m. for 3 h, and then resuspended it in PBS. The purity and fluorescence intensity of derivatized virus was determined by analytical size-exclusion fast protein liquid chromatography using a superose-6 column. After purification, we determined the relative concentrations of virus and dye by absorbance spectroscopy. We determined virus concentrations by measuring the absorbance at 260 nm; virus at 0.1 mg/ml gives a standard absorbance of 0.8. The average molecular weight of the CPMV virion was 5.6 × 106. We obtained fluorochrome concentration by measurement of absorbance at its emission maximum, using an extinction coefficient of 150,000 for A555, 72,000 for fluorescein, 71,000 for A488. We determined average labeling of CPMV-dye conjugates by dividing the moles of dye by the moles of CPMV. Images were created with the RasMol31 and DeepView32 programs using oligomer coordinates generated by the VIPER website33 from protein data bank file 1NY7.
Fluorescence quantification
We spotted 10 μl of a 100 μg/ml solution of fluorescent substrate on a glass slide and placed a cover slip over it. Multiple fields were digitally captured by a Hammamatsu ORCA-ER 12-bit camera at ×4, ×10 and ×20 magnification on a Zeiss Axioplan2ie-MOT upright fluorescent microscope using the appropriate filter set. We quantified image intensities using the average field intensity function of the OpenLab acquisition software (Improvision), and subtracted the background values. Data were collected and averaged over a minimum of five fields per objective per sample.
CPMV injections in adult mice
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute. We injected CPMV-A555 conjugate and/or FITC dextran (10 kDa, Molecular Probes) (50 μg −1 mg in 0.1–0.2 ml PBS) in the tail vein of CD-1 mice and allowed them to circulate for 5 min up to 72 h. We adjusted the amounts of fluorescein dextran control and dye-labeled CPMV so that equivalent levels of fluorescence were injected based on fluorescence measurements in Figure 1e. We fixed tissues in 4% paraformaldehyde for 4 h and froze them in Tissue Tek OCT embedding medium (Sakura Finetek) before sectioning them. We mounted the 20-μm cryosections with Vectashield mounting medium (Vector Laboratories) before examination under the Zeiss Axioplan2ie-MOT. In the experiments that used PEG-coated fluorescent CPMV, we injected adult mice in the tail vein with 250 μg or500 μg of either CPMV-FITC or CPMV-PEG-FITC, and collected tissues after 1 h.
Injection, culture and imaging of mouse embryos
We injected mouse embryos at E9.5–E15.5 using a MM-33 micromanipulator (Fine Science Tools), microinjection needles drawn from borosilicate glass capillaries and a micro-adjustable syringe pump (Braintree Scientific). We isolated staged embryos (morning of vaginal plug counted as E0.5) with the yolk sac and placenta intact to preserve the embryonic vasculature and blood flow. We cultured embryos in chamber slides using media as previously described28. We injected 10 μg of CPMV-A555 or an equivalent number of 0.04 μm fluorescent nanospheres (Molecular Probes) through a small venule on the surface of the yolk sac, and visualized whole embryos in vivo in culture media under the Zeiss Axioplan2ie-MOT upright microscope using ×1.25, ×10, ×20 and ×40 objectives.
Fluorescence imaging of chick embryo vasculature
We received fertilized White Leghorn chicken eggs from SPAFAS (North Franklin) and incubated them in a humidified incubator at 38 °C. At day 4, we removed eggshells, and incubated embryos under shell-less conditions, in a covered dish placed in a humidified air incubator at 38 °C and 60% humidity29. We injected chick embryos at 10 d of development with 50 μg of CPMV-A555 using a microinjection needle into a small venule in the CAM, and visualized their extraembryonic vasculature under a Zeiss Axioplan2ie-MOT upright microscope. For the studies using PEG-coated fluorescent CPMV, we injected embryos at 10 d of development with 200 μl of a solution containing 50 μg of CPMV-PEG-FITC and 50 μg of CPMV-A555.
