Abstract
Wild type green fluorescent protein (wt-GFP) and the variant S65T/H148D each exhibit two absorption bands, A and B, which are associated with the protonated and deprotonated chromophores respectively. Excitation of either band leads to green emission. In wt-GFP, excitation of band A (~390 nm) leads to green emission with a rise time of 10–15 picoseconds, due to excited state proton transfer (ESPT) from the chromophore hydroxyl group to an acceptor. This process produces an anionic excited state intermediate I* that subsequently emits a green photon. In the variant S65T/H148D, the A band absorbance maximum is red-shifted to ~415 nm and as detailed in the accompanying papers (1, 2), when the A band is excited, green fluorescence appears with rise time shorter than the instrument time resolution (~170 fs). Based on steady state spectroscopy and high resolution crystal structures of several variants described herein, we propose that in S65T/H148D, the red shift of absorption band A and the ultrafast appearance of green fluorescence upon excitation of band A is due to a very short (≤ 2.4 Å), and possibly low barrier, hydrogen bond between the chromophore hydroxyl and introduced Asp148.
Wt-GFP exhibits two bands in absorption spectra, peaked at 395 and 475 nm and designated A and B respectively [see reviews (3–5)]. While band A is attributed to absorption of the protonated chromophore, band B is due to absorption of the deprotonated chromophore (6, 7). Excitation of either band leads to green emission; however upon excitation of band A, the intensity of green emission rises on a time scale of several tens of picoseconds. This process is dramatically slowed upon deuteration of the sample, suggesting that excited state proton transfer (ESPT) from the neutral form of the chromophore generates an intermediate anionic excited state, which subsequently emits a green photon (6–9). On the basis of the crystal structures, a pathway has been proposed to allow proton transfer between the chromophore phenol moiety, through a water molecule and Ser205, to Glu222 as the terminal proton acceptor (10–12). In wt-GFP, the intensities of absorbance bands A and B are largely insensitive to pH over the range 4–10.
Point mutations can lead to increased sensitivity to pH and the formation of new ESPT pathways within GFP. For example, dual emission GFPs (deGFPs) switch between a form that emits blue light at low pH and a form that emits green light at high pH. Time resolved spectroscopy and structural studies of deGFPs suggested that the dual emission behavior results from pH-dependent formation of alternative ESPT pathways (13–15). Mutations can also eliminate the fluorescence from band A, for example, the S65T variant (a precursor of a popular molecular label EGFP (16)), has a strong pH dependence of fluorescence with pKa about 6.0. At pH < 6.0 the protonated form of the chromophore predominates, leading to strong absorbance from band A, however fluorescence emission from the protonated chromophore is vanishingly weak.
The subject of this and the accompanying papers (1, 2) is the unusual photophysical behavior of the double mutant GFP S65T/H148D, the construction of which was previously described (17). As briefly described in that work, the additional mutation His148 -> Asp upon the S65T background restores green fluorescence from band A (17). As with the variant S65T, the excitation and emission spectra of S65T/H148D are pH sensitive, consistent with a chromophore pKa of about 8.0. As the pH is varied, two-state behavior is observed with respect to green emission from either band A or B, indicating that the two species directly interconvert. Thus this mutant finds application as a ratiometric pH sensor (18).
GFP S65T/H148D exhibits unusual photophysical properties. Absorbance band A is maximal at 415 nm (17), thus it is red shifted by ~20 nm compared to wt-GFP or the S65T variant. In time resolved spectroscopic studies, both weak blue (~460 nm) and strong green (~510 nm) emission are observed upon excitation of band A at 400 nm, however both blue and green emission are characterized by a rise time that is shorter than the instrument response function (IRF, ~170fs). As detailed in the accompanying papers (1, 2), the emission kinetics are only weakly sensitive to deuterium substitution.
Here, we report on mutagenic and high resolution crystallographic studies which demonstrate that the unusual photophysical properties of S65T/H148D variant are related to a very short hydrogen bond (≥ 2.4 Å) between the chromophore hydroxyl and Asp148.
Materials and Methods
Mutagenesis, protein expression and crystallization
Mutagenesis was performed using the QuikChange method (Stratagene). Protein was expressed in E. coli strain JM109 (DE3), by use of the PRSETB His tagged expression system. Protein was purified by Ni2+ affinity chromatography over Ni-NTA agarose (Qiagen, Chatsworth, CA) and then buffer exchanged with PD-10 Sephadex columns (Amersham Pharmacia) into 50 mM HEPES (pH 7.9). Crystals of S65T/H148D were obtained after two months by hanging drop vapor diffusion (2 μl protein solution (50 mM HEPES pH7.9, A280=20.3) were mixed with 2 μl well solution (100 mM MgCl2, 100mM NaOAc pH 5.6, 26% PEG 4000)). For the high pH structure of S65T/H148D, crystals were soaked in 100 mM MgCl2, 100mM CAPS pH 10.0, 26% PEG 4000 overnight. S65T/H148N crystallized in 50 mM Li2SO4, 100 mM TRIS pH 9.5, 30% PEG 4000 after one week. To obtain crystals of S65T/H148N at low pH, crystals were soaked in 200 mM Ca(OAc)2, 100 mM MES pH 5.0, 20% PEG 8000. GFP H148D crystallized in 100 mM MgCl2, 100 mM Tris pH 9.0, 30% PEG 1550 overnight.
Spectroscopy
Absorbance spectra (400 μg/ml protein in 100 mM buffers) were recorded with a HP 8453 UV-visible spectrophotometer. Protein pKas were determined as described previously (13). Fluorescence spectra were taken with a Perkin-Elmer LS-55 fluorometer after diluting protein samples to 30–100 μg/ml in 2 ml 100 mM buffers of citric acid, HEPES, or CHES as appropriate. The quantum yields (QY) of emission were measured by referencing to the standards 9-aminoacridine and fluorescein. As controls, QY were determined for wt-GFP and S65T as well (Table 1). These values are in good agreement with previous results (4, 5).
Table 1.
