Abstract
It has been shown that the heparin-degrading endosulfatase, sulfatase 1 (SULF1), functions as a liver tumor suppressor, but the role of the related sulfatase, sulfatase 2 (SULF2), in liver carcinogenesis remains to be elucidated. We investigated the effect of SULF2 on liver tumorigenesis. Expression of SULF2 was increased in 79 (57%) of 139 hepatocellular carcinomas (HCCs) and 8 (73%) of 11 HCC cell lines. Forced expression of SULF2 increased HCC cell growth and migration, whereas knockdown of SULF2 using short hairpin RNA targeting SULF2 abrogated HCC cell proliferation and migration in vitro. Because SULF1 and SULF2 desulfate heparan sulfate proteoglycans (HSPGs) and the HSPG glypican 3 (GPC3) is up-regulated in HCC, we investigated the effects of SULF2 on GPC3 expression and the association of SULF2 with GPC3. SULF2-mediated cell growth was associated with increased binding of fibroblast growth factor 2 (FGF2), phosphorylation of extracellular signal-regulated kinase and AKT, and expression of GPC3. Knockdown of GPC3 attenuated FGF2 binding in SULF2-expressing HCC cells. The effects of SULF2 on up-regulation of GPC3 and tumor growth were confirmed in nude mouse xenografts. Moreover, HCC patients with increased SULF2 expression in resected HCC tissues had a worse prognosis and a higher rate of recurrence after surgery.
Conclusion
In contrast to the tumor suppressor effect of SULF1, SULF2 has an oncogenic effect in HCC mediated in part through up-regulation of FGF signaling and GPC3 expression.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer death world-wide. 1 Risk factors for HCC include chronic hepatitis and cirrhosis secondary to hepatitis B virus or hepatitis C virus infection, alcoholic cirrhosis, nonalcoholic steatohepatitis, hemochromatosis, alpha-1-antitrypsin deficiency, and dietary aflatoxin exposure.2 Survival of patients diagnosed with advanced HCC is dismal. Only a small proportion of HCCs are detected at an early stage at which potentially curative therapies are feasible. Because of frequent de novo and acquired resistance of HCCs to chemotherapy, standard chemotherapeutic agents are not effective against HCC.3 Recent trials have proven the efficacy of the multi-targeting Raf and receptor tyrosine kinase inhibitor sorafenib in advanced HCC.4 Therefore, there is considerable interest in identifying additional novel gene targets for therapy against HCC.
The recently identified human sulfatase 2 (SULF2) gene at chromosome 20q13 encodes a protein with heparin-degrading endosulfatase activity.5 Cell-surface signaling by several heparin-binding growth factors, including fibroblast growth factor (FGF), heparin-binding epidermal growth factor, hepatocyte growth factor, and vascular endothelial growth factor, requires the binding of both the growth factor and its cognate cell-surface receptor tyrosine kinase to sulfated heparan sulfate glycosaminoglycans. 6,7 For these growth factors, sulfation of particular saccharide moieties of heparan sulfate proteoglycans (HSPGs) is required for growth factor signaling. It has been shown that desulfation of HSPGs by the related protein, sulfatase 1 (SULF1), inhibits binding of the growth factor to its receptor, abrogates growth factor signaling, and has a tumor suppressor effect.8–15 In contrast to SULF1, it has been reported that SULF2 messenger RNA (mRNA) is up-regulated in human breast cancers,16 and suppression of SULF2 expression decreases tumorigenesis of pancreatic cancer cell lines.17 Thus, although SULF2 and SULF1 share the same HSPG substrate, it appears that they have different functional effects.18 This model is supported by recent evidence that human SULF2 mobilizes growth factors and chemokines from heparin and consequently facilitates growth factor signaling. 19
Because our initial studies suggested that SULF2 has an oncogenic effect, we hypothesized that SULF2 increases HCC bcell growth by release of growth factors from cell surface or extracellular matrix HSPG storage sites, thereby enhancing binding of FGF2 and other receptor tyrosine kinase ligands to their corresponding receptors and leading to cell growth. Therefore, we studied the expression of SULF2 in HCCs and examined the role of SULF2 in modulating growth of HCCs. Glypicans are a family of HSPGs that are bound to the cell surface by a lipid glycosyl-phosphatidylinositol anchor. Glypican 3 (GPC3) is markedly overexpressed in a high proportion of HCCs and promotes the growth of HCCs.20–22 We therefore also investigated the potential interactions between SULF2, FGF2 signaling, and GPC3. The following questions were addressed: Is SULF2 expression increased in HCC? Does SULF2 expression affect growth and migration of HCC cells? Does SULF2 affect FGF2 binding to the cell surface? Does SULF2 modulate expression of GPC3? Does SULF2 predict prognosis of HCC patients?
Materials and Methods
Chemicals and Antibodies
Complete Mini Protease Inhibitor Mixture, Protein G Sepharose, and 4′6-diamidino-2-phenylindole dihydrochloride were purchased from Sigma Chemical Co. (St Louis, MO); anti-GPC3 antibody was purchased from BioMosaics (Burlington, VT); anti-phospho-Akt ser 473 and anti-total AKT antibodies were purchased from Cell Signaling (Beverly, MA); and anti-actin antibody, horseradish peroxidase–conjugated mouse immunoglobulin G, and rabbit immunoglobulin G were purchased from Santa Cruz (Santa Cruz, CA). Enhanced chemiluminescence reagents were from Pierce (Rockford, IL). The plasmid vector pSS-H1p was provided by Dr. Daniel D. Billadeau, and vector pG-SUPER was provided by Dr. Shen Cheng. We generated a rabbit polyclonal antibody against SULF2 using a peptide from the SULF2 coding sequence (amino acids 421–444: HKRDNDKVDAQEENFLPKYQRVKD, Genbank accession number NM_018837). The antibody was purified with the Amino Link Column (Pierce).
HCC Cell Lines
The following HCC cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured as recommended: HepG2, Hep3B, Huh-7, SkHep1, PLC/PR5, SNU182, SNU387, SNU398, SNU423, SNU449, and SNU475. Normal primary human hepatocytes were provided by CellzDirect (Pittsboro, NC).
Isolation of Total RNA, Semiquantitative Reverse-Transcription Polymerase Chain Reaction (RT-PCR), and Quantitative Real-Time Polymerase Chain Reaction (PCR)
Total RNA was extracted from 11 HCC cell lines with the RNeasy kit (Qiagen, Valencia, CA). Complementary DNA (cDNA) synthesis was performed with Superscript II RNase H− reverse transcriptase (Life Technologies, Bethesda, MD) to transcribe 2 µg of total RNA primed with 1 µL of 500 µg/mL random hexamers. For semiquantitative RT-PCR, the primers used for SULF2 were SULF2-F (5′-CTGAATCCCCACATCGTCC-TC-3′) and SULF2-R (5′-GTCCACCTTGTCATTGTCTCTCTTGT-3′), yielding a 225–base pair PCR product; for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), they were GAPDH-F (5′-ACCACAGTCCATGCCATCAC-3′) and GAPDH-R (5′-TCCACCACCCTGTTGCTTGTA-3′). RT-PCR was performed as previously reported.11 For quantitative real-time PCR analysis, an ABI TaqMan assay (HS00378697) was used in an ABI 7300 system with the following profile: 95°C for 10 minutes followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. SULF2 mRNA levels were normalized by comparison to 18S ribosomal RNA levels in the same samples. Each measurement was performed in quadruplicate; standard curves were prepared from synthesized SULF2 and 18S standards.
