Abstract
Insight into how the mammalian genome is structured in vivo is key to understanding transcriptional regulation. This is especially true in complex domains in which genes are coordinately regulated by long-range interactions between cis-regulatory elements. The regulation of the H19/Igf2 imprinted region depends on the presence of several cis-acting sequences, including a methylation-sensitive insulator between Igf2 and H19 and shared enhancers downstream of H19. Each parental allele has a distinct expression pattern. We used chromosome conformation capture to assay the native three-dimensional organization of the H19/Igf2 locus on each parental copy. Furthermore, we compared wild-type chromosomes to several mutations that affect the insulator. Our results show that promoters and enhancers reproducibly co-localize at transcriptionally active genes, i.e. the endodermal enhancers contact the maternal H19 and the paternal Igf2 genes. The active insulator blocks traffic of the enhancers along the chromosome, restricting them to the H19 promoter. Conversely, the methylated inactive insulator allows the enhancers to contact the upstream regions, including Igf2. Mutations that either remove or inhibit insulator activity allow unrestricted access of the enhancers to the whole region. A mutation that allows establishment of an enhancer-blocker on the normally inactive paternal copy diminishes the contact of the enhancer with the Igf2 gene. Based on our results, we propose that physical proximity of cis-acting DNA elements is vital for their activity in vivo. We suggest that enhancers track along the chromosome until they find a suitable promoter sequence to interact with and that insulator elements block further tracking of enhancers.
INTRODUCTION
The organization of the genome into functional domains ensures that the appropriate temporal and tissue-specific patterns of gene expression are achieved. Within a certain domain, expression patterns are determined by the interaction of several regulatory elements, including promoters, enhancers and insulators. Epigenetic modifications, such as DNA methylation, can modify the interactions between genes and their regulatory sequences. In the case of imprinted genes, two alleles with parental-specific epigenetic modifications coexist and result in distinct allelic expression patterns.
At the H19/Igf2 locus, common enhancers activate H19 on the maternal chromosome and Igf2 on the paternal chromosome (1–4). The allele-specific pattern is due to the epigenetic switch of an insulator/transcriptional activator lying upstream of H19 (5), designated as the differentially methylated domain (DMD). On the maternal chromosome, this region binds to CTCF (6–10), with two outcomes: H19 expression is initiated in the blastocyst (11,12) and the enhancers driving this transcription are blocked from activating Igf2 promoters on that chromosome. On the paternal chromosome, the region is hypermethylated (11,13), CTCF does not bind and the enhancers can access the Igf2 promoters to drive transcription paternally. Subsequent methylation of the H19 promoter inhibits its expression.
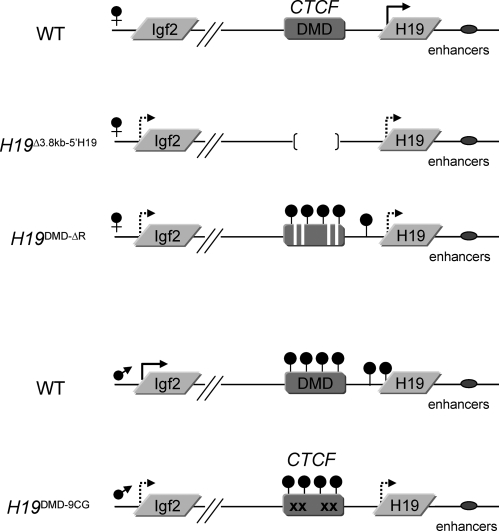
We have developed several mouse models to investigate the requirement of the DMD for proper imprinting regulation and to determine the role of CTCF binding to the DMD by generating loss of function and gain of function mutations (Fig. 1). The H19Δ3.8–5′H19 line has 3.8 kb upstream of the H19 gene encompassing the DMD deleted (14), resulting in biallelic Igf2 expression when the allele is inherited maternally. In the H19DMD-ΔR mutation, only the CTCF binding sites within the DMD have been deleted (12). We have shown that in this line, the maternal insulator cannot be established, and that transcriptional initiation of H19 is delayed. In the H199CG-DMD line, 9 CG dinucleotides within the CTCF binding sites have been mutagenized (10), and although methylation is established when inherited paternally, CTCF is able to bind to the DMD and establish an ectopic enhancer-blocker, resulting in decreased levels of Igf2.
Figure 1.