For the intravital localization studies, we injected chick embryos at 10 d with 50 μg of CPMV-A555. After a period of 15, 30 or 60 min, we slowly perfused embryos with 2 ml of a 1 mg/ml solution of Hoechst 33258 and visualized under the Zeiss Axioplan2ie-MOT microscope.
TEM
We injected chick embryos with 50 μg of CPMV-A555. We excised sections of CAM at 5 and 15 min and fixed them in 3% glutaraldehyde/1% paraformaldehyde in 0.1 M cacodylate buffer for 5 h. We post-fixed them in 1% osmium tetroxide, 0.1 M cacodylate buffer for 1 h. We visualized sections under a Philips CM100 Electron Microscope at the Electron Microscopy Core Facility at The Scripps Research Institute.
Intravital retained fluorescence assay
We injected chick embryos at 10 d of development with either CPMV-A555, CPMV-PEG-FITC, 0.04 μm fluorescent ‘orange’ nanospheres (Molecular Probes), rhodamine lectin (L. culinaris agglutinin, Vector Pharmaceuticals) or fluorescein dextran (10 kDa, Molecular Probes) into a small venule in the CAM. Reagent amounts were adjusted to equivalent fluorescence. We then incubated embryos under the Zeiss Axioplan2 microscope and imaged them every 5 min for 4 h. We sampled regions of interest within the vasculature at each time point for average fluorescence intensity, subtracted background values, and graphed the resulting values against initial fluorescence intensity.
CAM tumor angiogenesis mapping
We generated tumor onplants by overlaying two gridded plastic meshes and embedding them into 30 μl of 2.2 mg/ml collagen20. We embedded HT1080 tumor cells in the collagen at 50,000 cells per onplant. To generate cells lines ubiquitously expressing GFP, we infected cultured cells with pLXSN-GFP (Clontech) and selected cells by neomycin selection without clonal propagation. We passed selected cells in vivo to maintain a metastatic phenotype. We placed onplants on or introduced them under the surface of the CAM of 10-d-old shell-less embryos using a modified version of the CAM angiogenesis assay protocol20. We immediately injected embryos with 50 μg of CPMV-A555 and visualized the preexisting vasculature surrounding the tumor-cell bolus.
For the tumor angiogenesis studies, we injected 10,000–50,000 tumor cells as a bolus below the CAM surface in 10-d embryos. We injected embryos with 50 μg of CPMV-A555 immediately after introduction of the tumor cells and then again after 24 h of incubation. We visualized the tumor cells and surrounding vasculature using a Zeiss Axioplan2 upright microscope. For the vascular mapping study, we injected tumor cells without the GFP construct as a bolus into chick embryos at 10 d of development, and returned them to the incubator. On day 12, we intravenously injected 50 μg of CPMV-A555. After a further 24 h of incubation (day 13), we injected 50 μg of CPMV-A488 and visualized tumors under the fluorescent microscope. We captured a Z-stack of images of the tumor and the surrounding vasculature and flattened it into a single image. We collected image stacks and analyzed them using the software packages OpenLab and Volocity (Improvision).
Localization of internalized CPMV in lysosomal and endosomal compartments
We obtained bone marrow–derived dendritic cells from C57BL/6 (H-2b) female mice as previously described30. After 10 d of differentiation, we collected cells in suspension. We added differentiated dendritic cells to six-well plates and allowed them to attach to a cover slip placed in the wells. After a 1-h incubation, we washed wells to remove unbound cells and added CPMVA555 particles (13 μg/ml) to the medium and incubated the mixture at 37 °C. We washed cultures 2 h later to eliminate free CPMV particles. We allowed dendritic cells to internalize the CPMV particles for an additional 4 h at 37 °C. Then we fixed cells with 2% formaldehyde (Polysciences) in PBS, permeabilized them with 0.2% saponin (Sigma) in PBS and labeled them with rabbit antibodies specific to the Golgi apparatus (β-COP) or lysosomes (Lamp2), both from Affinity BioReagents. We stained cells with an rabbit-specific secondary antibody coupled with A488 (Molecular Probes) to detect the organelles and with Toto-3 iodide (Molecular Probes) to visualize the nucleus. We took micrographs at ×63 magnification with a BIORAD 1024 confocal microscope and analyzed them with Image J software.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by US National Institutes of Health grants R01 HL65738 and R21 HL72270 (to H.S.), AI47823, CA112075 and N01-CO-27181 (to M.M.), R01 CA55852 and R01 CA105412 (to J.Q.) and NSERC PDF-313420-2005 (to J.L.). The authors thank M. Wood (Scripps Microscopy Core Facility), X. Zhang, E. Rockenstein and N. MacLean (University of California San Diego) for their technical assistance.