Establishment of SULF2 Stable Transfectant Clones
Recombinant plasmids expressing full-length SULF2 cDNA cloned into the pcDNA3.1 expression plasmid (Invitrogen) were used. Hep3B cells were transfected with either SULF2-expressing plasmid DNA or pcDNA3.1 vector DNA and Lipofectamine Plus reagent (Invitrogen).11 The cells were placed under selection pressure in 400 µg/mL Geneticin (Invitrogen) for 15 to 20 days. Geneticin-resistant clones were isolated and expanded. Stable transfections of the Huh7 cell line using plasmids expressing short hairpin RNA (shRNA) sequences targeting the SULF2 mRNA cloned into the vector pSS-H1p were also performed. The target sequences used for SULF2 shRNA constructs were shRNA-a (AAGTACGTCCACAACCACA) and shRNA-b (AATGTGACTGTCACAAAAT). Constructs containing scrambled target sequences were used as controls. Several stable clonal transfectants were generated from each cell line.
Sulfatase Activity Assay
Ten thousand cells per well were seeded into 96-well plates in phenol red–free Roswell Park Memorial Institute 1640 medium containing 10% fetal calf serum. After 24 hours, the medium was replaced by fresh medium (200 µL/well) and preincubated with 10 µM estrogen-3-O-sulfamate (Sigma Chemicals) at 37°C for 1 hour to inhibit steroid sulfatase. 4-methylumbelliferyl sulfate (4-MUS; Sigma, St. Louis, MO; 0.5 mM) was then added and incubated at 37°C for an additional 4 hours. The supernatant (100 µL) was transferred to white plates (Microfluor; Dynex, Chantilly, VA) containing a 50-µL stop solution (1 M trishydroxymethylaminomethane-HCl, pH 10.4). The plates were read in a Fluoroskan II fluorescence microtiter plate reader (Titertek, Salzburg, Austria) at excitation and emission wavelengths of 355 and 460 nm, respectively.23 All experiments were carried out in triplicate for each sample and repeated at least three times.
Cell Proliferation Assays
For cell-counting assays, SULF2-positive or SULF2-negative cells were plated into 6-well plates at 105 cells per well and incubated in 10% fetal bovine serum (FBS) for 5 to 9 days. Viable cells identified by trypan blue exclusion were counted every 24 hours. For 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays, cells were plated into 96-well plates at 3000 cells per well and incubated in 10% FBS for 48 hours. Cell viability was then assessed by the MTT-reducing capacity.24 The bromodeoxyuridine (BrdU) incorporation assay was performed according to the manufacturer’s instructions (Roche, Indianapolis, IN). Each experiment was performed in six replicates at least three times.
Migration Assay
SULF2-positive or SULF2-negative Huh7 and Hep3B cells were plated onto 6-well plates and grown to confluency. Wounds were made with a 20-µL pipette tip. The wounds were photographed with a phase-contrast microscope at 0 and 24 hours. Cell migration was quantitated by the measurement of the width of the wounds. The experiments were repeated at least three times.
Immunocytochemistry and Confocal Microscopy
SULF2-positive or SULF2-negative Huh7 and Hep3B cells were seeded on glass cover slips in 6-well plates and incubated for 24 hours. Immunocytochemistry and confocal microscopy were performed as previously reported with antibodies against SULF2 and GPC3.11
Western Immunoblotting
SULF2-positive or SULF2-negative Huh7 and Hep3B cells were cultured for 24 hours and harvested for analysis. Whole cell lysates were prepared as previously reported.11 Equal amounts of protein (20 µg/lane) were separated on a 4%–12% trishydroxymethylaminomethane glycine gel (Invitrogen) and transferred to Hybond-ECL nitrocellulose filters (Bio-Rad, Richmond, CA). Blots were probed with polyclonal or monoclonal antibodies against SULF2, GPC3, p-AKTser473, and p-ERK44/42. The levels of total ERK, total AKT, and actin were also measured to control for equal loading. Immunoblots were developed with enhanced chemiluminescence.
Analysis of Biotinylated FGF2 Binding by Flow Cytometry
Recombinant human FGF2 (R&D) was biotinylated with the Pierce EZ-Link MicroSulfo-NHS-LC biotinylation kit. Eight microliters of a 9 mM Sulfo-NHS-LC-Biotin solution in dimethyl sulfoxide was used to label 25 µg of FGF2 in a total volume of 300 µL. Incubation was for 1 hour at room temperature. Excess biotin was removed with a Zeba desalt spin column with a final recovery volume of the biotinylated FGF2 of 500 µL (3.5 µM).
Hep3B cells stably transfected with a SULF2-expressing plasmid vector (clone Hep3B SULF2-5) or with empty vector (clone Hep3B Vector) were cultured for 24 hours and then collected and washed twice in ice-cold phosphate-buffered saline (PBS) with 3% heat-inactivated FBS/0.02% sodium azide. Cells were incubated with 10 nM biotinylated FGF2 at 4°C for 45 minutes. Cells were then stained with 5 µg/mL R-phycoerythrin–conjugated streptavidin (Jackson ImmunoResearch Laboratories, West Grove, PA) at 4°C for 30 minutes in the dark. The cells were washed and resuspended in 500 µL of PBS, and cell cycle analysis was performed with a FAC-Scan analyzer (BD Biosciences, San Jose, CA).
In Vivo Mouse Experiments. SULF2-negative Hep3B Vector and SULF2-positive Hep3B SULF2-5 cells (5 × 105) were each suspended in 150 µL of PBS and inoculated subcutaneously into the right flanks of ten 4- to 6-week-old male BALB/c nu/nu nude mice (National Cancer Institute/National Institutes of Health). Tumors were measured every 3 days, and mice were sacrificed when the tumor volume reached 4000 mm3.25 The experiment was repeated with an additional 10 mice for each cell line. Paraffin sections from xenografts were stained with hematoxylin and eosin and examined by light microscopy. Immunohistochemistry was performed with primary antibody against SULF2 or GPC3 as previously described.26 Negative controls were set up by the replacement of the primary antibody with 1% bovine serum albumin/trishydroxymethylaminomethane-buffered saline. All negative-control specimens showed no nonspecific staining. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Mayo Clinic (Rochester, MN).