Effects of substitution of residue 148 on the GFP
| Mutant | Absorbance band(nm) | Emission banda(nm) | pKa | Quantum yieldb | |||
|---|---|---|---|---|---|---|---|
| A | B | A | B | A | B | ||
| S65T | 394 | 489 | 459/508 | 510 | 5.95±0.02c | 0.02±0.01 | 0.67±0.01 |
| S65T/H148N | 393 | 488 | 459/510 | 511 | 6.37±0.02 | 0.01±0.01 | 0.45±0.02 |
| S65T/H148E | 397 | 484 | 460/510 | 511 | 7.17±0.05 | 0.04±0.01 | 0.24±0.02 |
| S65T/H148D | 415 | 487 | 510 | 510 | 7.95±0.02c | 0.21±0.04 | 0.32±0.03 |
| H148D | 400 | 469 | 503 | 505 | > 10.4 | 0.15±0.02 | 0.40±0.02 |
| wild type GFP | 395 | 475 | 460/509 | 505 | N.A. | 0.75±0.03 | 0.75±0.03 |
The emission maxima resulting from excitation at the peak of either absorbance band A or B.
Quantum yields of bands A and B were measured at pH 4.5 or 5 and 11, respectively.
Taken from reference (17)
Data collection and structure solution
Diffraction data sets were collected from single flash-frozen crystals of S65T/H148N at pH 5.0 and 9.5 (100K) using ADSC-Q315 detector on beam line 8.2.2 at the Advanced Light Source (Berkeley, CA). Diffraction data for S65T/H148D at pH 5.6 and 10.0, H148D at pH 9.0 were collected on a Quantum-315 CCD on beam line 14-BM-C at the Advanced Photon Source (Argonne, IL). Data were reduced using HKL2000 (HKL Research) or d*TREK (Molecular Structure Corp.). Molecular replacement was performed with EPMR (19) using the GFP S65T (PDB entry 1EMA) as a search model, providing unambiguous identification of the space group and unique solutions to the rotation and translation problems. Rigid body refinement was performed using TNT (20) and model building was conducted with O (21) or Coot (22) in several stages of increasing resolution. SHELX-97 (23) was used in the final stages of refinement for S65T/H148D at pH 10.0, S65T/H148N at pH 9.5 and H148D at pH 9.0. In order to calculate the standard uncertainties of the very short hydrogen bond between the chromophore and Asp148 (see Results), the low pH S65T/H148D model was also submitted to automated refinement by SHELX.
Results
Steady-state spectra
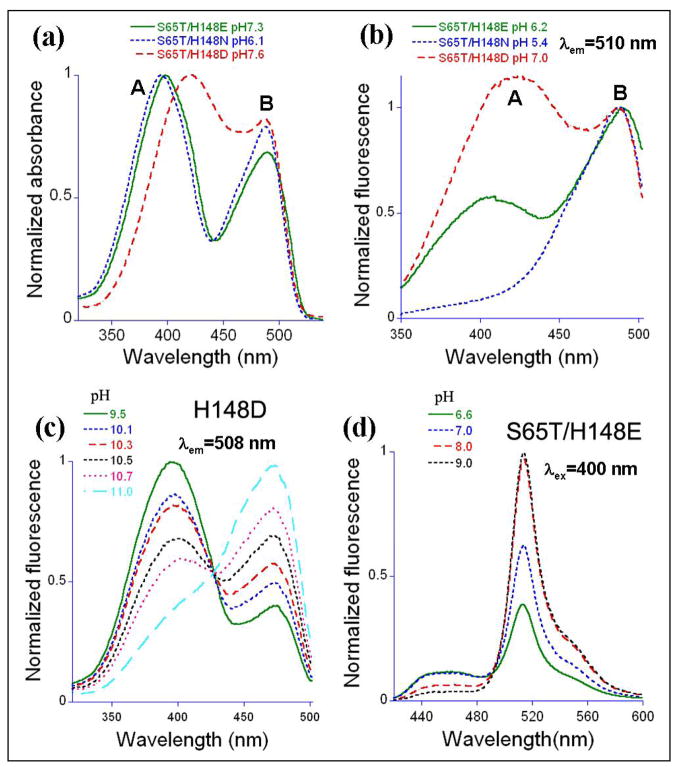
GFP S65T/H148D displays two pH titratable (pKa ~ 8.0) bands in absorption and excitation spectra (Fig. 1a and b)(17). Compared to the S65T variant, band A is broad and red shifted to 415 nm (by ~20 nm). However, the B band maximum remains at ~ 490 nm, essentially unchanged from that of S65T. Excitation of band A leads to relatively strong green emission (QY = 0.21), which is very weak in the S65T variant (QY ~ 0.02). Evidently, upon band A excitation, the H148D mutation partially reverses the severe reduction in fluorescence efficiency attributed to the S65T mutation. The emission maximum is approximately the same (509 +/− 1 nm) for all variants except for GFP/H148D, which emits at ~503 nm.
Figure 1.
Normalized absorbance, excitation and emission spectra. (a) Absorbance spectra of S65T/H148E/N/D at pH 7.3/6.1/7.6, respectively. (b) Excitation spectra of S65T/H148E/N/D by monitoring fluorescence at 510 nm at pH 6.2/5.4/7.0, one unit lower than the respective variants’ pKas. (c) Excitation spectra of H148D with detection at 508 nm as a function of pH. (d) Emission spectra of S65T/H148E with excitation at 400 nm as a function of pH.
To investigate the role of Asp148 on the spectroscopic properties we first mutated it to asparagine, which is isosteric but cannot carry a charge. The S65T/H148N variant has properties that are very similar to those of S65T (Table 1). Absorbance spectra reveal two pH-titratable bands (Fig. 1a) with pKa ~ 6.4, suggesting that a partial charge on Asp148 could be responsible for the apparent chromophore pKa of 8.0 in S65T/H148D. The Asn 148 substitution restores the band A absorbance maximum to ~ 395 nm, whereas the band B maximum remains at ~ 490 nm. S65T/H148N is essentially nonfluorescent (QY ~ 0.01) upon excitation of band A (Fig. 1b). Excitation of band B however, leads to efficient green fluorescence with QY = 0.45.