Representation of the Igf2/H19 loci and the mutants used in this study, including their methylation and expression patterns. Arrows represent expression, hatched arrows represent reduced levels of expression and filled circles indicate DNA methylation. The H19Δ3.8 kb-5′H19 allele has a 3.8 kb deletion that removes the DMD. The H19DMD-ΔR allele lacks the four core binding sites for CTCF within the DMD. The H19DMD-9CG allele replaces 9CGs within the CTCF binding sites of the DMD.
These experiments highlighted the critical importance of CTCF in the control of expression patterns in the H19/Igf2 locus. Studies at the β-globin locus had suggested that CTCF functions by organizing the three-dimensional structure of the locus, inducing the formation of chromatin loops that establish independent domains of transcriptional activity (15). At the H19/Igf2 locus, recent studies have investigated different aspects of the interactions between the regulatory elements (16–18). Some discrepancies have emerged from these reports. Kurukuti et al. suggest that on the maternal chromosome, the insulator interacts with a matrix attachment region and a differentially methylated region at the Igf2 locus to generate a tight loop around the maternal Igf2 gene, which results in silencing. Yoon et al. report that the DMD forms direct associations with the enhancers and the inactive Igf2 promoters. Whereas the former studies indicate that the insulator forms a physical impediment to activation of Igf2, the latter posits that enhancer-blocking is the result of direct interaction with the enhancers and inactive promoter, generating a transcriptionally unproductive association. The findings on the paternal-specific associations also conflict. Kurukuti et al. maintain that both H19 and Igf2 are accessible to interaction with the enhancers on the paternal chromosome, whereas Yoon et al. only tested association between the enhancers and the active Igf2 promoter.
While each of these studies yielded insight into the organization of the H19/Igf2 locus, the discrepancies in their results have left some unanswered questions with respect to the mechanisms of enhancer-blocking and transcriptional activation. We wished to carry out a detailed scan across the entire H19/Igf2 region to determine the parental-specific chromatin loop organization associated with the active transcriptional status as well as repression mediated by enhancer-blocking activity or methylation. We focused on the behavior of the endodermal enhancers in neonatal liver, and our array of mouse models provided an opportunity to compare wild-type chromosomes with alleles containing a variety of mutations in the CTCF sites. We used Chromosome Conformation Capture (3C) (19) in order to identify the long-range interactions between the promoters, the endodermal enhancers and the DMD.
In the wild-type mouse, we find that on the maternal chromosome, the enhancers make contacts throughout the region including the H19 coding unit and promoter, up to the DMD. On the paternal chromosome, the enhancers are engaged exclusively by the Igf2 promoter and have no detectable interaction with the H19 gene. Upon deletion of the maternal DMD, we find that the maternal enhancers interact in an unrestricted way with both H19 and Igf2 regions. This suggests that the DMD is required to confine the interaction of enhancers to the H19 region. Similarly, when the CTCF binding sites are absent on the maternal allele, the enhancers have unlimited access to the whole region, showing that CTCF is necessary for enhancer-blocking activity. When the H19DMD-9CG allele is inherited paternally, the enhancers are mainly found at the H19 region. Physical presence beyond the paternally established insulator is limited, indicating that CTCF binding in the mutant DMD restricts the access of the enhancers to the Igf2 promoter.
RESULTS
We used 3C technology to investigate the interactions that occur between the endodermal enhancers and the H19 and Igf2 genes in the neonatal liver. The 3C assay employs cross-linking, restriction digestion, ligation and PCR amplification of ligated fragments to determine the frequency of physical contacts between different regions along the chromosome (19). In our experimental approach, the samples were generated by crosses between wild-type or mutant C57BL/6J and B6(CAST7) mice. The alleles assayed are depicted in Figure 1. We analyzed neonatal livers, the tissue with the highest expression of H19 and Igf2. Nuclei were treated with formaldehyde to crosslink proteins that mediate association between the endodermal enhancers that lie downstream of H19 and other regulatory elements. The crosslinked chromatin was digested with PsuI, separating fragments that contained the regulatory elements of the H19/Igf2 locus. PsuI digestion generated appropriately spaced fragments that fully contained the regulatory elements of the H19/Igf2 locus. There was no allelic or regional bias in the digestion pattern (data not shown). DNA ligase was added to link the sequences that co-localized in the nucleus, independently of their location along the chromosome. After reversing the crosslinks and removing the proteins, DNA sequences associated with the endodermal enhancers were identified by PCR. In each reaction, the PCR product was the correct size. Restriction fragment length polymorphisms between the strains used allowed us to distinguish the parental origin of the ligation products. None of the products were observed in the absence of ligation of the chromatin. At least two PCRs were carried out for each of the two independent 3C experiments. Representative results are shown throughout.