Analysis of Human HCCs
RNA from HCC and adjacent benign tissue from 139 individuals (61 Chinese and 78 white) who had surgical resection for HCC was analyzed at the National Cancer Institute with the Qiagen Human Array-Ready Oligo Set (version 2.0), which contains 70-mer probes for 21,329 genes. A Cy-5/Cy-3 dye-swap strategy was used as previously described.26–28 Expression ratios of each gene were averaged from duplicate experiments. The clinical and pathological features for the 139 HCCs are presented in Supplementary Table 2 of Lee et al.27 The median age of the individuals was 57; 73.3% were male. Of the 78 white individuals, 17 underwent liver transplantation, and 9 received palliative treatment. Data from these 26 individuals were not included in the analysis of survival; hence, survival was analyzed for the remaining 113 patients. Recurrence data were available for 66 of the 139 patients and were used for analysis of recurrence. The median duration of follow-up was 23.4 months; 74 individuals died during follow-up. Immunohistochemical staining for Ki-67 (Novocastra Laboratories, Newcastle upon Tyne, United Kingdom) as a measure of cell proliferation and with the ApoTag peroxidase in situ apoptosis detection kit (Serologicals Corp., Norcross, GA) was performed as previously described.26
Statistical Analysis
All data represent at least three independent experiments using cells from separate cultures and are expressed as the mean ± standard error of the mean. Differences between groups were compared with an unpaired two-tailed t test. Differences between the Kaplan-Meier curves of HCC patients with up-regulated or down-regulated SULF2 expression in the HCC tissue in comparison with the adjacent benign tissue were analyzed with the log rank test.
Results
SULF2 Is Up-Regulated in Human HCC Cell Lines
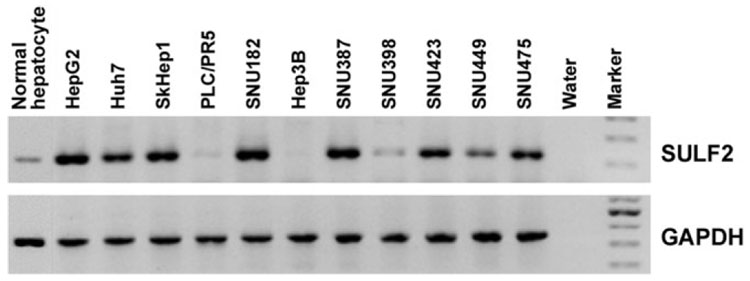
SULF2 mRNA expression was evaluated in 11 HCC cell lines by RT-PCR and real-time RT-PCR; 8 of the 11 HCC cell lines expressed SULF2 at high levels in comparison with primary human hepatocytes (Fig. 1).
Fig. 1. SULF2 mRNA expression in normal primary hepatocytes and 11 HCC cell lines measured by RT-PCR.
GAPDH was used as a loading control.
Expression of SULF2 Promotes HCC Cell Proliferation and Migration
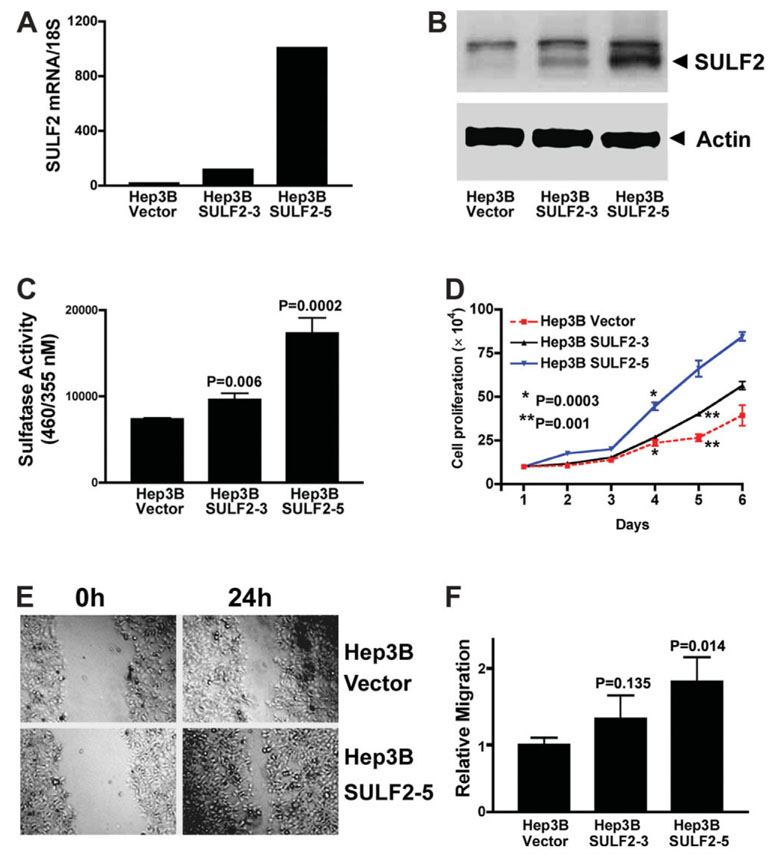
Of the 11 HCC cell lines, Hep3B is the only cell line with almost complete absence of SULF2 mRNA. We transfected a SULF2 plasmid construct or control plasmid vector into Hep3B cells and isolated 18 clones stably expressing SULF2. Quantitation of SULF2 expression by real-time PCR showed that SULF2 transfection increased the relative SULF2 mRNA copies adjusted for 18S ribosomal RNA from 10 copies in Hep3B Vector cells to 82 copies in Hep3B SULF2 clone 3 (Hep3B SULF2-3) and 1008 copies in Hep3B SULF2 clone 5 (Hep3B SULF2-5; Fig. 2A). Western immunoblotting confirmed that Hep3B SULF2-5 expresses SULF2 protein at a high level and that Hep3B SULF2-3 expresses SULF2 at an intermediate level (Fig. 2B). Increased sulfatase enzyme activity in the SULF2-expressing stable clones 3 and 5 was also confirmed (Fig. 2C). To investigate the effect of forced expression of SULF2 on HCC cell growth, we quantitated cell proliferation by cell counting, the MTT cell viability assay, and cell cycle analysis. Hep3B SULF2-5 cells grew significantly faster than Hep3B Vector cells (Fig. 2D). Expression of SULF2 also increased the S phase population from 19.8% ± 3.1% in Hep3B Vector cells to 28.5% ± 2.7% in Hep3B SULF2-5 cells as measured by flow cytometry (P < 0.05). These results were also confirmed with the MTT assay (data not shown). To determine whether forced expression of SULF2 increases HCC cell migration, we measured cell migration using the scratch wound–healing assay in cell monolayers. Forced expression of SULF2 increased Hep3B SULF2-5 cell migration in comparison with Hep3B Vector cells (Fig. 2E,F). Clone Hep3B SULF2-3 expresses a lower level of SULF2 than clone Hep3B SULF2-5 and showed smaller effects on cell proliferation and migration (Fig. 2). We also tested cell proliferation and migration of other high SULF2-expressing clones and obtained results similar to those shown for Hep3B SULF2-5 (data not shown). In summary, therefore, SULF2 enhances growth and migration of Hep3B cells in vitro.