The possible effect of a negative charge at the 148 position was studied by substitution with glutamate. The variant S65T/H148E also shows two bands in absorption spectra (Fig. 1a) with the band A maximum again consistent with wild type at ~395 nm. Green emission from band A is weak with QY = 0.04, whereas emission from band B has QY = 0.24 (Table 1). Nevertheless, S65T/H148E is a new dual emission GFP [deGFP (13)], in that it shows both blue and green emission upon excitation at 400 nm, with relative intensities depending strongly on pH (Fig. 1d). Blue emission at 460 nm is increased at low pH, while the green emission is increased at high pH. The pKa (7.2) of S65T/H148E is very close to physiological pH and is intermediate between those of S65T/H148D (8.0) and S65T/H148N (6.4) Thus, this mutant might find application as a pH indicator in living cells.
For these mutations, the change in side chain polarity near the chromophore (specifically at positions 148 and/or 222) correlates well with an increase in the chromophore pKa, as exhibited by the series (S65T-> S65T/H148N-> S65T/H148E, S65T/H148D-> H148D) (Table 1).
We also characterized the role of the S65T mutation in the S65T/H148D variant by reverting Thr65 to serine. The generated variant H148D behaves in many ways like wt-GFP. The A band maximum is 400 nm in both absorption and excitation spectra (for emission at 510 nm). Excitation of either the A or B band leads to green emission, as in wt-GFP (Fig. 1c). Band A dominates at pH below 9 and can be decreased by increasing pH higher than 9, with apparent pKa greater than 10.5. This variant is ratiometric by excitation (Fig. 1c), similar to S65T/H148D (17). Thus, the mutation His148->Asp on the wild-type background is shown to confer ratiometric pH-sensitive behavior, which might conceivably find applications in high-pH environments; however the pKa is very high.
We conclude from the mutagenesis and the steady-state spectroscopic analysis that in S65T/H148D, the H148D mutation is required for both the ~20 nm red shift of band A, and for green emission from band A. The S65T mutation is required for the red shift, acting synergistically with H148D, however S65T is not essential for green emission upon excitation of band A.
Crystal structures
As detailed in the accompanying papers (1, 2), the photophysics of green emission from S65T/H148D (upon band A excitation) is unusual for its rise time of less than 170 fs (IRF). In order to investigate the structural basis for this remarkable behavior, crystal structures of S65T/H148D, S65T/H148N and H148D were solved, with the former two at both low and high pH. All variants crystallized isomorphously with S65T GFP (24), with one protomer in the asymmetric unit. Data collection and refinement statistics for the atomic models are presented in Table 2. In all crystals, electron density is weak for residues 230–238, so these residues were not modeled. For the same reason, residues not modeled in S65T/H148D were 1–3, 189–190 (low pH) and 1, 147–148, 155–157 (high pH); in S65T/H148N were 1–3, 156–157 (low pH) and 1 (high pH); in H148D were 1–3, 147–148, 155–159 (pH 9.0). The refinement and atomic model statistics are excellent. PROCHECK (25) reveals that there are no residues in either the disallowed or “generously allowed” regions of the Ramachandran diagram.
Table 2.
Data collection and refinement statistics
| S65T/H148D(pH 5.6) | S65T/H148D(pH 10.0) | S65T/H148N(pH 5.0) | S65T/H148N(pH 9.5) | H148D(pH 9.0) | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) | 51.1,63.0,68.7 | 50.9,62.6,69.9 | 51.2,62.5,68.9 | 50.7,62.4,68.6 | 50.7,62.6,68.6 |
| Resolution (Å) | 50–1.50(1.55–1.50) | 50–1.24(1.28–1.24) | 34–1.40(1.45–1.40) | 10–1.20(1.24–1.20) | 50–1.36(1.41–1.36) |
| Rsym or Rmerge (%) | 4.9 (42.3) | 4.4 (32.9) | 5.5 (50.8) | 5.3 (33.4) | 4.9 (31.9) |
| I/σI | 35.0 (2.1) | 42.9 (2.7) | 11.0 (2.0) | 17.5 (3.9) | 55.7 (8.4) |
| Completeness (%) | 94.7 (68.6) | 93.7 (61.8) | 98.1 (85.9) | 94.5 (93.3) | 96.0 (86.9) |
| Redundancy | 3.2 (2.2) | 3.5 (2.2) | 6.4 (3.9) | 6.5 (6.2) | 3.7 (3.6) |
| Refinement | |||||
| Resolution (Å) | 6–1.50 | 10–1.24 | 6–1.40 | 10–1.20 | 10–1.36 |
| No. reflections | 34,193 | 59,945 | 43,438 | 61,533 | 45,807 |
| Rwork/Rfree | 17.2/24.4 | 15.47/20.4 | 17.7/24.1 | 15.2/20.9 | 15.1/21.6 |
| No. atoms | |||||
| Protein | 1769 | 1773 | 1799 | 1808 | 1797 |
| Water | 294 | 249 | 237 | 324 | 267 |
| B-factors (Å2) | |||||
| Protein | 27.0 | 18.1 | 28.7 | 19.0 | 21.9 |
| Water | 40.1 | 32.0 | 43.8 | 37.4 | 36.5 |
| R.m.s. deviations | |||||
| Bond length (Å) | 0.017 | 0.013 | 0.020 | 0.013 | 0.011 |
| Bond angle (Å) | 1.8(°) | 0.031 | 1.9 (°) | 0.031 | 0.029 |
Values in parenthesis indicate statistics for the highest resolution shell.