Allele-specific chromosome looping at the H19/Igf2 locus
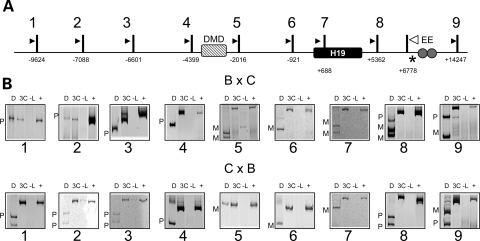
In the wild-type mouse, H19 is expressed from the maternal allele, whereas Igf2 is active on the paternal chromosome. Our results from the 3C assay were identical for reciprocal crosses of C57BL/6J × B6(CAST7) and indicated that the maternal enhancers contacted the entire H19 region, including the promoter, correlating with the active transcriptional status of the maternal H19 gene (Fig. 2, see maternal bands in panels 5–7). However, on the maternal chromosome, the enhancers were not found beyond the DMD. This suggests that the presence of an insulator assembled in this region on the maternal copy physically bars the enhancers from moving upstream.
Figure 2.
Analysis of long-range interactions at the H19 locus. (A) Schematic of the H19 locus, including the position of the coding sequence (black box), the differentially methylated domain (hatched box, DMD) and the endodermal enhancers (circles, EE). Vertical bars indicate PsuI restriction sites, numbers beneath the sites are the distance relative to the H19 transcriptional start site, and the resulting digestion fragments are numbered (1–9). Arrowheads represent the location of PCR primers for 3C analysis. The white arrowhead is the reverse primer for all reactions, which test for ligation of fragments 1–9 to the EE fragment. An asterisk indicates a restriction polymorphism that distinguishes C57BL/6J and B6(CAST7) alleles. Samples were from wild-type progeny of C57BL/6J and B6(CAST7) mice (B × C, C × B), with the female indicated first. (B) Representative gel image of 3C analysis at the H19 locus. Shown are the 3C PCR products: D, digested with NlaIII, which distinguishes paternal (P) from maternal alleles (M); 3C, non-digested PCR product; -L, no ligase control; +, positive control. The endodermal enhancers on the maternal chromosome associate with fragments in the H19 region up to the DMD, beyond which only fragments from the paternal chromosome are observed.
Whereas the enhancers were absent from the regions upstream of the DMD on the maternal chromosome, they interacted freely with those regions on the paternal allele (Fig. 2, see paternal bands in panels 1–4). This suggests that upon inactivation of the DMD by methylation, the enhancers are no longer restricted to the H19 region.
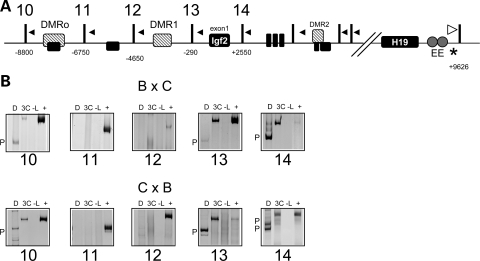
The paternally expressed Igf2 gene has four promoters and exhibits differential promoter usage throughout development (20). Restriction digestion with PsuI yields three fragments containing Igf2 promoter sequences: fragment 10 contains the placental promoter, fragment 13 contains promoter 1 and fragment 14 contains promoters 2 and 3 (Fig. 3A). In the 3C assay, the paternal enhancers associated with the three fragments containing the Igf2 promoters (Fig. 3, see paternal bands in panels 10, 13 and 14), with the strongest signal at promoters 2 and 3. In fact, promoter 3 is the main origin of transcripts arising in neonatal liver. The signal at the placental promoter is either very weak or absent, consistent with the lack of expression from this promoter in liver. These results confirm that enhancer–promoter associations are required to achieve RNA expression. Interestingly, no physical proximity between the enhancers and the H19 promoter was detected on the paternal chromosome (Fig. 2, panels 5–7), suggesting that the enhancers are somehow excluded from the region, possibly due to the hypermethylation on this allele.
Figure 3.