Fig. 2. Forced expression of SULF2 increases HCC cell proliferation and migration.
(A) Plasmids expressing the full-length SULF2 mRNA or the control empty vector pcDNA3.1 were stably transfected into Hep3B cells. SULF2 mRNA expression in different clones was confirmed with quantitative real-time RT-PCR in the three indicated clones. Compared to Hep3B Vector cells, Hep3B SULF2-3 cells showed an intermediate level of SULF2 mRNA expression, whereas Hep3B SULF2-5 cells showed a high level of SULF2 mRNA expression. (B) Western immunoblotting was performed on whole cell lysates from Hep3B Vector, Hep3B SULF2-3, and Hep3B SULF2-5 cells with antibodies against SULF2; the blots were then stripped and reprobed with antibodies against actin to control for loading. (C) Nonsteroid sulfatase activity was detected in the indicated clones after inhibition of steroid sulfatases with estrone-3-O-sulfamate. (D) One hundred thousand Hep3B Vector, Hep3B SULF2-3, or Hep3B SULF2-5 cells were plated into 6-well plates, and 3 wells of each cell clone were harvested, stained with trypan blue, and counted each day. The mean and standard error of triplicate measurements are plotted (P < 0.01 at days 4, 5, and 6, Hep3B SULF2-5 versus Hep3B Vector). (E) Scratch wounds were made in confluent cell culture monolayers with a 20-µL pipette tip; photomicrographs of the wounds were taken at 0 and 24 hours thereafter. (F) Relative migration of the wound edge was quantitated and showed a significantly increased rate of migration in Hep3B SULF2-5 cells when compared to Hep3B Vector cells (P = 0.013).
Knockdown of SULF2 Decreases Cell Proliferation and Migration
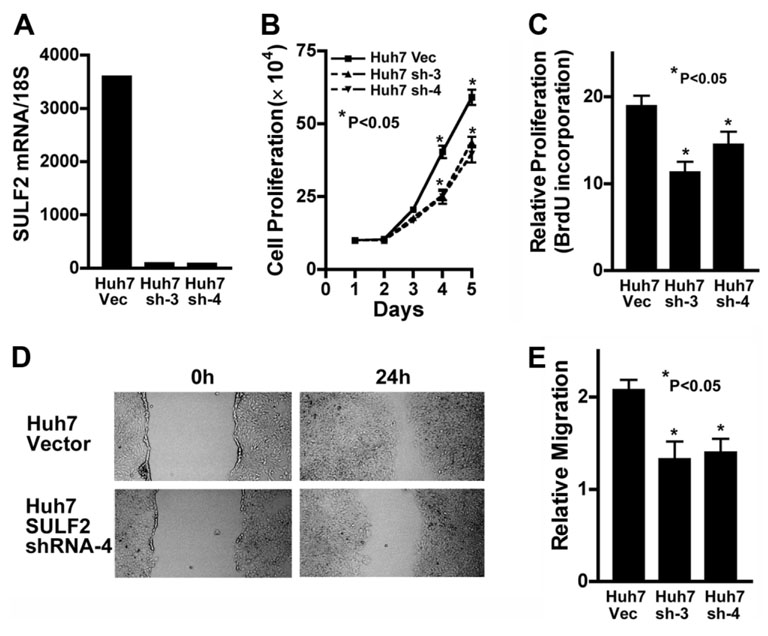
The HCC cell lines Huh7 and SNU182 constitutively express high levels of SULF2 (Fig. 1). Huh7 cells do not express SULF1, whereas SNU182 cells also express SULF1.11 To investigate the effects of down-regulation of SULF2 on cell growth and migration, we stably knocked down SULF2 in Huh7 cells using a plasmid construct expressing an shRNA targeting the SULF2 mRNA. SULF2 mRNA expression in the stable clones was quantitated by real-time RT-PCR. SULF2 mRNA was significantly decreased by expression of the SULF2-targeting shRNA in Huh7 cells in comparison with the control vector (Fig. 3A). Cell proliferation was evaluated by cell counting and by BrdU incorporation. Decreased SULF2 expression inhibited cell growth and decreased BrdU incorporation in Huh7 cells (P < 0.05; Fig. 3B,C). Knockdown of SULF2 also decreased cell migration in Huh7 cells (Fig. 3,E). Similar results were obtained for SNU182 cells (data not shown).
Fig. 3. Knockdown of SULF2 decreases HCC cell proliferation and migration.
(A) Plasmid constructs expressing an shRNA targeting the SULF2 mRNA or the empty vector plasmid pSS-H1p were stably transfected into Huh7 cells, which expressed high levels of SULF2 mRNA. SULF2 mRNA levels were quantitated by real-time RT-PCR in Huh7 Vector (Huh7 Vec) and the two SULF2 shRNA clones, designated Huh7 SULF2shRNA-3 (Huh7 sh-3) and Huh7 SULF2shRNA-4 (Huh7 sh-4). SULF2 mRNA levels were profoundly suppressed by the shRNA plasmid. (B) Huh7 Vec, Huh7 sh-3, and Huh7 sh-4 cells were plated into 6-well plates at 1 × 105 cells per well for the indicated time periods, and the total number of viable cells from each well was counted after trypan blue staining (P < 0.05 at days 4 and 5). (C) Huh7 Vec, Huh7 sh-3, and Huh7 sh-4 cells were plated into 96-well plates at 3000 cells per well. After 24 hours, cell proliferation was evaluated by the BrdU incorporation assay. Knockdown of SULF2 mRNA significantly decreased BrdU incorporation of both shRNA clones (P < 0.05). (D,E) Scratch wounds were made in confluent cell culture monolayers with a 20-µL pipette tip; photomicrographs of the wounds were taken at 0 and 24 hours thereafter. Relative migration of the wound edge was quantitated and showed a significantly decreased rate of migration in Huh7 sh-3 and Huh7 sh-4 cells when compared to Huh7 Vec cells (P < 0.05).