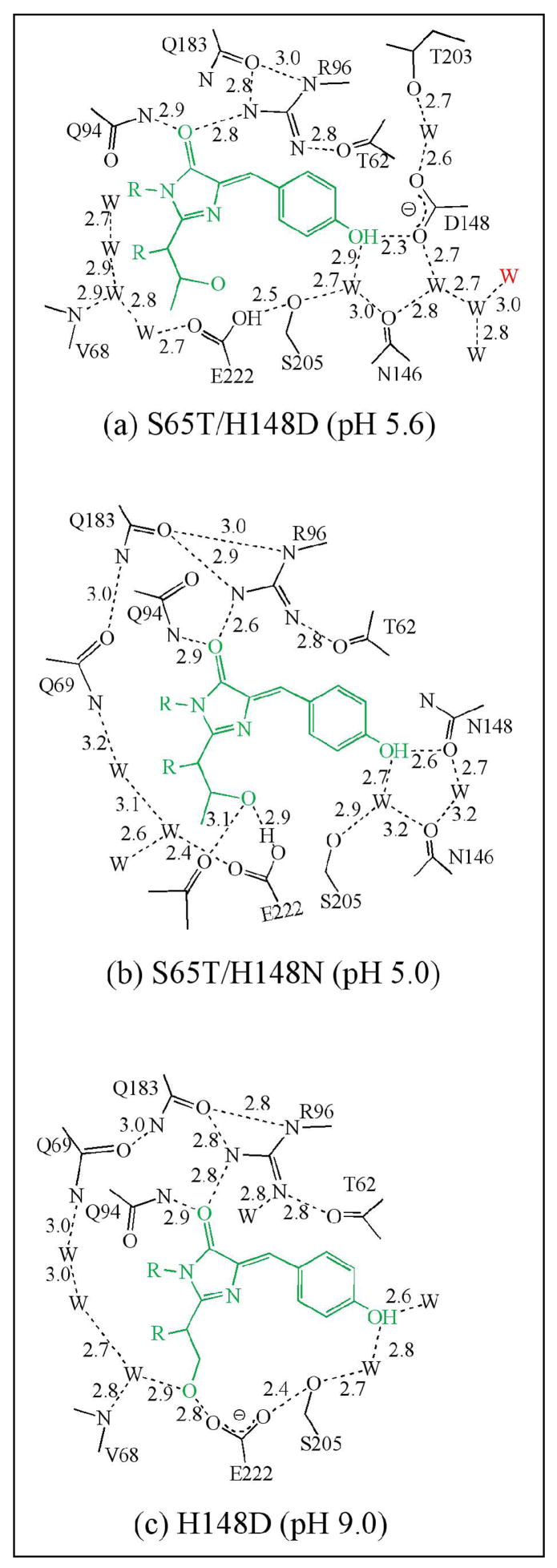
Upon superposition of the α-carbons with GFP S65T, rms deviations are observed to be less than 0.40 Å for all structures. The atomic models are thus all very similar, which indicates that the protein matrix is largely insensitive toward variation at position 148. Schematic diagrams (Fig. 2a, b, c) of the chromophore environment in each case demonstrate that the overall environment is similar to that of S65T or wt-GFP. Nevertheless, interesting and significant local effects are observed as detailed in the following sections.
Figure 2.
Schematic diagram of chromophore environment of (a) S65T/H148D (pH 5.6), (b) S65T/H148N (pH 5.0) and (c) H148D (pH 9.0). Hydrogen bonds are shown in black dashed lines, labeled with approximate lengths in Å. The water shown in red in panel (a) is exterior to the protein. The protonation state of the chromophore was assigned on the basis of results from absorption spectroscopy.
S65T/H148D
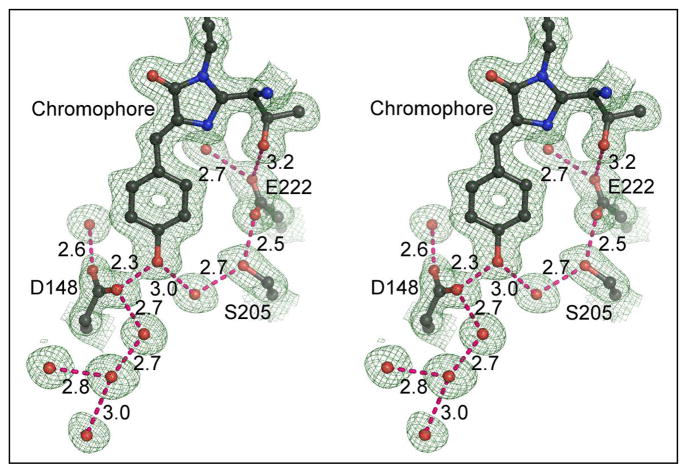
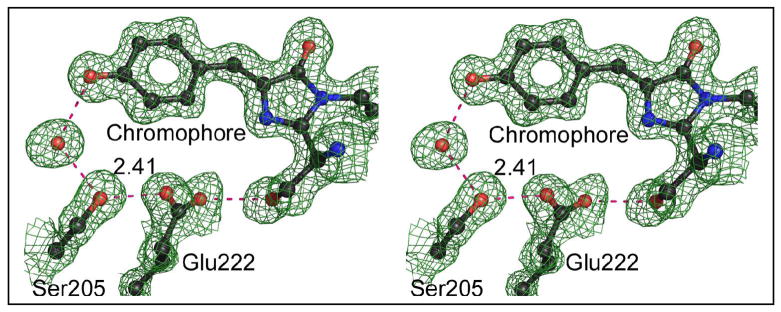
The crystal structure of this protein at pH 5.6 was refined at 1.5 Å resolution. A portion of the final 2Fo-Fc map (Fig. 3) reveals that a very short hydrogen bond, 2.32 Å(as determined after refinement using isotropic temperature factors with TNT), forms between phenol oxygen of the chromophore and a carboxylate oxygen of Asp148. After SHELX refinement with anisotropic temperature factors, the length of this hydrogen bond is estimated to be 2.41 ± 0.03 Å (estimated standard error). Given that the model errors in the two refinement procedures are different, the most probable value lies somewhere between these two estimates. Asp148 adopts the least favored χ1 rotamer (~60°, see Fig. 3), which according to the rotamer library of COOT (22), is seen in only 2% of cases. Two symmetrically placed water molecules are hydrogen bonded with the two carboxylate oxygens of Asp148 separately. In addition, a hydrogen-bonded water chain is attached to Asp148, leading to the exterior of the protein. Finally, a hydrogen bond network (chromophore OH->H2O->Ser205->Glu222) is formed, restoring the potential ESPT pathway found in wt-GFP (10, 11). As this ESPT pathway is disrupted by the S65T mutation, it appears that one consequence of the H148D mutation is indeed to reverse some of the changes caused by the S65T mutation, even though these sites are spatially quite distinct in the molecule.
