Abstract
We identified a new Drosophila gene, peter pan (ppan), in a screen for larval growth–defective mutants. ppan mutant larvae do not grow and show minimal DNA replication but can survive until well after their heterozygotic siblings have pupariated. We cloned the ppan gene by P-element plasmid rescue. ppan belongs to a highly conserved gene family that includes Saccharomyces cerevisiae SSF1 and SSF2, as well as Schizosaccharomyces pombe, Arabidopsis, Caenorhabditis elegans, mouse, and human homologues. Deletion of both SSF1 and SSF2 in yeast is lethal, and depletion of the gene products causes cell division arrest. Mosaic analysis of ppan mutant clones in Drosophila imaginal disks and ovaries demonstrates that ppan is cell autonomous and required for normal mitotic growth but is not absolutely required for general biosynthesis or DNA replication. Overexpression of the wild-type gene causes cell death and disrupts the normal development of adult structures. The ppan gene family appears to have an essential and evolutionarily conserved role in cell growth.
INTRODUCTION
The developmental program of cell proliferation in Drosophila has been well characterized (Foe et al., 1993; Edgar and Lehner, 1996). By the end of embryogenesis, cell proliferation has ceased, and most cells in the embryo are arrested in the G1 phase of the cell cycle (Hartenstein and Campos-Ortega, 1985). These cells remain quiescent through the first 8–12 h of larval development, until the first G1–S transitions begin in response to feeding (Britton and Edgar, 1998). Two different kinds of cell cycles are present in larval tissues; endoreplication and mitotic cycles. Endoreplication occurs in most larval tissues and is responsible for larval growth, whereas adult progenitor cells in the nervous system and the primordial tissues called imaginal discs undergo mitotic replication. The G1 arrest in early larvae may be like the G0 seen in serum-deprived tissue culture cells and can be maintained by allowing the larvae to hatch on media containing an energy source (sucrose) but lacking amino acids. Under these conditions the animal supports virtually no DNA replication or growth (Britton and Edgar, 1998). Exit from the G1 arrest of the first larval instar is an event in Drosophila development that provides a unique opportunity to study the relationships among nutrition, growth, and the cell cycle.
We took advantage of this opportunity by performing a genetic screen for larval growth–defective (LGD) mutants. We hypothesized that LGD mutants would be good candidates to have defects in growth or initiation of DNA replication. This hypothesis was based on the observation that first instar larvae blocked in G1 with DNA replication inhibitors remain small because they cannot undergo the DNA endoreplication required for most of Drosophila larval growth (Edgar, unpublished observation). Here we describe the characterization and cloning of the mutant peter pan (ppan), identified by its phenotype of first instar growth arrest and failure to undergo larval DNA replication. Our studies of the ppan mutant allow us to confirm some of the screen’s original assumptions and to define a conserved novel gene family required for normal growth and DNA replication in Drosophila larvae.
MATERIALS AND METHODS
Fly Stocks
l(3)6B6 was obtained from Y.N. Jan, and l(3)02231 was obtained from the Bloomington Stock Center. UAS-ppan, CASPR-ppan.xba I, CASPR-ppan.eco RI, and CASPR-ppan.pst I lines were generated by P-element–mediated transformation. UAS-p35 was a gift from B. Hay, and en-GAL4 was a gift from A. Brand. M(3)95APlac92 (Andersson et al., 1994) and MS 1096-GAL4 (Capdevila and Guerrero, 1994) have been previously described.
UAS and Casper Transgenes
The CASPR-ppan.xba I, CASPR-ppan.eco RI, and CASPR-ppan.pst I lines were generated by ligation of XbaI, EcoRI, and PstI genomic fragments into the XbaI, EcoRI, and PstI sites of the vector CASPR. The UAS-ppan construct was generated by excision of the ppan cDNA from pNB using SalI and XbaI. This fragment was subcloned into the vector pSL1180 (Pharmacia, Piscataway, NJ) and re-excised with EcoRI, and the resulting fragment was ligated into the EcoRI site in the polylinker of pUAST.
Yeast Strains
The yeast strain H50–16C (Mata leu2-3112, trp1-1, ura3-1, ade2-1, his3-11,15, can1-100, ssf1-1::HIS3, ssf2::TRP1 [pGAL-SSF1.14, LEU2]) was transformed to ura− with a yeast expression vector containing the ppan cDNA.
Histology
Larvae were fed 5-bromodeoxyuridine (BrdU) at a concentration of 100 μg/ml, dissected, and fixed in 6% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) in PBS for 1 h at room temperature. Immunohistochemistry was performed using standard techniques (Patel, 1994). Incorporated BrdU was detected using anti-BrdU (1:100) 1° antibody (Becton Dickinson, San Jose, CA) and HRP-coupled goat anti-mouse (1:350) 2° antibody (Jackson ImmunoResearch, West Grove, PA). Discs were also fixed in 6% paraformaldehyde and washed in PBS and 0.1% Tween 20 (Sigma, St. Louis, MO). Clonal markers were visualized using anti-myc (1:50) 1° antibody (Oncogene Science, Uniondale, NY) and preabsorbed goat anti-mouse FITC-conjugated (1:500) antibody (Jackson ImmunoResearch). Cell nuclei and cell outlines were visualized by incubation of fixed discs in DAPI (0.05 mg/ml in PBS) and rhodamine-conjugated phalloidin (Molecular Probes, Eugene, OR) for 1 h at room temperature. After washing, discs were mounted in Fluoroguard antifade reagent (Bio-Rad, Hercules, CA). Ovaries were dissected from mature females and fixed in 6% paraformaldehyde in PBS. Tissues were washed three times with PBS and 0.1% Tween 20 and permeabilized by 1-h room temperature incubation in PBS and 1% Triton X-100 (Sigma). Permeabilized ovaries were then incubated with DAPI and rhodamine-conjugated phalloidin as described above, washed, and mounted.