Footnotes
Supplementary information is available on the Nature Medicine website.
COMPETING INTERESTS STATEMENT
The authors declare that they have competing financial interests (see the Nature Medicine website for details).
References
- 1.McDonald DM, Choyke PL. Imaging of angiogenesis: from microscope to clinic. Nat. Med. 2003;9:713–725. doi: 10.1038/nm0603-713. [DOI] [PubMed] [Google Scholar]
- 2.Yang CS, et al. Nanoparticle-based in vivo investigation on blood-brain barrier permeability following ischemia and reperfusion. Anal. Chem. 2004;76:4465–4471. doi: 10.1021/ac035491v. [DOI] [PubMed] [Google Scholar]
- 3.Josephson L, Kircher MF, Mahmood U, Tang Y, Weissleder R. Near-infrared fluorescent nanoparticles as combined MR/optical imaging probes. Bioconjug. Chem. 2002;13:554–560. doi: 10.1021/bc015555d. [DOI] [PubMed] [Google Scholar]
- 4.Lee PJ, Peyman GA. Visualization of the retinal and choroidal microvasculature by fluorescent liposomes. Methods Enzymol. 2003;373:214–233. doi: 10.1016/S0076-6879(03)73014-6. [DOI] [PubMed] [Google Scholar]
- 5.Rizzo V, Steinfeld R, Kyriakides C, DeFouw DO. The microvascular unit of the 6-day chick chorioallantoic membrane: a fluorescent confocal microscopic and ultrastructural morphometric analysis of endothelial permselectivity. Microvasc. Res. 1993;46:320–332. doi: 10.1006/mvre.1993.1056. [DOI] [PubMed] [Google Scholar]
- 6.Jilani SM, et al. Selective binding of lectins to embryonic chicken vasculature. J. Histochem. Cytochem. 2003;51:597–604. doi: 10.1177/002215540305100505. [DOI] [PubMed] [Google Scholar]
- 7.Pardanaud L, Altmann C, Kitos P, Dieterlen-Lievre F, Buck CA. Vasculogenesis in the early quail blastodisc as studied with a monoclonal antibody recognizing endothelial cells. Development. 1987;100:339–349. doi: 10.1242/dev.100.2.339. [DOI] [PubMed] [Google Scholar]
- 8.Larson DR, et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 9.Kirchner C, et al. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 10.Goldbach R, Van Kammen A. Structure replication and expression of the bipartite genome of Cowpea mosaic virus. In: Davies J, editor. Molecular Plant Virology. CRC Press; Boca Raton, Florida: 1985. pp. 83–120. [Google Scholar]
- 11.Lomonossoff G, Shanks M. The nucleotide sequence of Cowpea mosaic virus B RNA. EMBO J. 1983;2:2253–2258. doi: 10.1002/j.1460-2075.1983.tb01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porta C, et al. Cowpea mosaic virus-based chimaeras. Effects of inserted peptides on the phenotype, host range, and transmissibility of the modified viruses. Virology. 2003;310:50–63. doi: 10.1016/s0042-6822(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 13.Chatterji A, et al. Chemical conjugation of heterologous proteins on the surface of Cowpea mosaic virus. Bioconjug. Chem. 2004;15:807–813. doi: 10.1021/bc0402888. [DOI] [PubMed] [Google Scholar]
- 14.Lin T, et al. The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- 15.Brennan FR, Jones TD, Hamilton WD. Cowpea mosaic virus as a vaccine carrier of heterologous antigens. Mol. Biotechnol. 2001;17:15–26. doi: 10.1385/MB:17:1:15. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas BL, et al. Characterization of the immune response to canine parvovirus induced by vaccination with chimaeric plant viruses. Vaccine. 2002;20:2727–2734. doi: 10.1016/s0264-410x(02)00200-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks. Wild-type cowpea mosaic virus. Chem. Biol. 2002;9:805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]