Figure 3.
Atomic model and (2Fo-Fc) electron density map of S65T/H148D at pH 5.6. Contour level 1σ. Dashed lines indicate hydrogen bonds, labeled with distances in Å. Blue spheres are nitrogen, red are oxygen.
The crystal structure of S65T/H148D at pH 10 was determined at 1.24 Å resolution. A dramatic structural change is observed in that the main chain segment comprising Asp148 and Ser147 is disordered at this pH. No significant electron density is observed in the final 2Fo-Fc map for these two residues (data not shown). We presume that the disorder is a result of a repulsive electrostatic interaction between the negatively charged chromophore and Asp148. As described below, this observation accounts for the overall pH dependence of fluorescence.
S65T/H148N
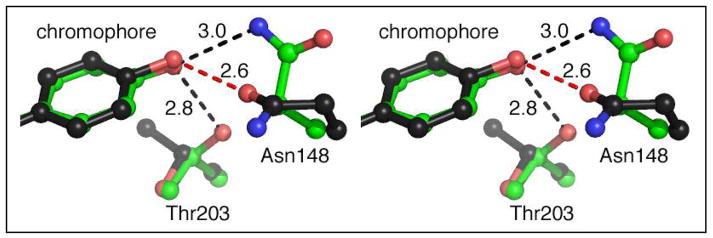
Crystal structures of this protein were determined at 1.4 (pH 5.0) and 1.2 Å resolution (pH 9.5). The low-pH crystal structure indicates that Glu222 is partially disordered and has two conformations; one of which is found in the S65T structure (Fig. 2b) (17), while the other conformation restores a potential ESPT pathway similar to that found in wt-GFP (chromophore OH->H2O->Ser205->Glu222) (10, 11). However, this pathway seems not to contribute to emission from band A since the protein is essentially nonfluorescent at low pH. We attribute the very low quantum yield of fluorescence to the nonplanarity of the chromophore in this variant (see below). We note that in GFP structural disorder in Glu222 is often observed, for example as seen in wt-GFP (26). The conformations of Asn148 and Thr203 are pH-dependent (Fig. 4), presumably due to the change in protonation state of the chromophore hydroxyl. Thr203 side chain hydroxyl forms a hydrogen bond with the chromophore phenol end only at high pH, as found in wt-GFP (10, 11). Based on this change in the hydrogen bonding pattern, it is reasonable to conclude that the chromophore hydroxyl is the group that dissociates at high pH.
Figure 4.
Stereo image of superposed atomic models showing structural changes in S65T/H148N at pH 5.0 (black bonds) and 9.5 (green bonds). Dashed lines indicate hydrogen bonds, labeled with distances in Å. Blue spheres are nitrogen, red are oxygen.
At the resolution of these diffraction data, nitrogen and oxygen atoms cannot be distinguished from one another, however based on the hydrogen bond pattern between Asn148 and the chromophore, the orientation of the side chain amide group of Asn148 could be inferred. At pH 5.0, the amide oxygen of Asn148 appears to accept a proton from the chromophore and the hydrogen bond is of typical length (2.62 Å). At pH 9.5, the side chain of Asn148 reorients, rotating by ~125° in χ1, so that the amide nitrogen donates a proton to the anionic hydroxyl oxygen of the chromophore.
H148D
Crystal structures of H148D were determined at pH 7 and 9 (apparent chromophore pKa > 10.5) and the atomic model of the high pH crystals could be refined at 1.36 Å resolution. The structure of H148D is very similar to that of wt-GFP although Asp148 is disordered at both pH values (data not shown), presumably due to a repulsive interaction between the negatively charged Asp148 and Glu222. The ESPT pathway from the phenol end of the chromophore to Glu222, as found in wt-GFP, survives upon the H148D mutation, and is shown in Fig. 5. The hydrogen bond between Glu222 and Ser205 has estimated length of 2.4 Å, which is short relative to all other nearby hydrogen bonds between 2.6 to 3.0 Å. At pH 7.0, the crystal structure of H148D shows no obvious differences to the crystal structure at pH 9 with Asp148 disordered (data not shown). Thus, the pKa of Asp148 appears to be much lower than 7, as expected.
Figure 5.
Stereo image of (2Fo-Fc) electron density map and atomic model of H148D at pH 9.0 showing potential ESPT pathway. Blue spheres are nitrogen, red are oxygen; contour level 1σ. Dashed lines indicate the hydrogen bonds, labeled with distances in Å. For other hydrogen bond lengths refer to Fig. 2c.
Nonplanar chromophore geometry
The chromophores in all GFP variants discussed show significant deviations from planarity, which is modeled by allowing the two torsion angles associated with the linkage between the 5- and 6-membered rings to deviate from 180° (27). The twist and tilt angles describing the distortion are significantly larger than those of the highly fluorescent proteins such as DsRed and GFP (27) (Table 3), but less than those found in the nonfluorescent chromoproteins KFP (27) and Rtms5 (28). The chromophore for both S65T/H148D and S65T/H148N is more bent at low pH than at high pH, which correlates with the dependence of the QY on pH (Table 1). This apparent correlation between relatively low QY and the chromophore non-coplanarity is also seen in monomeric red fluorescent proteins known as mFruits (29), suggesting a general relationship. The non-coplanarity of the chromophore is presumably due to the shape of protein cavity (30).
Table 3.