Clonal Analysis in Discs and Ovaries
Clonal analysis was performed as described by Xu and Rubin (1993). ppan clones in imaginal discs were induced by heat shock induction of flp recombinase in hsFLP; FRT πmyc ppan6B6/FRT larvae at 48, 72, and 96 h after egg deposition (AED). Clones were scored at 120 h AED in wandering third instar larvae. For the Minute experiments, clones were induced in hsFLP; FRT πmyc ppan6B6/FRT M(3)95APlac92 larvae and scored at 168 h AED. Mutant clones in discs were identified by antibody staining for πmyc. ppan mutant clones were generated in the germ line using an flp/FRT ovoD system as described by Chou et al. (1993). Ovaries were dissected, permeabilized, and stained with DAPI and rhodamine-conjugated phalloidin as described above.
Ectopic Overexpression
Ectopic overexpression of ppan in wing imaginal discs was achieved using the GAL4 expression system (Brand and Perrimon, 1993). The engrailed GAL4 driver was used to overexpress ppan in the posterior compartment of the discs, and the MS1096 GAL4 driver (Capdevila and Guerrero, 1994) was used to preferentially overexpress ppan on the dorsal side of the disc.
P-Element Reversion
Reversion of the P-element presumptively conferring the ppan phenotype was accomplished by crossing w; P[w+ l(3)6B6]/TM3 Sb flies with w; Δ2-3 Sb Ly/TM6 Ubx e flies carrying the Δ2-3 stable transposase source (Robertson et al., 1988). Dysgenic males of the genotype w/Y; P[w+ l(3)6B6]/Δ2-3 Sb Ly were then mated to w; P[w+ l(3)6B6]/TM3 Sb females. w; Sb+ Ly+ revertants were scored for viability to the adult stage, and several hundred were recovered. Twenty-one lethal excision mutants were also isolated from w; P[revertant}/TM3 Sb progeny of the above cross.
Molecular Biology
Plasmid rescue was performed on the P[w+ l(3)6B6] allele and a fragment corresponding to that rescued from P[w+ l(3)2231] was subcloned. This fragment was already mapped to a P1 genomic clone. Using the rescued fragment from the P[w+ l(3)6B6] as a probe, we isolated EcoRI and XbaI genomic clones (see Figure 3A). The EcoRI fragment is completely included in the XbaI fragment. Subfragments of these were in turn used to probe a plasmid-based cDNA library (Brown and Kafatos, 1988). Although four cDNAs were found to span the XbaI fragment, in both alleles, the P-element appears to be inserted into the 5′ end of the coding sequence of just one of those cDNAs (see Figure 3B).
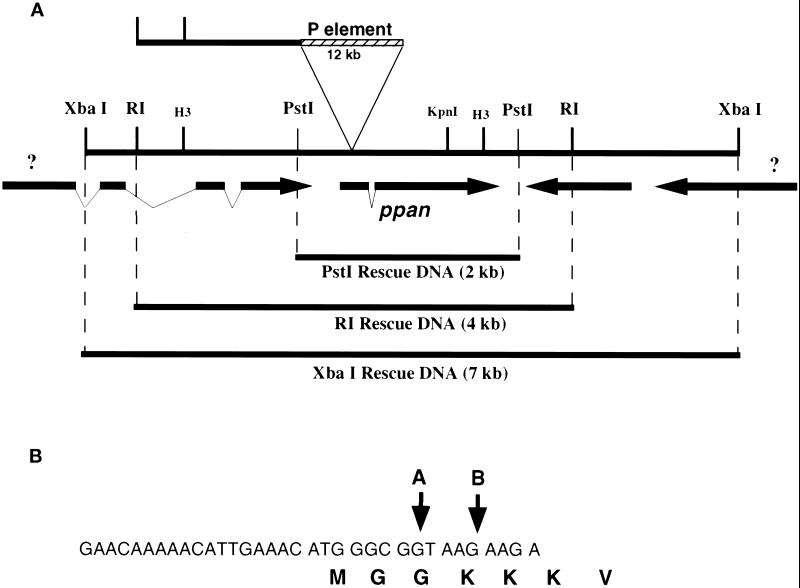
Figure 3.
ppan genomic region and P-element location. (A) Map of genomic region showing the location of the P-element insertion (top), restriction sites, cDNAs (arrows), and DNA fragments used for rescue of the ppan mutant (below). (B) Nucleotide map of P-element insertion sites as determined by sequencing of genomic DNA recovered by plasmid rescue. P-element insertions sites just inside the ppan ORF for P[w+ l(3) 6B6] (A) and P[w+ l(3) 02231] (B) are indicated by the arrows.
Three different genomic fragments from around the ppan locus (Figure 3A) were transformed into flies to confirm that disruption of the ppan cDNA results in the observed larval growth arrest phenotype. These fragments are 1) the 7-kb XbaI fragment, which includes ppan, another complete cDNA, and fragments of two other cDNAs; 2) the 4-kb EcoRI fragment, which includes ppan and parts of two other cDNAs; and 3) a 2-kb PstI fragment, which includes only ppan and a small piece of the 3′ end of an adjacent cDNA.