Analysis of long-range interactions between the endodermal enhancers and the Igf2 locus. (A) Schematic of the Igf2 locus, showing the exons (black boxes) and the differentially methylated regions (hatched boxes, DMRo, DMR1, DMR2). Vertical bars indicate PsuI restriction sites, numbers beneath the sites are the distance relative to the Igf2 exon 1 and the resulting digestion fragments from the region contacted by the endodermal enhancers are numbered (10–14). Downstream fragments showed no contact with the endodermal enhancers (data not shown). Arrowheads represent the location of PCR primers for 3C analysis. The white arrowhead at the endodermal enhancers downstream of H19 is the reverse primer for all reactions, which test for ligation of fragments 10–14 to the EE fragment. An asterisk indicates a restriction polymorphism that distinguishes C57BL/6J and B6(CAST7) alleles. Samples were from wild-type progeny of C57BL/6J and B6(CAST7) mice (B × C, C × B), with the female indicated first. (B) Representative gel image of 3C analysis of interactions of the endodermal enhancers with the Igf2 locus. Shown are the 3C PCR products: D, digested with HaeIII, which distinguishes paternal (P) from maternal alleles (M), 3C, non-digested PCR product; -L, no ligase control; +, positive control. The endodermal enhancers on the maternal chromosome associate predominantly with fragments originating in the paternal chromosome.
The DMD is required for appropriate maternal chromosome conformation
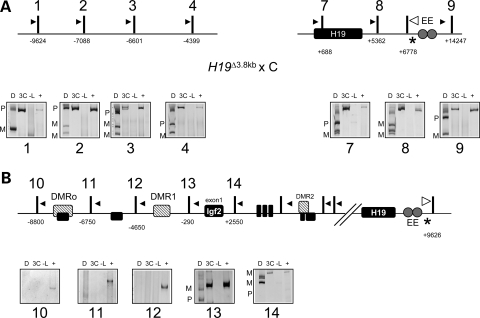
To prove that the restricted interaction of the endodermal enhancers on the maternal chromosome is dependent on the presence of the insulator, we used an allele that removed the DMD. When the H19Δ3.8 kb-5′H19 mutation is inherited maternally, the normally silent Igf2 gene is activated, resulting in biallelic expression (14). In contrast to behavior of the wild-type maternal chromosome, the analysis of the maternally inherited H19Δ3.8 kb-5′H19 allele showed contact between the endodermal enhancers and the regions upstream of the DMD (Fig. 4A, see maternal and paternal signals in panels 1–4).
Figure 4.
The H19 DMD is required to prevent interactions between the maternal endodermal enhancers and the Igf2 promoters. The 3C analysis was carried out on samples from a H19Δ3.8 kb-5′H19 × B6(CAST7) cross. (A) Top, schematic of the H19Δ3.8 kb-5′H19 allele (see Fig. 2 for details), with 3.8 kb upstream of the H19 promoter removed. Bottom, representative gel image of 3C PCR products: D, digested with NlaIII, which distinguishes paternal (P) from maternal (M) alleles; 3C, non-digested PCR product; -L, no ligase control; +, positive control. When the H19Δ3.8 kb-5′H19 allele is inherited maternally, the endodermal enhancers on the maternal chromosome interact with the whole H19 region. (B) Top, schematic of the Igf2 locus (see Fig. 3 for details). Bottom, representative gel image of 3C PCR products digested with HaeIII, which distinguishes paternal (P) from maternal (M) alleles.
In the absence of the DMD, the enhancers occupy Igf2 promoters 1–3 on both maternal and paternal chromosomes (Fig. 4B, see maternal and paternal bands in panels 13 and 14). These observations establish that the DMD is required for the appropriate chromosome conformation that restricts the maternal enhancers to the H19 promoter.
CTCF binding sites are essential in organizing the chromosome domains
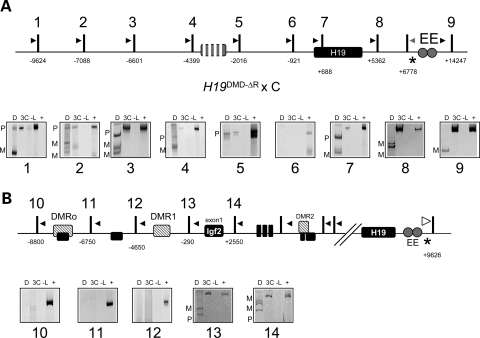
While the previous experiment showed that the DMD was required for appropriate interactions of the locus, it remained to be determined whether it was the CTCF binding sites within the region that were mediating these contacts. The H19DMD-ΔR mutation lacks the four core binding sites of CTCF in the DMD region. Mice inheriting the mutation maternally exhibit biallelic Igf2 expression and a delay in the onset of H19 expression (12). The 3C analysis showed that upon maternal inheritance of the H19DMD-ΔR allele, the endodermal enhancers were not confined to the H19 region, but rather displayed physical association throughout the H19 locus (Fig. 5A, see maternal and paternal bands in panels 1–3). This suggests that the binding of CTCF to its cognate sites is required to block the enhancers from associating with regions beyond the DMD.