Chromophore geometry in the GFPs, angles in degrees. See reference (27) for a definition of twist and tilt angles. The quantum yields were measured by excitation at maximum of absorbance band at specific pH
| Model | QY | Twist | Tilt |
|---|---|---|---|
| wt-GFP | 0.79 | 0.0 | 3.7 |
| DsRed | 0.79 | 0.9 | 0.2 |
| S65T/H148D (pH 5.6) | 0.21 | 9.5 | 9.6 |
| S65T/H148D (pH 10.0) | 0.32 | 3.2 | 9.0 |
| S65T/H148N (pH 5.0) | 0.01 | 7.6 | 13.5 |
| S65T/H148N (pH 9.5) | 0.45 | 1.3 | 7.7 |
| H148D (pH 9.0) | 0.15 | 0.4 | 12.8 |
Discussion
The double mutant GFP S65T/H148D is unusual with respect to a number of its photo- and biophysical properties. The introduction of the S65T mutation into wt-GFP leads to loss of fluorescence upon excitation of the A band, which is restored by the second site mutation H148D. Unlike wt-GFP, both mutants are strongly pH sensitive and display two-state titration behavior. S65T/H148D is ratiometric by excitation, which potentially makes this variant useful as a pH sensor for use in living cells. Furthermore, the second site mutation introduces a large red shift into the A band absorbance, without introducing a significant shift in the B band absorbance maximum or the fluorescence emission. Finally, the rise time of green emission is ultrafast, in contrast to wild type, where green emission upon excitation of the A band shows a rise time of tens of picoseconds.
Structural studies revealed that in S65T/H148D, Asp148 makes a very short (<2.4 Å) hydrogen bond with the chromophore hydroxyl oxygen, which suggests a strong, specific interaction. However it is possible that the effect of Asp148 is due to a nonspecific electrostatic interaction, that is, the effect results from the introduction of a negative charge adjacent to the chromophore. In order to examine these possibilities and untangle the various possible contributions of Asp148 and Thr65, we made a number of substitutions at these positions and compared their respective biophysical properties.
In the following we focus first on the structural basis of the ratiometric pH sensitivity and then on the possible relationships between the very short H-bond and the unusual photophysical properties of the S65T/H148D mutant. Several possible mechanisms accounting for the extremely short rise time of green fluorescence are briefly discussed. For more details, the reader is referred to the two the accompanying papers (1, 2).
Structural basis of the ratiometric pH sensitivity
pH-dependent transitions in the excitation spectra of S65T/H148D can be accurately modeled assuming a single-site titration with pKa ~ 8.0. The structural basis of the ratiometric pH sensitivity is suggested by the crystal structures of S65T/H148D at pH 5.6 and 10.0. At low pH, a well-ordered H-bond forms between the chromophore and Asp148, whereas at high pH, the chain segment comprised of Ser147 and Asp148 becomes highly disordered. It is expected that the pKa of Asp148 is much lower than 8.0 and thus it is reasonable to assign the apparent pKa of ~ 8.0 to direct titration of the chromophore hydroxyl group. This assignment is supported by evidence from structural analysis of S65T/H148N at low and high pH, where the pH-dependent rearrangement of Asn148 and Thr203 implicated the chromophore hydroxyl as the titrating group, with pKa about 6.4. The assignment is also consistent with earlier structural studies of the GFP variant S65T, in which the chromophore hydroxyl was identified as the titrating group (17).
A simple two-state model adequately explains the ratiometric response of S65T/H148D to titration. At low pH, the chromophore population is on average protonated and as in wt-GFP, ESPT can explain the strong green fluorescence from band A. At high pH, the anionic form of the chromophore predominates and gives rise to green fluorescence as expected. Due to charge-charge repulsion between Asp148 and the chromophore, a segment of the main chain comprising residues 147 and 148 becomes disordered. This disorder does not appear to substantially compromise fluorescence efficiency for excitation of the anionic chromophore.
However, the results from ultrafast spectroscopy suggest that for wt-GFP and S65T/H148D, the mechanisms of fluorescence emission upon excitation of the A bands are different.
Band A red shift
As described above, the red shift of band A of GFP S65T/H148D can be attributed to the interaction between the chromophore and Asp148. In order to determine whether the red shift is due to an electrostatic interaction, or is related to the formation of a short hydrogen bond, we introduced further mutations at positions 65 and 148 and characterized the resulting purified proteins.
Substitution of Asp148 with glutamate leads to normal A band absorbance of 397 nm, observable over the range of pH 5–9 (above pH 9, A band absorbance is weak or nonexistent). It is very likely that the introduced Glu148 is negatively charged over this range, thus we conclude that a presumed negative charge on Asp148 cannot be responsible for the red shift in the S65T/H148D mutation. Likewise, reversion of Thr65 to Ser, as found in wild type, leads to a variant containing the single mutation H148D. This variant has a normal A band absorbance maximum at 400 nm and an extremely high apparent pKa (> 10). This behavior is similar to wt-GFP, in which the negative charge of Glu222 is proposed to stabilize the protonated chromophore (10). Thus, it is likely that in H148D, Glu222 is also negatively charged. Structural studies revealed that the Asp 148 side chain is disordered, but is nevertheless in the immediate vicinity of the chromophore. It is very likely that Asp148 is anionic over the range of pH 9–11. Thus, the absence of a significant red shift is consistent with our previous conclusion that electrostatic effects alone cannot be responsible for the effect.
Finally, the mutant S65T/H148N was investigated, and crystal structures were determined at pH 5.0 and 9.5. Asn148 forms a hydrogen bond to the chromophore hydroxyl at either pH (Fig. 4), but in either case the hydrogen bond lengths are typical and the A band absorbance maximum is about 393 nm. However, the side chain of Asn148 does rearrange as a function of pH, consistent with a change in hydrogen bonding due to dissociation of the chromophore hydroxyl at high pH.