RESULTS
ppan Is a Larval Growth Arrest Mutant
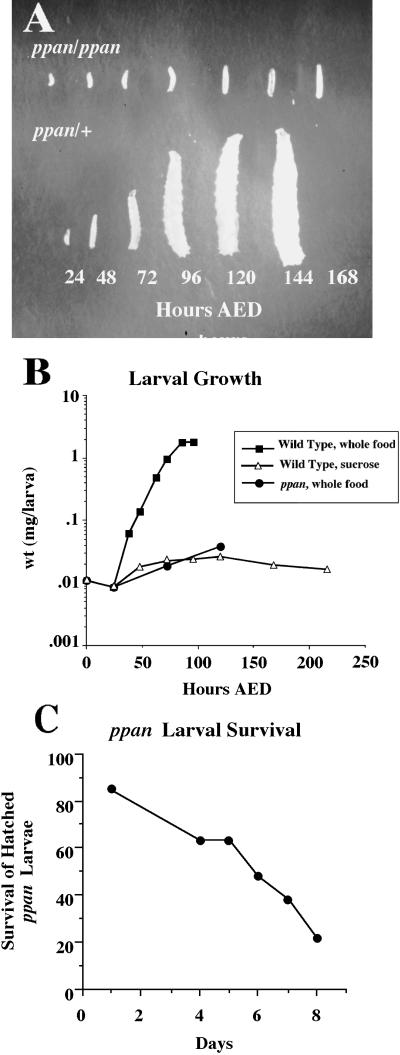
The initial ppan allele, P[w+ l(3)6B6], was identified in a visual screen of 352 P-element lethal mutants (courtesy of Y.N. Jan), based on its striking growth arrest phenotype. ppan is a recessive P-element mutant with biphasic lethality. Forty-five percent of the homozygotes die in late embryogenesis with no obvious morphological defects and a normal cuticle (our unpublished results). Fifty-five percent of homozygous larvae hatch and live for 4–8 d but remain very small (Figure 1A). These mutant larvae move and feed normally, and no morphological defects were noted. Mutant larvae were weighed at time points after hatching, and we found that their growth closely resembled that of sucrose-fed wild-type larvae, which remain arrested in the first instar without protein accumulation (Figure 1B). Twenty percent of the ppan larvae that hatch survive for at least 8 d, 3 d after their heterozygous siblings pupariate (Figure 1C). Thus, the ppan mutant is specifically deficient in growth, but this deficiency is not immediately lethal.
Figure 1.
Larval growth arrest phenotype and survival of ppan mutant larvae. (A) Heterozygous and homozygous ppan larvae at 24, 48, 72, 96, 120, 144, and 168 h AED. (B) Larval growth curve for normally fed wild-type (black squares), sucrose-fed wild-type (open triangles), and normally fed homozygous ppan (black circles) larvae. Groups of ≥20 larvae were weighed at various times after hatching, and an average weight per larva was determined. (C) Survival of hatched ppan larvae Mutant ppan larvae were counted for several days after hatching.
ppan Is Required for Endoreplication and Normal Mitotic Proliferation in the Whole Animal
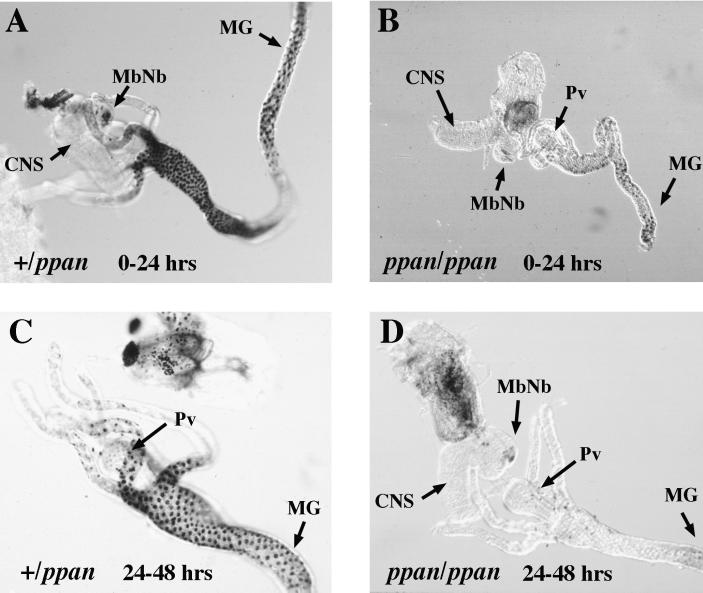
To determine whether the growth defect was accompanied by defects in larval DNA replication, analysis of the incorporation of the nucleotide analogue BrdU was performed on heterozygous and homozygous ppan larvae at 24-h intervals after hatching (Figure 2). At 0–24 h after hatching, the homozygous mutants showed greatly reduced BrdU labeling compared with heterozygous siblings. Between 24 and 48 h after hatching, there was no more DNA replication in the homozygotes except in the mushroom body neuroblasts in the brain, which continue proliferation even in nutrient-deprived larvae (Britton and Edgar, 1998). Thus, in the mutant ppan, there is little or no DNA replication in either endoreplicating tissues such as the gut or in mitotically proliferating tissues such as the brain and larval histoblasts. The presence of DNA replication in the mushroom body neuroblasts indicates that ppan is not essential for DNA replication per se.