SULF2 Increases FGF2 Binding to HCC Cells and Activates FGF Signaling
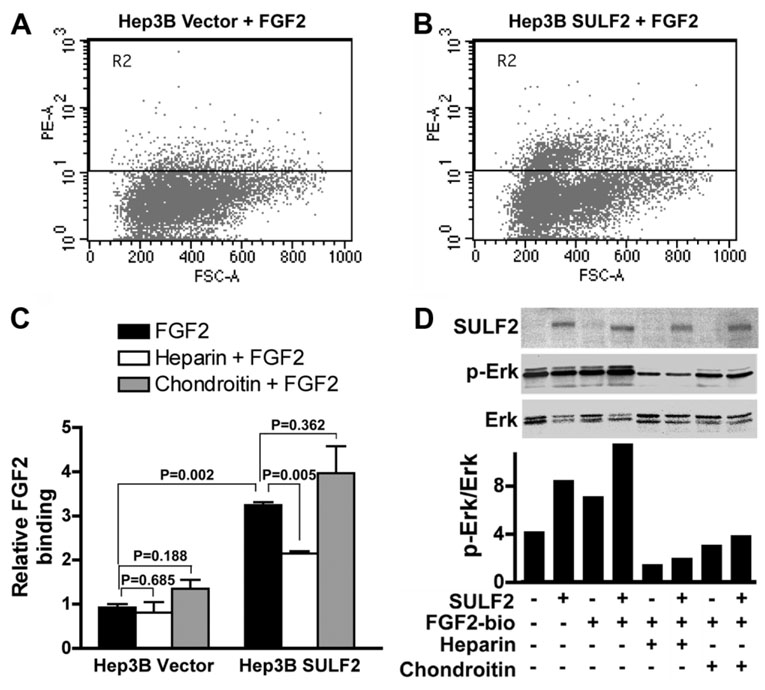
FGF2 signaling is important for HCC cell growth. We have previously confirmed that FGF2 increases FGF receptor phosphorylation and activates downstream phosphorylation of Erk and AKT in HCC cells.11 To determine whether SULF2 affects binding of FGF2 to HCC cells, we incubated Hep3B cells with or without SULF2 expression with biotinylated FGF2 ligand and assayed FGF2 binding by flow cytometry. SULF2 significantly enhanced FGF2 binding in Hep3B SULF2-5 cells versus Hep3B Vector cells (Fig. 4A–C; P = 0.002). To confirm the specificity of FGF2 binding to HSPGs, we determined whether heparin or chondroitin inhibits FGF2 binding. Heparin significantly decreased FGF2 binding to Hep3B SULF2-5 cells, whereas chondroitin did not (Fig. 4C). In parallel experiments, we prepared whole cell lysates and performed immunoblotting for SULF2, phosphorylated Erk, and total Erk. We found that SULF2 increased Erk phosphorylation (Fig. 4D), and these effects were amplified by stimulation with biotinylated FGF2 (Fig. 4D). However, the effect of biotinylated FGF2 on phosphorylation of Erk was decreased by addition of heparin but less so by chondroitin sulfate (Fig. 4D). Similar effects were also found on phosphorylation of AKT (data not shown).
Fig. 4. SULF2 increases FGF2 binding to the cell surface and activates the FGF signaling pathway.
(A–C) Hep3B Vector and Hep3B SULF2-5 cells were collected and incubated with 10 nM biotinylated FGF2 ligand with or without 50 ng/mL heparin or 20 ng/mL chondroitin for 45 minutes at 4°C. After staining with streptavidin, 20,000 cells were counted by flow cytometry. (D) Hep3B Vector and Hep3B SULF2-5 cells were plated into 6-well plates and stimulated with 1 ng/mL biotinylated FGF2 for 15 minutes after overnight starvation. Western immunoblotting was performed on whole cell lysates with antibody against SULF2, p-Erk, and Erk. Optical densities of p-Erk and Erk were quantitated, and the ratios of p-Erk to Erk were calculated. Similar results were found after reprobing of the blots with p-AKT and AKT (data not shown).
Expression of SULF2 Up-Regulates GPC3
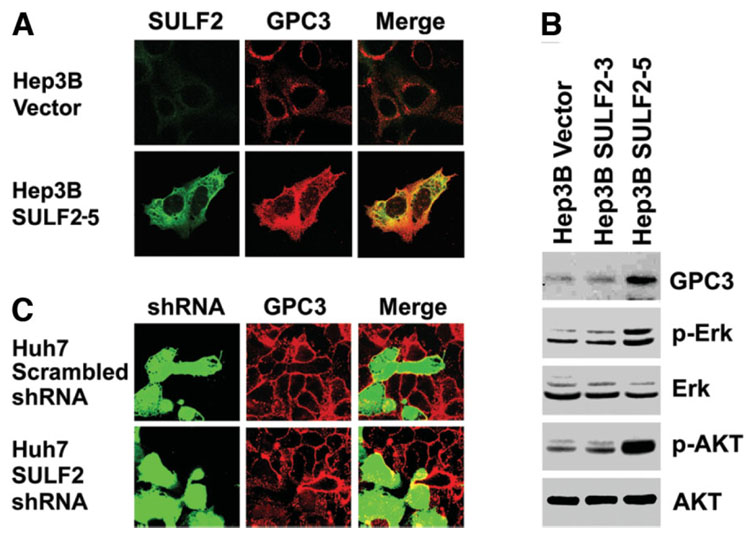
GPC3 is an HSPG that is anchored to the cell membrane. GPC3 has been shown to bind to Wnt3a at the cell surface, enhance Wnt signaling, and promote proliferation of HCC cells.20,21 To determine potential mechanisms by which SULF2 promotes HCC cell proliferation, we investigated the relationship between SULF2, GPC3, and receptor tyrosine kinase signaling in HCC cells. First, we evaluated SULF2-induced changes in expression of GPC3 by immunocytochemistry and confocal microscopy and also by western immunoblotting of lysates from SULF2-positive or SULF2-negative HCC cells. Forced expression of SULF2 increased GPC3 expression; SULF2 and GPC3 also colocalized at the cell surface of Hep3B SULF2-5 cells (Fig. 5A). The SULF2-induced changes in GPC3 were confirmed by western immunoblotting (Fig. 5B). Furthermore, we found that forced expression of SULF2 increased phosphorylation of both Erk and AKT (Fig. 5B).
Fig. 5. Expression of SULF2 up-regulates cell-surface GPC3 and activates the Erk and AKT signaling pathways.
(A) Hep3B Vector and Hep3B SULF2-5 cells were plated on cover slips in 6-well plates. Cells were stained with antibodies against SULF2 (green) and GPC3 (red) without permeabilization. SULF2 and GPC3 were both expressed at the cell surface and colocalized with each other. (B) The blots shown in Fig. 2B were stripped and reprobed with antibodies against GPC3, phospho-AKT, and phospho-Erk. Total AKT and total Erk were used as loading controls. (C) Huh7 cells were plated onto cover slips in 6-well plates and transiently transfected with GFP-expressing plasmids coexpressing either a scrambled shRNA sequence or an shRNA targeting SULF2 mRNA. After 24 hours, cells were immunostained for GPC3.
Next, we examined the effect of down-regulation of SULF2 on GPC3 expression. Huh7 cells express a high level of GPC3 on their cell surface. Cells were transfected with plasmids coengineered to express both shRNA and green fluorescent protein (GFP). After transfection of a plasmid construct expressing a control scrambled shRNA, there was no change in GPC3 expression at the surface of Huh7 cells; however, knockdown of SULF2 using an shRNA plasmid construct targeting the SULF2 mRNA down-regulated cell-surface GPC3 expression (Fig. 5C).