From the above, we conclude that in the variant S65T/H148D, the band A red shift is due to the short hydrogen bond formed between the chromophore and Asp148. One possible explanation for the red shift is that the short hydrogen bond promotes additional delocalization of the chromophore electron density over the carboxylate of Asp148, thus increasing the physical extent of the chromophore. If this interaction were to be maintained in the excited state, one would expect emission to be similarly red shifted, however it is normal at ~510 nm. On the basis of this observation, we additionally suggest that the chromophore-Asp148 interaction is altered or lost in the excited state.
Ultrafast rise time of green fluorescence
Upon excitation of the A band, the rise time of green fluorescence of S65T/H148D is faster than the instrument response function (170 fs). Weak blue fluorescence is also observable, however as detailed in the accompanying papers (1, 2), the kinetics of blue and green fluorescence suggest multiple emitting species, some of which may not interconvert via ESPT pathways. As pointed out above, in S65T/H148D an ESPT pathway similar to that found in wt-GFP is present (see Fig. 3). It seems unlikely that the restored ESPT pathway could be responsible for the ultrafast kinetics of green emission of S65T/H148D, since in wt-GFP, green fluorescence attributed to ESPT arises on the time scale of tens of picoseconds (6, 7). Moreover, the rise time is significantly slowed upon deuteration of the sample, which is not the case for S65T/H148D.
Thus, one must look elsewhere to explain the ultrafast rise time (< 170 fs) of green fluorescence. We again suggest that the short H-bond between the chromophore and Asp148 is responsible for this behavior. One possibility to consider is ESPT from the protonated chromophore phenol to Asp148 through the short H-bond. Such ultrafast proton transfer (with transfer times expected to be on the order of 10–20 fs) has previously been described, for example in solution between preformed, hydrogen-bonded HPTS…−OAc complexes (31). However, to our knowledge, there is no precedence for similar observations of ultrafast proton transfer within a protein molecule.
A second possibility is that the short hydrogen bond indeed has a low barrier, or no barrier, to proton transfer and that the proton is delocalized between the chromophore hydroxyl and the carboxylate of aspartate 148. Thus, effectively instantaneous proton transfer could be achieved as a result of collapse of the proton wave function.
As detailed in the accompanying papers (1, 2), the kinetics of blue and green fluorescence in S65T/H148D are quite complicated. This may be related to the presence of a minor species or proton transfer pathway not apparent in the crystal structure (up to 10–15% occupancy of a minor species would be difficult or impossible to detect in the electron density maps. It should be noted that the crystal contains no glycerol, while for low temperature spectroscopic experiments in the accompanying papers (1, 2), were conducted with the sample in ~ 60% glycerol solution. As an example of alternative species not detectable in the crystal structure, different protonation or tautomeric states of the titratable groups comprising the chromophore/Asp148 interaction may need to be considered. The reader is referred to the accompanying papers (1, 2) for kinetic analyses and more detailed discussions.
A low barrier hydrogen bond?
In order to investigate whether the short hydrogen bond between the chromophore and Asp148 is a so-called low barrier hydrogen bond (LBHB) (32, 33), we determined the 600 MHz jump-return 1H-NMR spectrum of GFP S65T/H148D (pH 5.6, 28 °C). However, the spectrum does not reveal any unusually downfield shifted resonances diagnostic of a solvent exchange-protected hydrogen in a LBHB (34, 35) (see Supplemental Figure S1). The resolved signals between 10 and 13 ppm all arise from nitrogen-bonded protons in imidazole or amide moieties, as evidenced by ~95 Hz scalar couplings in a uniformly 15N-labeled sample of the protein (L. McIntosh and M. Heller, data not shown). No additional downfield signals were detected upon reducing the temperature to 5 °C. The excitation maximum for the jump-return pulse sequence was ~15 ppm. We conclude from the NMR data that there is no evidence for an unusual downfield chemical shift related to any O–H…O hydrogen bond. It nevertheless seems possible that the very short hydrogen bond between the chromophore hydroxyl and Asp148 is a LBHB that is not protected from rapid solvent exchange. On the other hand, cases have been identified in which the chemical shift of a proton in a very short hydrogen bond is not as large as one might expect (36, 37). These observations were attributed to increased shielding of the proton by the approaching heavy atoms.
In conclusion, we suggest that the double mutant GFP-S65T/H148D and several variations thereof offer unique opportunities to study excited state proton transfer pathways in an environment characterized by an unusually short hydrogen bond. Furthermore, the environment can be perturbed by mutagenesis and the positions of the interacting groups may be determined at atomic resolution.
Supplementary Material
Figure S1, showing results from a 600 MHz jump-return NMR experiment to investigate whether unusual downfield shifts might be associated with the chromophore-Asp148 hydrogen bond. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank L. McIntosh and M. Heller, University of British Columbia, for obtaining the 1D- NMR spectra of S65T/H148D.
Abbreviations
- GFP
green fluorescent protein
- H-bond
hydrogen bond
- LBHB
low barrier hydrogen bond
- ESPT
excited state proton transfer
- IRF
instrument response function
- rms
root mean square
Footnotes
This work was supported by grants from the NIH (R01 GM42618 to S.J.R. and GM 27738 to S.G.B.). The atomic coordinates and structure factors have been deposited in the Protein Data Bank (entry 2DUF and 2DUE for GFP S65T/H148D at pH 5.6 and 10.0 respectively, 2DUG and 2DUH for GFP S65T/H148N at pH 5.0 and 9.5, 2DUI for GFP H148D at pH 9.0).