Figure 2.
ppan mutants show decreased DNA replication. ppan heterozygotes (A and C) and homozygotes (B and D) were fed the nucleotide analogue BrdU (100 μg/ml) during the periods 0–24 h (A and B) and 24–48 h (C and D). Internal organs were dissected and histochemically stained to detect incorporated BrdU (black). MG, midgut; Pv, proventriculus; MbNb, mushroom body neuroblasts.
Isolation of the ppan Gene
Having established ppan to be an interesting mutant with defects in larval growth and DNA replication, we set about to identify the gene defective in the mutant. The P-element lethal allele P[w+ l(3)6B6] was the first identified, and a second allele, P[w+ l(3)2231], was found in the Drosophila Genome Project database. Both P[w+ l(3)6B6] and P[w+ l(3)2231] were mapped by the Drosophila Genome Project to the right arm of the third chromosome (93B10-11) using the P[w+ l(3)2231] allele. The P[w+ l(3)6B6] allele is the subject of most of the work described in this paper, but the phenotype of the P[w+ l(3)2231] allele is indistinguishable from that of P[w+ l(3)6B6]. To be sure that the P-element insertion is responsible for the LGD phenotype and the lethality, the P[w+ l(3)6B6] was mobilized. In doing so, we reverted the phenotype and the lethality, confirming that there is no mutation elsewhere on the chromosome that caused the observed growth defects.
We used the plasmid rescue technique to isolate the gene defective in the mutant ppan (Cooley et al., 1988). Plasmid rescue was performed on the P[w+ l(3)6B6] allele, and a rescued fragment was used to isolate genomic DNA fragments (Figure 3A), which were in turn used to probe a plasmid-based cDNA library. Four cDNAs were found to span the largest genomic fragment (an XbaI fragment), but in both alleles, the P-element is inserted into the 5′ end of the coding sequence of just one of those cDNAs (Figure 3B). As these P-elements are inserted in the putative coding region of the ppan gene, it is likely that these mutants are null for function.
To confirm that disruption of the ppan cDNA is in fact responsible for the larval growth arrest phenotype, flies were transformed with three different genomic fragments from the ppan locus (Figure 3A). A single copy of any of these constructs was able to rescue homozygous ppan mutants. The smallest fragment (a 2-kb PstI fragment) includes only ppan and a small piece of the 3′ end of an adjacent cDNA. Surprisingly, this fragment contains only 175 bp of DNA 5′ to the putative initiating methionine that are not a part of the 3′ region of the adjacent cDNA. Thus it appears that a transcriptional regulatory region sufficient for function is very small and compact. The presence of such a small regulatory region suggests that ppan transcription is not subject to complex regulation.
ppan Expression
Only 55% of homozygous ppan larvae hatch. The homozygotes that die as embryos appear normal. This indicates that the maternally derived ppan gene product is sufficient to support most or all of embryonic development. Examination of ppan expression by Northern blot indicated that ppan mRNA is loaded into the oocyte and that mRNA levels decrease by ∼50% during embryogenesis. RNA in situ hybridization performed on embryos showed that ppan mRNA is ubiquitously expressed and that RNA levels are highest in the early embryo. This ubiquitous distribution is consistent with the small regulatory region we have observed.
ppan Is Similar to the SSF1 and SSF2 Genes in Saccharomyces cerevisiae
Sequencing of the ppan cDNA identified it as a Drosophila homologue of the yeast genes SSF1 and SSF2 (Figure 4). ppan shares 28% amino acid identity with the yeast genes and 35% amino acid identity with the Caenorhabditis elegans homologue. SSF1 and SSF2 were identified in a screen for multicopy suppressors of a yeast mating pathway defect. The mating pathway defect was a temperature-sensitive mutation in the Gβ subunit of the heterotrimeric G-protein that couples the mating factor receptor to its downstream effects on cell cycle and cell morphology. Overexpression of SSF1 increased the mating efficiency of the mutant strain at the restrictive temperature. A second gene, SSF2, was found to share 94% amino acid identity and functional interchangeability with SSF1. Deletion of both yeast genes is lethal, and depletion of the gene products causes cell division arrest (Yu and Hirsch, 1995). SSF1 and SSF2 are believed to be localized in the nucleus (Kim and Hirsch, 1998). Analysis of ppan sequence using the sequence analysis program PSORT (Nakai and Kanehisa, 1992; Horton and Nakai, 1997) suggests that ppan has a 0.987 probability of being nuclearly localized. Murine, human, Arabidopsis, C. elegans, and Schizosaccharomyces pombe homologues have also been identified (Figure 4). Thus, these genes are likely to have an evolutionarily conserved role in growth and cell cycle progression.
Figure 4.
Comparison of amino acid sequences of ppan with putative homologues from other species. Homologues were first aligned with the program Clustal-W and then hand aligned. The names and sources of the sequences is as follows: DM, D. melanogaster ppan, sequencing of both strands of a cDNA; CE, C. Elegans cDNA, GenBank accession number AF043700; SP, S. Pombe cDNA, GenBank number Z98531; SSF1, S. cerevisiae cDNA, GenBank number P38789; SSF2, S. cerevisiae cDNA, GenBank number Q12153; MM, Mus musculus, compilation of expressed sequence tag fragments AA451276, AA475332, AA068339, AA237717, AA517621, AA270523, AA756790, AA028364, AA575760, AA239726, AA561626, and AA265569; HS, Homo sapiens, compilation of expressed sequence tag fragments N34073, N40373, AI147481, AI1084732, AA321112, AA300789, and AA258103; AT, A. thaliana.