Figure 5.
The CTCF sites are necessary for the enhancer-blocking activity of the DMD. The 3C analysis was carried out on samples from a H19DMD-ΔR × B6(CAST7) cross. (A) Top, schematic of the H19DMD-ΔR allele, in which the CTCF sites in the DMD have been deleted (see Fig. 2 for details). Bottom, representative gel image of 3C PCR products: D, digested with NlaIII, which distinguishes paternal (P) from maternal (M) alleles; 3C, non-digested PCR product; -L, no ligase control; +, positive control. (B) Top, schematic of the Igf2 locus (see Fig. 3 for details). Bottom, representative gel image of 3C PCR products digested with HaeIII, which distinguishes paternal (P) from maternal (M) alleles.
Consistent with the biallelic expression of Igf2, the enhancers contact the Igf2 promoters equally on both parental chromosomes (Fig. 5B, see maternal and paternal bands in panels 13 and 14). This behavior is equivalent to that of the H19Δ3.8kb-5′H19 allele, in which the DMD has been completely deleted. Thus, binding of CTCF in the DMD is necessary to abolish maternal associations between the enhancers and the Igf2 promoters.
Methylation diminishes the strength of the insulator function
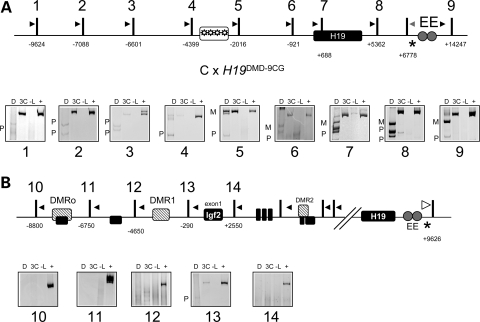
Binding of CTCF to its recognition sites within the DMD is methylation-sensitive. Therefore, the paternally hypermethylated DMD does not exhibit enhancer-blocking activity. The H19DMD-9CG mutation is a parent-of-origin-specific gain-of-function mutation, in which an insulator is assembled on the paternal DMD (10). As a result, paternal Igf2 mRNA levels are reduced, and the mice display a growth restriction phenotype. Furthermore, the normally silent paternal allele of H19 is activated. The 3C analysis showed that the endodermal enhancers interacted with the H19 promoter biallelically, consistent with the biallelic expression of H19 in this mutant (Fig. 6A, panels 5–7). Beyond the DMD, only very faint signals were observed, indicating reduced paternal interactions (Fig. 6A, panels 1–4). Furthermore, the strength of interaction between the enhancer and the Igf2 promoters was diminished or undetectable (Fig. 6B, panels 13 and 14). These results indicate that the ectopic insulator established on the paternal DMD is partially successful in restricting the enhancers to the H19 region.
Figure 6.
The H19DMD-9CG insulator is weaker than the wild-type one. The 3C analysis was carried out on samples from a B6(CAST7) × H19DMD-9CG cross. (A) Top, schematic of the H19DMD-9CG allele, in which nine CpGs in the CTCF binding sites in the DMD have been replaced with alternative nucleotides (see Fig. 2 for details). Bottom, representative gel image of 3C PCR products: D, digested with NlaIII, which distinguishes paternal (P) from maternal (M) alleles; 3C, non-digested PCR product; -L, no ligase control; +, positive control. (B) Top, schematic of the Igf2 locus (see Fig. 3 for details). Bottom, representative gel image of 3C PCR products digested with HaeIII, which distinguishes paternal (P) from maternal (M) alleles.
DISCUSSION
Imprinted gene expression is unique in that two disparately regulated alleles co-exist in the same nuclear environment. Moreover, the epigenetic status of the alleles depends on their parental origin. The imprinted H19 /Igf2 locus has been the focus of extensive studies. Differential DNA methylation and allele-specific chromatin modifications have been described in detail (13,21–23). Several targeted mutations in the mouse have helped delimit and elucidate the function of important cis-acting elements (5,6,10,24–26). The model that has emerged and is strongly supported by these studies proposes that on the maternal chromosome, a CTCF-mediated enhancer-blocker is assembled at a region upstream of H19, and as a result, enhancers lying downstream of H19 cannot access the Igf2 promoters and thus activate the H19 promoter. On the paternal chromosome, methylation inherited from the male gamete inhibits binding of CTCF, enabling the interaction between the enhancers and the Igf2 gene.