Knockdown of GPC3 Decreases FGF2 Binding
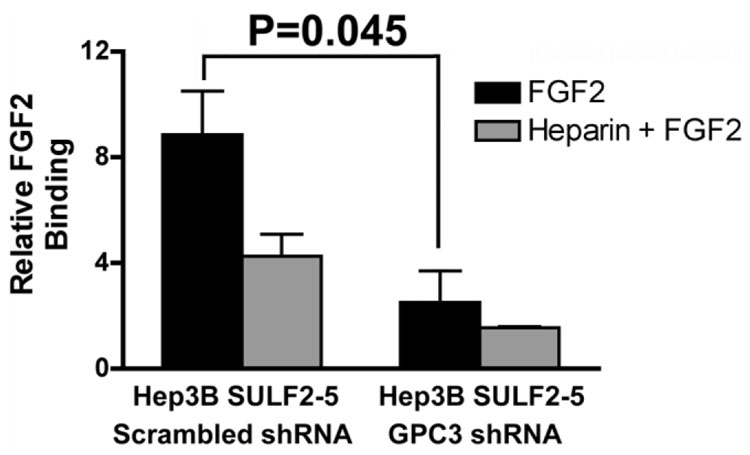
To explore whether GPC3 is required for enhancement of FGF signaling by SULF2, we examined the effects of down-regulation of GPC3 in Hep3B SULF2-5 cells. After transfection of a pG Super-based plasmid construct coexpressing GFP and either a scrambled shRNA or GPC3-targeting shRNA, we repeated the FGF2 binding assay as before. Cells were sorted by gating for GFP, and FGF binding of transfected cells was compared. We found that knockdown of GPC3 significantly decreased FGF2 binding to Hep3B SULF2-5 cells (Fig. 6). Therefore, GPC3 plays an important role in SULF2-enhanced FGF signaling in vitro.
Fig. 6. Knockdown of GPC3 inhibits FGF2 binding.
Plasmid constructs expressing a GFP-coexpressing plasmid with an shRNA targeting the GPC3 mRNA or a scrambled shRNA were transiently transfected into Hep3B SULF2-5 cells. Cells were collected and incubated with 10 nM biotinylated FGF2 ligand with or without 50 ng/mL heparin for 45 minutes at 4°C. After staining with streptavidin, 20,000 cells were counted by flow cytometry. FGF2 binding was compared in GFP-gated cells.
SULF2 Promotes Tumorigenesis in Nude Mice and Up-Regulates GPC3 In Vivo
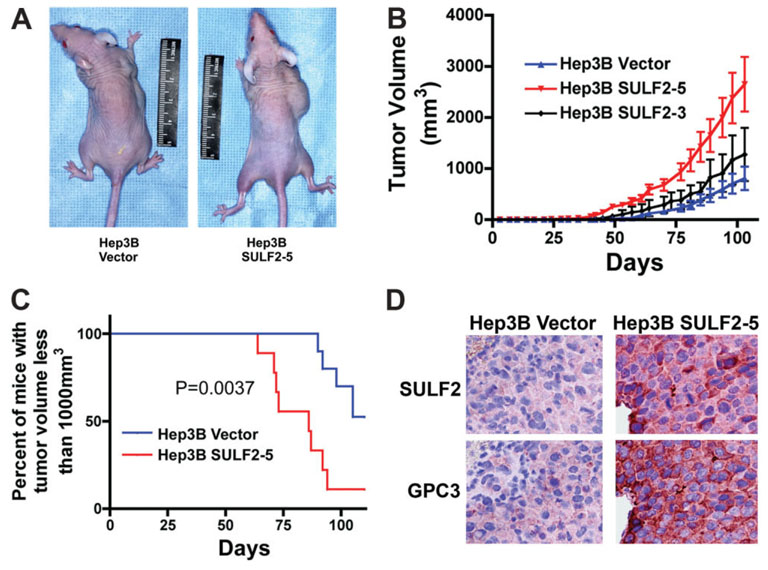
To confirm the in vitro effects of forced expression of SULF2 in vivo, we inoculated 5 × 105 Hep3B Vector and Hep3B SULF2-5 cells subcutaneously into 10 nude mice each and observed the growth of the tumors over time. SULF2 expression significantly increased tumor growth, decreasing the median time to reach a tumor size of 1000 mm3 by 28 days. (Fig. 7A–C). To confirm the SULF2-induced changes of GPC3 in vivo, we performed immunohistochemistry using antibodies against SULF2 and GPC3 in consecutive sections of xenografts derived from Hep3B Vector and Hep3B SULF2-5 cells. We found that, similar to our in vitro results, SULF2-induced up-regulation of GPC3 also occurred in vivo (Fig. 7D). Therefore, promotion of the tumor growth of Hep3B xenografts by SULF2 was associated with enhanced expression of GPC3 in vivo.
Fig. 7. SULF2 promotes tumor growth and up-regulates GPC3 in vivo in HCC xenografts in nude mice.
(A) 5 × 105 Hep3B Vector (left mouse) or Hep3B SULF2-5 cells (right mouse) in 150 µL of PBS were inoculated subcutaneously into the right frank of 10 mice for each cell clone. (B,C) Tumor growth was monitored with calipers. SULF2 expression significantly enhanced tumor growth in vivo. (D) Successive sections from paraffin-embedded xenografts from Hep3B Vector (left panel) and Hep3B SULF2-5 cells (right panel) were immunostained with antibodies against SULF2 and GPC3, respectively; nuclei were counterstained with hematoxylin.