Expression of the Drosophila Homologue in SSF1, SSF2 Mutant Yeast Can Suppress the Yeast Growth Defect
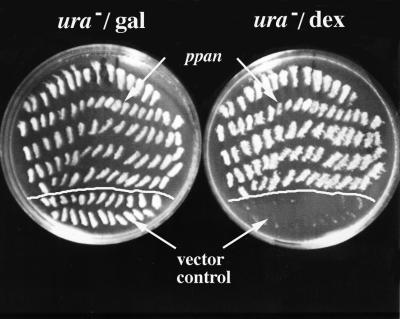
Given that the ppan gene is conserved across species, we examined whether the molecular function is also conserved by testing whether expression of ppan could rescue loss of SSF1 and SSF2 in S. cerevisiae. We used a strain of yeast in which SSF1 and SSF2 were deleted and SSF1 had been added back under the control of galactose promoter (Yu and Hirsch, 1995). Cells were depleted of the SSF1 gene products by switching the yeast from media containing galactose to media containing dextrose. In the dextrose-containing media, the cells arrested after 12 h. This strain was transformed with either a ura-marked plasmid containing the ppan cDNA under the transcriptional control of the GAPDH promoter or the same plasmid without the cDNA as a control. ura+ colonies from both transformations were replated onto ura− dextrose plates (Figure 5). Only the colonies transformed with the ppan cDNA were able to grow; the control plasmid transformed colonies were not. Thus, the ppan gene complements SSF1 and SSF2 function.
Figure 5.
The ppan gene complements ssf1 and ssf2 function. The yeast strain H50[hyphem]16C is deficient for ssf1 and ssf2 but has the ssf1 gene added back on a plasmid under the control of a galactose promoter. Thus these cells can only grow in media containing galactose. These cells were transformed to ura− galactose plates with either the empty pAB23BXN plasmid or the same plasmid with ppan subcloned into it under the transcriptional control of the strong constitutive GAPDH promoter. The streaked colonies from both the control and ppan transformations were replated onto ura− dextrose plates. The ppan expressing colonies (pAB23BXN-ppan) were able to grow, whereas the control colonies (pAB23BXN) were not.
ppan Mutant Cells Have a Growth Defect
Although the lack of growth and DNA endoreplication in ppan mutant larvae demonstrates that ppan is required for these processes in the whole animal, it does not address whether the ppan gene product is required cell autonomously. BrdU-labeling experiments showed no DNA replication in the mitotically replicating brain of ppan mutant larvae, except in the mushroom body neuroblasts, which continue to divide just as in amino acid–deprived larvae. However, in wild-type larvae growth of the endoreplicating larval tissues is required to start replication in the mitotic tissues such as the brain (Britton and Edgar, 1998). Therefore, the lack of BrdU labeling in homozygous ppan larval brains may be due to the absence of growth and endoreplication in other tissues.
To characterize the proliferative capability of ppan mutant cells, clonal analysis was performed in imaginal discs using the FLP/FRT mitotic recombination system (Xu and Rubin, 1993). This technique allowed us to generate clones of homozygous ppan mutant cells in a ppan heterozygous background. We induced mutant clones at various developmental stages and then scored the marked clones in imaginal discs from wandering stage larvae (120 h AED) and in adult eyes (Table 1). We also inspected adult flies carrying clones of mutant cells for appendage and cuticle abnormalities. The fate of ppan mutant cells depended on when the clones were induced (Table 1). When clones were induced early in disc development (24–48 h AED) we were unable to detect homozygous ppan mutant clones, even though their adjacent twin spots (homozygous wild-type sister clones) were present. When clones were induced late in disc development (110–114 h AED), we saw small ppan clones with larger twin spots. Close inspection of the anterior wing margin revealed occasional defects in some chemo- and mechanosensory bristles. These defects included deletion and disruption of these bristles. Thus, homozygous ppan clones induced early are eliminated by “cell competition” (Simpson, 1979), whereas late induced clones can survive but grow slowly and often differentiate defectively in the adult. These results indicate that ppan-deficient cells have a cell-autonomous growth disadvantage and a compromised ability to produce differentiated structures.
Table 1.
Mosaic analysis: ppan mutant cells have a growth defect
| HS (h AED) | Genotype | ppan clone survival (L3 discs) | Morphology/clone survival (adults) |
|---|---|---|---|
| 96 | FRT ppan/FRT | +++ | Normal/many small clones |
| 72 | FRT ppan/FRT | + | Normal/many small clones |
| 48 | FRT ppan/FRT | − | Normal/no surviving clones |
| 36 | FRT ppan/FRT | − | Normal/no surviving clones |
| 144 | FRT ppan/FRT Minute | +++ | Rough eyes, wing and cuticle defects/ND |
| 120 | FRT ppan/FRT Minute | +++ | Rough eyes, wing and cuticle defects/ND |
| 96 | FRT ppan/FRT Minute | ++ (>30 cells) | Pupal lethal/ND |
| 72 | FRT ppan/FRT Minute | ++ (>50 cells) | Pupal lethal/ND |
Data summary for studies of ppan clones allowed to grow for various times after heat shock (HS) induction of flp recombinase (see MATERIALS AND METHODS). For analysis in L3 discs, larvae were fixed at 120 h AED (wild-type background) and at 168 h AED (Minute background). Mutant clones in discs were identified using the πmyc marker and in adult eyes by pigmentation derived from the w+ marker in P[w+ ppan6B6]. +++, mutant clones equivalent in size to wild-type sister clones (“twin spots”); ++ and +, mutant clones smaller than twin spots; −, no detectable mutant clones; ND, clone survival was not determined.