- 18.Chatterji A, et al. New addresses on an addressable virus nanoblock; uniquely reactive Lys residues on cowpea mosaic virus. Chem. Biol. 2004;11:855–863. doi: 10.1016/j.chembiol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Gref R, et al. Poly(ethylene glycol)-coated nanospheres: potential carriers for intravenous drug administration. Pharm. Biotechnol. 1997;10:167–198. doi: 10.1007/0-306-46803-4_6. [DOI] [PubMed] [Google Scholar]
- 20.Seandel M, Noack-Kunnmann K, Zhu D, Aimes RT, Quigley JP. Growth factor-induced angiogenesis in vivo requires specific cleavage of fibrillar type I collagen. Blood. 2001;97:2323–2332. doi: 10.1182/blood.v97.8.2323. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J, Lin T, Lomonossoff G. Presentation of heterologous peptides on plant viruses: genetics, structure, and function. Annu. Rev. Phytopathol. 1997;35:67–86. doi: 10.1146/annurev.phyto.35.1.67. [DOI] [PubMed] [Google Scholar]
- 22.Lin T, Porta C, Lomonossoff G, Johnson JE. Structure-based design of peptide presentation on a viral surface: the crystal structure of a plant/animal virus chimera at 2.8 Å resolution. Fold. Des. 1996;1:179–187. doi: 10.1016/s1359-0278(96)00030-2. [DOI] [PubMed] [Google Scholar]
- 23.Lomonossoff GP, Johnson JE. The synthesis and structure of comovirus capsids. Prog. Biophys. Mol. Biol. 1991;55:107–137. doi: 10.1016/0079-6107(91)90003-b. [DOI] [PubMed] [Google Scholar]
- 24.Rae C, et al. Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology. 2005;343:224–235. doi: 10.1016/j.virol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Tsourkas A, et al. In vivo imaging of activated endothelium using an anti-VCAM-1 magnetooptical probe. Bioconjug. Chem. 2005;16:576–581. doi: 10.1021/bc050002e. [DOI] [PubMed] [Google Scholar]
- 26.White JG, Amos WB, Fordham M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J. Cell Biol. 1987;105:41–48. doi: 10.1083/jcb.105.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dessens JT, Lomonossoff GP. Cauliflower mosaic virus 35S promoter-controlled DNA copies of cowpea mosaic virus RNAs are infectious on plants. J. Gen. Virol. 1993;74:889–892. doi: 10.1099/0022-1317-74-5-889. [DOI] [PubMed] [Google Scholar]
- 28.Jones EA, et al. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis. 2002;34:228–235. doi: 10.1002/gene.10162. [DOI] [PubMed] [Google Scholar]
- 29.Zijlstra A, et al. Collagenolysis-dependent angiogenesis mediated by matrix metalloproteinase-13 (collagenase-3) J. Biol. Chem. 2004;279:27633–27645. doi: 10.1074/jbc.M313617200. [DOI] [PubMed] [Google Scholar]
- 30.Hahm B, Arbour N, Oldstone MB. Measles virus interacts with human SLAM receptor on dendritic cells to cause immunosuppression. Virology. 2004;323:292–302. doi: 10.1016/j.virol.2004.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 32.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy VS, et al. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 2001;75:11943–11947. doi: 10.1128/JVI.75.24.11943-11947.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.