SULF2 Is Up-Regulated in Human HCC and Predicts a Worse Prognosis
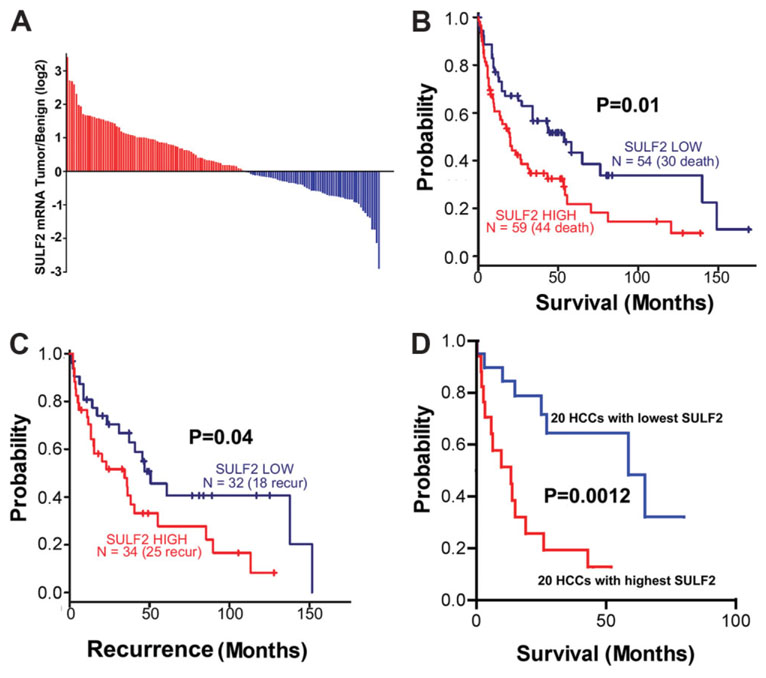
To investigate the role of SULF2 in HCC development and progression, we abstracted the SULF2 mRNA expression results from microarray experiments performed on benign and tumor tissues from 139 HCC patients from four different geographic regions. There was increased SULF2 expression in the tumor compared to the benign tissues in 79 (57%) of the HCCs (Fig. 8A). Survival analysis showed that patients with tumors expressing high levels of SULF2 had a significantly worse prognosis (P = 0.01; Fig. 8B) and a more rapid rate of tumor recurrence after surgery (P = 0.04; Fig. 8C). To assess the prognostic value of mRNA gene expression profiles in HCCs, Lee et al.27,28 analyzed the global gene expression patterns of 91 human HCCs and identified two distinct subclasses (A and B) of HCC that are highly associated with patient survival. Patients with a cluster A gene expression pattern have a worse prognosis than patients in cluster B. We therefore compared the survival curves and the association between SULF2 expression and the A and B prognostic subclasses in the 20 patients with the highest tumor SULF2 level and in the 20 patients with the lowest tumor SULF2 expression. We found that not only did patients with the highest SULF2 expression have shorter survival (P < 0.01; Fig. 8D) but there was also a significant association of high SULF2 expression with the poor prognosis cluster A subclass (P < 0.0001; Table 1). The association of high SULF2 expression with subclass A was maintained when all 139 HCCs were analyzed; 45 of 79 (57%) patients with up-regulated SULF2 were in subclass A, whereas 17 of 60 (28%) patients with down-regulated SULF2 were in subclass A (P = 0.0008, chi-square test). The results of immunohistochemical analysis of cell proliferation and apoptosis were obtained for 30 of the HCCs, 14 with higher SULF2 than adjacent benign tissue and 16 with lower SULF2 expression than the adjacent benign tissue. As with the entire group of HCCs, subclass A tumors were more frequent in the high SULF2 group (13 of 14) and less frequent in the low SULF2 group (2 of 16; P = 0.00001, chi-square test). HCCs with high SULF2 expression had a significantly higher Ki-67 proliferation index (28.0 versus 15.9, P = 0.007, two-sample t test) and a significantly lower apoptosis index (0.49 versus 0.96, P = 0.0001, two-sample t test) than those with low SULF2 expression. SULF2 expression therefore correlated with increased proliferation and decreased apoptosis. Information on microvascular density was not available; however, for the 76 HCCs with data on gross vascular invasion, there was a trend toward an increased frequency of vascular invasion in the high SULF2 group (39 of 50, 78%) compared to the low SULF2 group (15 of 26, 58%; P = 0.06, chi-square test). Thus, SULF2 is frequently up-regulated in human HCCs and predicts a more aggressive tumor phenotype. Examination of the list of genes that correlate most closely to the tumor expression of SULF2 revealed the presence of a number of tyrosine kinase signaling and Wnt pathway genes.
Fig. 8. SULF2 mRNA expression is increased in approximately 60% of HCCs.
High SULF2 is associated with decreased patient survival and more rapid recurrence after surgical resection. (A) Results of oligonucleotide microarray analysis performed on 139 pairs of tumor and benign samples from primary HCCs, plotted as a log 2–fold change of SULF2 mRNA expression, tumor over benign. (B,C) Survival and tumor recurrence after surgical resection in 139 patients with HCCs. Differences between the Kaplan-Meier curves of patients with up-regulated expression of SULF2 and those with down-regulated expression of SULF2 with the log rank test. (D) Survival after surgical resection in the 20 patients with the highest tumor/benign SULF2 mRNA expression and in the 20 patients with the lowest tumor/benign SULF2 mRNA expression. Increased SULF2 is associated with a worse survival and more rapid recurrence after surgical resection of the primary tumor.
Table 1.
Association Between SULF2 Expression and the Cluster A and Cluster B Prognostic Gene Expression Signatures in the 20 Patients with the Highest SULF2 mRNA Expression and the 20 Patients with the Lowest SULF2 mRNA Expression
| Cluster A | Cluster B | |
|---|---|---|
| Highest SULF2 | 18 (90%) | 2 (10%) |
| Lowest SULF2 | 3 (15%) | 17 (85%) |
Cluster A showed worse prognosis; cluster B showed better prognosis. P < 0.0001.
Discussion
Recent advances in cancer biology suggest that a limited number of pathways are responsible for initiating and maintaining dysregulated cell proliferation, which is the major cellular alteration responsible for the cancer phenotype. New agents in development target several of these critical pathways, including the mitogen-activated protein kinase pathway, the phosphoinositide 3-kinase/Akt pathway, the Wnt/betacatenin signaling pathway, and the sonic hedgehog pathway. Many of these pathways have ligands for which cell-surface proteoglycans act as coreceptors. Studies of the newly discovered family of heparin-degrading endosulfatases, SULF1 and SULF2, suggest that HSPGs in the extracellular matrix or on the cell surface can sequester growth factor ligands and cytokines in a sulfation-dependent manner and release the factors when desulfated by heparin-degrading endosulfatases. The principal findings of the present study include the following: (1) up-regulation of the heparin-degrading endosulfatase SULF2 is found in approximately 60% of HCCs and 73% of HCC cell lines, (2) SULF2 functions as an oncogenic protein promoting growth and migration of HCC cells, (3) SULF2 increases FGF2 binding to HCC cells, (4) SULF2 upregulates the HSPG GPC3 at the cell surface, (5) the effect of SULF2 on FGF binding is decreased by knockdown of GPC3, and (6) up-regulation of SULF2 correlates with a worse prognosis in HCC patients.
The human sulfatases SULF1 and SULF2 have been proposed to mediate effects on carcinogenesis through desulfation of cell-surface HSPGs.9 We have previously found that SULF1 functions as a tumor suppressor in HCC, head and neck cancer, and ovarian cancer.9,10,11,29 SULF1 has also been shown to inhibit tumor proliferation in vitro and in vivo in pancreatic cancer.12,30 However, the functional role of SULF2 in cancer has not been completely elucidated. It has been reported that like SULF1, SULF2 also inhibits the growth of multiple myeloma cells.8 In contrast, SULF2 is up-regulated and has been shown to be mutated in breast cancer, although the functional effects of these mutations are not yet determined. 16,31 Increased expression of SULF2 is also associated with increased resistance of the MCF-7 cell line to chemotherapy.32 Furthermore, silencing of SULF2 was recently shown to decrease proliferation and survival of pancreatic cancer cells.17
In the present study, we found that unlike SULF1, SULF2 functions as an oncogenic protein in HCC. Gene expression analysis of 139 pairs of HCC and adjacent benign tissues showed that SULF2 was upregulated in 57% of HCCs. Up-regulation of tumor SULF2 was associated with a worse prognosis and higher risk of recurrence. In a previous in vitro study, we showed that 9 of 11 HCC cell lines had loss of expression of SULF111; in contrast, we found that SULF2 was expressed at high levels in 8 of the same 11 HCC cell lines. We transfected SULF2 constructs into SULF2-negative Hep3B HCC cells and knocked down SULF2 in SULF2-expressing Huh7 HCC cells and found that SULF2 promotes proliferation and migration of HCC cells. The tumorigenic effect of SULF2 was also confirmed in vivo in nude mice.