To further study the growth and proliferation properties of ppan mutant imaginal cells, we induced clones in the genetic background of heterozygous Minute l(3)95A larvae. Minute mutants share the phenotype of dominant growth delay and recessive cell lethality. M l(3)95A is mutant in ribosomal protein rpS3 (Andersson et al., 1994). Although homozygous ppan clones induced during early disc development (24–48 h AED) in a wild-type background were outcompeted and eliminated, clones induced at the equivalent stage in a slow-growing Minute background were easily detected and reached significant size (>50 cells). Nevertheless, wild-type clones induced at the same time were much larger and sometimes filled a complete compartment (Garcia-Bellido et al., 1973; Simpson, 1979). This clearly demonstrates that ppan mutant cells grow much more slowly than wild-type cells. Without the competition of the wild-type cells, homozygous ppan cells can continue to divide and are not eliminated but grow more slowly than wild-type cells. We also found that early induction of ppan clones in the Minute background resulted in pupal lethality. Thus, the presence of large clones of ppan cells severely disrupts pupal development.
Three major conclusions may be drawn from these experiments. First, ppan is not absolutely required for mitotic proliferation. Second, although growth and mitotic proliferation do proceed in ppan mutant cells, these processes are slow, and the cells have a growth disadvantage. Finally, clones of ppan mutant cells that do survive through larval life result in either disruptions of adult structures such as mechano- and chemosensory bristles or in pupal lethality. Thus, although still able to grow and proliferate at a reduced rate, ppan mutant cells are deficient in a specific cellular function essential for differentiation.
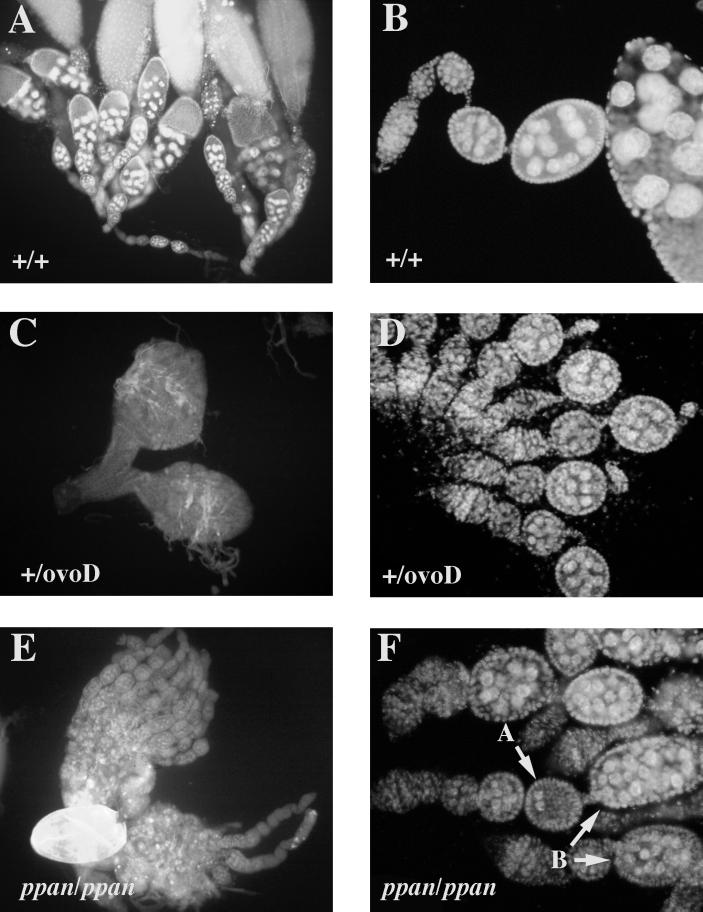
ppan Is Required for Oogenesis
We thought we might gain further insight about the molecular function of ppan by examining the phenotypes of ppan mutant cells in the female germ line. ppan mutant clones were generated in the germ line using an FLP/FRT ovoD system (Chou et al., 1993). ppan mutant clones in the ovary were never (0%) able to produce viable eggs. However, upon examination of the ovaries in which the mutant ppan clones were generated, we found that the ppan mutant oocytes progressed further than either the homozygous or heterozygous ovoD oocytes, sometimes even allowing the deposition of malformed, deflated eggs (Figure 6, A–C). Interestingly, ppan mutant oocytes often had incorrect numbers of nurse cells. Wild-type oocytes typically have 15 nurse cells (Figure 6, A and B), whereas ppan mutant oocytes exhibit a variety of nurse cell numbers ranging from 0 to >30 (Figure 6, E and F). This phenotype is indicative of a defect early in oogenesis when cysts pinch off from the germarium. However, judging from the size of the mutant cysts and the nuclei within, germ line proliferation, DNA endoreplication, and overall growth were not completely blocked.
Figure 6.