References
- 1.Shi X, Abbyad P, Shu X, Kallio K, Kanchanawong P, Childs W, Remington SJ, Boxer SG. doi: 10.1021/bi7009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiderman P, Genosar L, Huppert D, Kallio K, Remington SJ, Solntsev KM, Tolbert LM. doi: 10.1021/bi7009053. [DOI] [PubMed] [Google Scholar]
- 3.Remington SJ. Structural basis for understanding spectral variations in green fluorescent protein. Bioluminescence and Chemiluminescence, Pt C. 2000;305:196–211. doi: 10.1016/s0076-6879(00)05489-6. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Zimmer M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chemical Reviews. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 6.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent protein: Multiple states and proton transfer. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lossau H, Kummer A, Heinecke R, Pollinger Dammer F, Kompa C, Bieser G, Jonsson T, Silva CM, Yang MM, Youvan DC, MichelBeyerle ME. Time-resolved spectroscopy of wild-type and mutant Green Fluorescent Proteins reveals excited state deprotonation consistent with fluorophore-protein interactions. Chemical Physics. 1996;213:1–16. [Google Scholar]
- 8.Creemers TM, Lock AJ, Subramaniam V, Jovin TM, Volker S. Three photoconvertible forms of green fluorescent protein identified by spectral hole-burning. Nature structural biology. 1999;6:557–560. doi: 10.1038/9335. [DOI] [PubMed] [Google Scholar]
- 9.Kennis JTM, Larsen DS, van Stokkum IHM, Vengris M, van Thor JJ, van Grondelle R. Uncovering the hidden ground state of green fluorescent protein. Biophys J. 2004;86:168A–168A. doi: 10.1073/pnas.0404262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, Wlodawer A. The structural basis for spectral variations in green fluorescent protein. Nature structural biology. 1997;4:361–365. doi: 10.1038/nsb0597-361. [DOI] [PubMed] [Google Scholar]
- 12.Stoner-Ma D, Jaye AA, Matousek P, Towrie M, Meech SR, Tonge PJ. Observation of excited-state proton transfer in green fluorescent protein using ultrafast vibrational spectroscopy. J Am Chem Soc. 2005;127:2864–2865. doi: 10.1021/ja042466d. [DOI] [PubMed] [Google Scholar]
- 13.Hanson GT, McAnaney TB, Park ES, Rendell MEP, Yarbrough DK, Chu SY, Xi LX, Boxer SG, Montrose MH, Remington SJ. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1 Structural characterization and preliminary application. Biochemistry. 2002;41:15477–15488. doi: 10.1021/bi026609p. [DOI] [PubMed] [Google Scholar]
- 14.McAnaney TB, Park ES, Hanson GT, Remington SJ, Boxer SG. Green fluorescent protein variants as ratiometric dual emission pH sensors. 2 Excited-state dynamics. Biochemistry. 2002;41:15489–15494. doi: 10.1021/bi026610o. [DOI] [PubMed] [Google Scholar]
- 15.McAnaney TB, Shi X, Abbyad P, Jung H, Remington SJ, Boxer SG. Green fluorescent protein variants as ratiometric dual emission pH sensors. 3 Temperature dependence of proton transfer. Biochemistry. 2005;44:8701–8711. doi: 10.1021/bi050132a. [DOI] [PubMed] [Google Scholar]
- 16.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 17.Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 18.De Giorgi F, Lartigue L, Bauer MK, Schubert A, Grimm S, Hanson GT, Remington SJ, Youle RJ, Ichas F. The permeability transition pore signals apoptosis by directing Bax translocation and multimerization. Faseb J. 2002;16:607–609. doi: 10.1096/fj.01-0269fje. [DOI] [PubMed] [Google Scholar]
- 19.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallographica Section D-Biological Crystallography. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 20.Tronrud DE, Teneyck LF, Matthews BW. An Efficient General-Purpose Least-Squares Refinement Program for Macromolecular Structures. Acta Crystallographica Section A. 1987;43:489–501. [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A 47. 1991:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Sheldrick GM, Schneider TR. SHELXL: High-resolution refinement. Macromolecular Crystallography, Pt B. 1997;277:319–343. [PubMed] [Google Scholar]
- 24.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 26.van Thor JJ, Zanetti G, Ronayne KL, Towrie M. Structural events in the photocycle of green fluorescent protein. J Phys Chem B. 2005;109:16099–16108. doi: 10.1021/jp051315+. [DOI] [PubMed] [Google Scholar]
- 27.Quillin ML, Anstrom DA, Shu XK, O’Leary S, Kallio K, Chudakov DA, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: Dark-state structure at 1.38 angstrom resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 28.Prescott M, Ling M, Beddoe T, Oakley AJ, Dove S, Hoegh-Guldberg O, Devenish RJ, Rossjohn J. The 2.2 A crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation. Structure. 2003;11:275–284. doi: 10.1016/s0969-2126(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 29.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 30.Maddalo SL, Zimmer M. The role of the protein matrix in green fluorescent protein fluorescence. Photochemistry and photobiology. 2006;82:367–372. doi: 10.1562/2005-04-11-RA-485. [DOI] [PubMed] [Google Scholar]
- 31.Rini M, Magnes BZ, Pines E, Nibbering ETJ. Real-time observation of bimodal proton transfer in acid-base pairs in water. Science. 2003;301:349–352. doi: 10.1126/science.1085762. [DOI] [PubMed] [Google Scholar]
- 32.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. Journal of Biological Chemistry. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 33.Cleland WW, Kreevoy MM. Low-Barrier Hydrogen-Bonds and Enzymatic Catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 34.Mock WL, Morsch LA. Low barrier hydrogen bonds within salicylate mono-anions. Tetrahedron. 2001;57:2957–2964. [Google Scholar]
- 35.Zhao QJ, Abeygunawardana C, Talalay P, Mildvan AS. NMR evidence for the participation of a low-barrier hydrogen bond in the mechanism of Delta(5)-3-ketosteroid isomerase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8220–8224. doi: 10.1073/pnas.93.16.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hibbert F, Emsley J. Hydrogen Bonding and Chemical Reactivity. Advances in physical organic chemistry. 26:255–380. [Google Scholar]
- 37.Mildvan AS, Harris TK, Abeygunawardana C. Nuclear magnetic resonance methods for the detection and study of low-barrier hydrogen bonds on enzymes. Methods in enzymology. 1999;308:219–245. doi: 10.1016/s0076-6879(99)08012-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, showing results from a 600 MHz jump-return NMR experiment to investigate whether unusual downfield shifts might be associated with the chromophore-Asp148 hydrogen bond. This material is available free of charge via the Internet at http://pubs.acs.org.