To determine the specific mechanistic role of SULF2 in HCCs, we investigated the effects of SULF2 and the HSPG GPC3 on FGF signaling in liver carcinogenesis. We found that SULF2 up-regulated GPC3 expression and FGF2 ligand-receptor binding and signaling. Consistent with this result, knockdown of SULF2 in the SULF2-overexpressing Huh7 HCC cell line down-regulated cell membrane GPC3 expression. Thus, the effects of SULF2 appear to be mediated in part through increased GPC3 expression. Further study will explore the machinery by which SULF2 up-regulates GPC3.
In summary, we have made the novel observations that SULF2 is overexpressed in the majority of HCCs and that up-regulation of SULF2 correlates with worse prognosis of HCC patients. In addition, our results suggest that SULF2 functions as an oncogenic protein in HCCs in part through activation of FGF signaling and the downstream AKT/mitogen-activated protein kinase pathway and increased expression of GPC3, with consequent promotion of HCC cell proliferation and migration in vitro, resulting in enhancement of HCC tumor growth in vivo. We are now pursuing studies to determine the exact mechanism by which SULF2 regulates GPC3 and to determine how SULF2 modulates tyrosine kinase activation at the cell surface. These unique observations raise important questions about the molecular mechanisms underlying the disparate effects of SULF1 and SULF2 on cell growth. Because both molecules appear to have 6-O-endosulfatase activity on HSPGs, we hypothesize that they subserve their differing effects through the nonenzymatically active C-terminal domains of the proteins, which have different structures and may target the sulfatases to different subtypes and/or subdomains of HSPGs. Experiments specifically designed to examine this hypothesis are currently in development.
Acknowledgment
The authors thank Dr. Shen Cheng for the provision of the pG-SUPER vector, Dr. Daniel D. Billadeau for the provision of the pSSH1p plasmid vector, Patrick L. Splinter and Linda M. Murphy for technical assistance, Erin M. Bungum and Stacy L. Roberson for secretarial assistance, and Dr. Gregory J. Gores and Dr. Rosebud O. Roberts for critical review of the manuscript.
Supported by the Mayo Clinic and the Mayo Clinic Cancer Center, by National Institutes of Health grant CA100882, by The Richard M. Schulze Family Foundation, and by the Miles and Shirley Fiterman Center for Digestive Diseases at the Mayo Clinic, Rochester, MN (to L.R.R.).
Abbreviations
- BrdU
bromodeoxyuridine
- cDNA
complementary DNA
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GPC3
glypican 3
- HCC
hepatocellular carcinoma
- HSPG
heparan sulfate proteoglycan
- mRNA
messenger RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RT-PCR
reverse-transcription polymerase chain reaction
- shRNA
short hairpin RNA
- SULF1
sulfatase 1
- SULF2
sulfatase 2
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Roberts LR, Gores GJ. Emerging drugs for hepatocellular carcinoma. Expert Opin Emerg Drugs. 2006;11:469–487. doi: 10.1517/14728214.11.3.469. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Alfa GK, Schwartz L, Ricci S, Amadori D, Santoro A, Figer A, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:4293–4300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- 5.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pye DA, Vives RR, Hyde P, Gallagher JT. Regulation of FGF-1 mitogenic activity by heparan sulfate oligosaccharides is dependent on specific structural features: differential requirements for the modulation of FGF-1 and FGF-2. Glycobiology. 2000;10:1183–1192. doi: 10.1093/glycob/10.11.1183. [DOI] [PubMed] [Google Scholar]
- 7.Pye DA, Vives RR, Turnbull JE, Hyde P, Gallagher JT. Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J Biol Chem. 1998;273:22936–22942. doi: 10.1074/jbc.273.36.22936. [DOI] [PubMed] [Google Scholar]
- 8.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, et al. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 9.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 10.Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, et al. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 11.Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, Montoya DP, et al. hSulf1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126:231–248. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, Felix K, et al. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer. 2005;4:14. doi: 10.1186/1476-4598-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 14.Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, et al. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66:6025–6032. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 15.Shridhar V, Sen A, Chien J, Staub J, Avula R, Kovats S, et al. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002;62:262–270. [PubMed] [Google Scholar]
- 16.Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, et al. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7:1001–1010. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS ONE. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP., Jr Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 2006;281:4969–4976. doi: 10.1074/jbc.M511902200. [DOI] [PubMed] [Google Scholar]
- 19.Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, et al. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capurro MI, Shi W, Sandal S, Filmus J. Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma growth. J Biol Chem. 2005;280:41201–41206. doi: 10.1074/jbc.M507004200. [DOI] [PubMed] [Google Scholar]
- 21.Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 22.Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, Chen CT, et al. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2004;315:950–958. doi: 10.1016/j.bbrc.2004.01.151. [DOI] [PubMed] [Google Scholar]
- 23.Wolff B, Billich A, Brunowsky W, Herzig G, Lindley I, Nussbaumer P, et al. Microtiter plate cellular assay for human steroid sulfatase with fluorescence readout. Anal Biochem. 2003;318:276–284. doi: 10.1016/s0003-2697(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 24.Betz CS, Lai JP, Xiang W, Janda P, Heinrich P, Stepp H, et al. In vitro photodynamic therapy of nasopharyngeal carcinoma using 5-aminolevulinic acid. Photochem Photobiol Sci. 2002;1:315–319. doi: 10.1039/b109817a. [DOI] [PubMed] [Google Scholar]
- 25.Lai JP, Yu C, Moser CD, Aderca I, Han T, Garvey TD, et al. SULF1 inhibits tumor growth and potentiates the effects of histone deacetylase inhibitors in hepatocellular carcinoma. Gastroenterology. 2006;130:2130–2144. doi: 10.1053/j.gastro.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 26.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 27.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. HEPATOLOGY. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 29.Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, et al. Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26:4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 30.Abiatari I, Kleeff J, Li J, Felix K, Buchler MW, Friess H. Hsulf-1 regulates growth and invasion of pancreatic cancer cells. J Clin Pathol. 2006;59:1052–1058. doi: 10.1136/jcp.2005.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 32.Chekhun VF, Lukyanova NY, Kovalchuk O, Tryndyak VP, Pogribny IP. Epigenetic profiling of multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals novel hyper- and hypomethylated targets. Mol Cancer Ther. 2007;6:1089–1098. doi: 10.1158/1535-7163.MCT-06-0663. [DOI] [PubMed] [Google Scholar]