Germ line mosaic analysis. ppan is required to produce a viable oocyte. DAPI staining of the nuclei is shown in wild-type ovaries (A; magnification, 10×), wild-type ovarioles (B; 20×), ovoD/+ ovaries (C; 10×), ovoD/+ ovarioles (D; 20×), ppan/ppan ovaries (E; 10×), and ppan/ppan ovarioles (F; 20×). Arrows in F indicate cysts that deviate from the normal number of nurse cells (15).
ppan Overexpression Studies
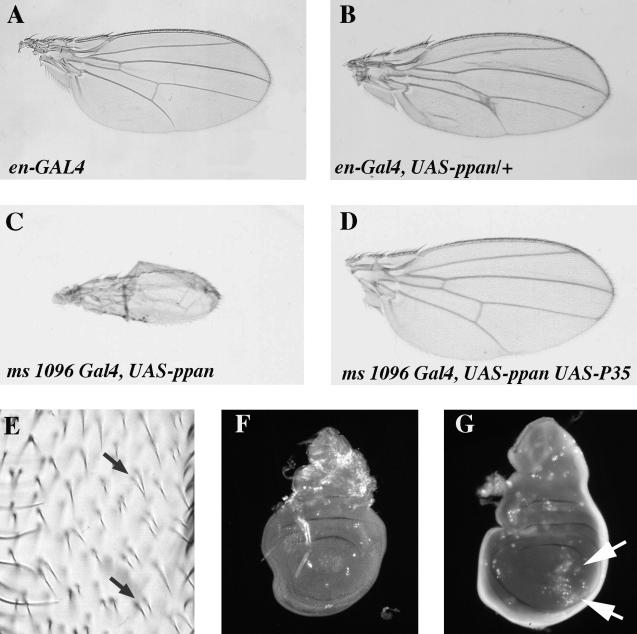
Another approach to understanding the molecular function of ppan is to examine the effects of ppan overexpression. Using the GAL4 expression system (Brand and Perrimon, 1993), ppan was overexpressed in wing imaginal discs. Overexpression of ppan in the engrailed pattern in the posterior compartment of the wing imaginal disc caused the posterior part of the wing to take on a crinkled appearance and disrupted normal wing vein formation. The crinkled appearance was accompanied by increased opacity (Figure 7B). The disruption of wing vein formation consisted of ectopic wing vein formation, thickening of wing veins, and loss of wing veins.
Figure 7.
Ectopic overexpression of ppan causes cell death and disrupts wing development. Results of ppan overexpression in the posterior compartment of the imaginal wing disc using the engrailed GAL4 driver: (A) en-GAL4; (B) en-GAL4, UAS-ppan. Results of ppan overexpression in the imaginal wing disc using the ms 1096-GAL4 driver, which expresses GAL4 preferentially in the dorsal portion of the disc: (C) ms 1096 GAL4, UAS-ppan; (D) ms 1096 GAL4, UAS-ppan, UAS-P35. (E) Closeup of irregular and duplicated wing trichomes in ms 1096-GAL4, UAS-ppan/+ flies. Visualization of cell death in imaginal discs using acridine orange: (F) wild-type; (G) en-GAL4, UAS-ppan. Arrows indicate abnormal cell death in the posterior wing compartment.
We also overexpressed ppan using the MS1096 GAL4 driver (Capdevila and Guerrero, 1994), which expresses GAL4 strongly on the dorsal side of the wing imaginal disc and weakly on the ventral side. Flies having one copy of this GAL4 driver and one copy of UAS-ppan construct had wings that were slightly curved upward (our unpublished results). Closer examination of the trichomes on these wings showed that there were fewer trichomes on the dorsal side of the wing than on the ventral side. In some spots on the dorsal side there were multiple fused trichomes (Figure 7E). Although it is possible that each trichome comes from individual smaller cells, the even spacing suggests the multiple fused trichomes originate from single cells. Normally each trichome derives from an individual cell, and thus we infer that overexpressing ppan with the MS1096 GAL4 driver reduced the number of cells on the dorsal side of the wing. This may account for the curved wing phenotype. Increasing the number of copies of either the MS1096 GAL4 driver or the UAS-ppan transgene ultimately resulted in the near complete elimination of the wing (Figure 7C).
To further study the effects of ppan overexpression, we examined cell death in imaginal discs overexpressing ppan. Acridine orange staining of apoptotic cells showed increased numbers of apoptotic cells in regions of the imaginal disc overexpressing ppan using en-GAL4 (Figure 7G). Thus, overexpression of ppan causes cell death, which in turn may be responsible for the observed phenotypes in the wings. In fact, even the most severe ppan overexpression wing phenotype can be rescued by coexpression of the baculovirus P35 protein (Figure 7D), which blocks cell death by inhibition of Caspases (Hay et al., 1994). Clearly we can conclude that imaginal disc cells are sensitive to levels of ppan and that increased expression of ppan decreases cell viability.
DISCUSSION
Here we describe the characterization and cloning of the LGD mutant ppan. Close examination of the phenotype of ppan mutant larvae and clones of ppan mutant cells revealed that ppan is an essential gene required for a variety of developmental and cellular functions. Recent work in yeast has provided some clues to the molecular function of ppan. From studies of the ppan homologues SSF1 and SSF2, Kim and Hirsch (1998) propose that SSF1 and SSF2 are nuclear proteins that affect mating efficiency by altering mating projection formation. Mating projection formation in yeast was prevented by depletion of SSF gene products and increased by overexpression of SSF1. Both mating projection formation and bud formation during vegetative growth are forms of polarized cell growth that involve significant actin cytoskeletal reorganization (Leberer et al., 1997). A role for SSF1 and SSF2 in a specialized form of cell growth rather than general growth is supported by the observation that yeast cells depleted of SSF gene products continue to increase in size but are unable to form mating projections in response to mating factor (Kim and Hirsch, 1998). Another laboratory found SSF1 to be a weak suppressor of a temperature-sensitive tor2 mutant (Schmidt and Hall, personal communication). TOR2, a target of FKBP–rapamycin complexes and a putative phosphatidylinositol-3 kinase, functions both in nutrition-stimulated activation of protein synthesis and also in cell cycle–dependent polarized distribution of the actin cytoskeleton (Helliwell et al., 1998).
These studies in yeast prompted us to investigate whether ppan, like SSF1 and SSF2, might also function in polarized cell growth. The biology of Drosophila and the genetic techniques available allowed us to study the requirements for ppan in a variety of organismal and cellular processes. In the ovary, we found that ppan mutant clones exhibited early defects in cytoarchitecture and nurse cell segregation. One step in this process that may require polarized growth, or at least actin cytoskeleton rearrangements, is the pinching off of cysts from the germarium. Despite defects in this process, ppan mutant cysts grew in size, and the nurse cells appeared to undergo multiple rounds of endoreplication before finally degenerating. Thus the ppan phenotype in the fly ovary bears some similarity to that found in the SSF depletion experiments in yeast, in which the mutant yeast grew in size but failed to form mating projections. The cytoarchetectural defects in ppan mutant ovaries might be attributed to defects in polarized growth or defective regulation of the actin cytoskeleton.
In other situations, however, loss of ppan function did not result in phenotypes readily explained by defects in polarized cell growth. For instance, actin staining of ppan mutant cells in imaginal discs did not reveal the expected defects in cell morphology or polarization. The fate of ppan mutant cells in imaginal discs depended on when the mutant clones were induced and the genetic background in which the analysis was performed. Early induced mutant clones in a wild-type background were lost, whereas those induced late, or in a Minute background, grew significantly. Our interpretation of these results is that ppan is not absolutely required for mitotic proliferation but that its absence confers on the mutant cells a general growth delay that results in their elimination. Cell clones deficient in ppan behaved quite differently from those deficient in genes required for cell cycle progression, such as string, CyclinE, and Cdc2, which cease proliferation but exhibit continued cell growth (Neufeld et al., 1998), or from those deficient in protein synthesis, such as rpS3 (l(3) M95A), which neither proliferate nor grow (Andersson et al., 1994). In addition to these growth defects, we found that the survival of ppan clones into the pupa resulted in the disruption of adult structures or in lethality. Thus it seems that ppan is required for some aspects of cell differentiation as well as for cellular growth.
The requirement for ppan in the germ line seemed to follow a similar pattern. ppan mutant ovaries produce no viable eggs, and thus the gene is required for normal oogenesis. However, a careful analysis of the mutant phenotype revealed that growth and DNA replication were not completely arrested. These observations seem paradoxical in light of the phenotype of homozygous ppan larvae, which exhibit a total inability to grow and never initiate global DNA replication. To reconcile these cellular and organismal phenotypes, we suggest that in ppan mutant larvae defects in a specialized cellular function, perhaps in a specific larval tissue, may feed back on and block growth and DNA replication throughout the animal. In mutant imaginal disc cells and oocytes these defects do not appear to be so closely monitored. In summary, at the organismal level ppan is required for initiation of larval growth and DNA replication, progression of oogenesis, and maturation of some imaginal tissues into adult structures. At the cellular level ppan is not absolutely required for growth, mitosis, or DNA endoreplication, but its absence does confer a growth delay, and it is also required for some aspects of normal cell differentiation. Identification of ppan as a homologue of the yeast genes SSF1 and SSF2 provides a useful framework in which to study the molecular, cellular, and organismal role of this gene.
ACKNOWLEDGMENTS
We thank Mireille Galloni for comments on the manuscript and Dr. J. P. Hirsch for communicating unpublished data. This work was supported by National Institutes of Health grants GM-17983 (to J.C.M.), HG-00176 (to M.S.G.), and GM-51186 (to B.A.E.). B.A.E. is a Lucille P. Markey and Rita Allen Scholar. The ppan accession number is AF102805.
REFERENCES
- Andersson S, Saeboe-Larssen S, Lambertsson A, Merriam J, Jacobs-Lorena M. A Drosophila third chromosome Minute locus encodes a ribosomal protein. Genetics. 1994;137:513–520. doi: 10.1093/genetics/137.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;128:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-B, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehner CF. Developmental control of cell cycle regulators: a fly’s perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- Foe VE, O’Dell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In: Bate, Martinez-Arias, editors. The Development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 149–300. [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalization of the wing disc of Drosophila. Nat New Biol. 1973;243:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA. Fate-mapping in wild-type Drosophila melanogaster. I. The spatiotemporal pattern of embryonic cell divisions. Arch Dev Biol. 1985;194:805–816. [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Howald I, Barbet N, Hall MN. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbor classifier. Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Kim J, Hirsch JP. A nuclear protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics. 1998;149:795–805. doi: 10.1093/genetics/149.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Thomas DY, Whiteway M. Pheromone signaling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AFA, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Patel N. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. San Diego: Academic Press; 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson Sclitz D, Benz WK, Engels WR. A stable source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. Parameters of cell competition in the compartments of the wing disc of Drosophila. Dev Biol. 1979;69:182–193. doi: 10.1016/0012-1606(79)90284-7. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yu Y, Hirsch JP. An essential gene pair in Saccharomyces cerevisiae with a potential role in mating. DNA Cell Biol. 1995;14:411–418. doi: 10.1089/dna.1995.14.411. [DOI] [PubMed] [Google Scholar]