The development of 3C technology has allowed us to detect physical interactions between distant regions of the chromosome, supporting a model in which intervening DNA is looped out to accommodate the functional association between cis-acting regulatory elements (19). We have used 3C at the H19/Igf2 locus to examine the interactions between the endodermal enhancers, the H19 and Igf2 promoters, and the DMD on each of the parental alleles. In the 3C assay, predicted interactions between regulatory elements are systematically tested by examining the relative frequency of physical proximity between them. Although physical proximity does not necessarily translate into functional significance, in conjunction with the expression patterns and using mutations in the regulatory elements comparatively, the association of DNA sequences can yield insight into molecular mechanisms of transcriptional activation and repression.
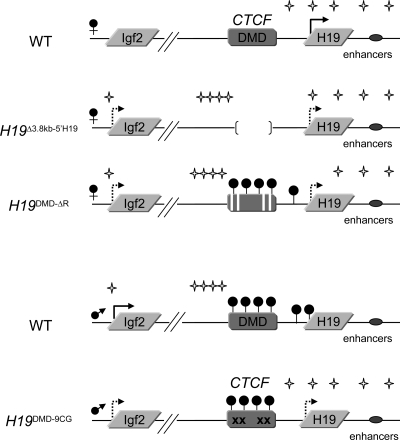
We have analyzed wild-type mice and three mouse models in which mutations affecting the insulator activity have been targeted to the DMD. Our findings are summarized in Figure 7. We find that in the wild-type mouse, interactions between the endodermal enhancers and the H19 and Igf2 promoters reflect the imprinted expression pattern, i.e. they occur in an allele-specific way. On the maternal chromosome, the enhancers contact the active H19 promoter, whereas on the paternal chromosome, the enhancers associate with the paternally expressed Igf2 promoters. Furthermore, in all of our targeted mouse models, biallelic expression correlates with enhancer localization at both paternal and maternal promoters. In the paternally inherited H19DMD-9CG allele, CTCF is able to bind its cognate sites within the DMD. Methylation is established normally in the germ line, but is partially lost by the blastocyst stage, possibly because the presence of CTCF impedes access of the maintenance DNA methyltransferase. As a result, H19 is expressed from the normally silent paternal allele at 35% the levels of the maternal allele in neonatal liver. Concordantly, our 3C results show the interactions of the endodermal enhancers with both the maternal and paternal H19 promoters. In the H19Δ3.8kb-5′H19 and H19DMD-ΔR mutants, both of which display biallelic Igf2 expression, the enhancers are in proximity to both maternal and paternal Igf2 promoters. Thus, direct physical contact between the enhancers and the promoters correlate with the gene expression, supporting the view that the interactions are functional. This is consistent with findings in a previous report (18).
Figure 7.
Summary of the parental-specific interactions of the endodermal enhancers along the H19/Igf2 locus in neonatal livers of wild-type and targeted mice. Mutations are described in the legend of Figure 1 and detailed in the text. Arrows represent expression, hatched arrows represent low levels of expression, and filled circles indicate methylation. Stars represent regions of contact, as detected by the 3C assay.
A major finding of this study is that the enhancer-blocking activity of insulators does not allow physical interactions to occur between the enhancers and regions beyond the insulator. Specifically, in the wild-type mouse, the presence of the DMD on the maternal chromosome restricts the association of the endodermal enhancers to the H19 promoter and gene body. No contacts are detected beyond the DMD. In contrast, in the mutants in which the DMD is either removed (H19Δ3.8kb-5′H19) or inactivated by deletion of the CTCF binding sites (H19DMD-ΔR), the enhancers are capable of forming associations with the regions upstream of the DMD. Indeed, on the wild-type paternal chromosome, where the insulator activity is inhibited by DNA methylation, the enhancers freely interact with the region starting immediately upstream of the DMD. Together, these data suggest that the association between enhancers and promoters does not occur by random collision, but rather by a progressive scanning activity on the part of the enhancers, which then form a stable interaction with the first suitable element they encounter, i.e. the H19 promoter on the maternal chromosome and the Igf2 promoter on the paternal chromosome (27). The fact that insulators must be located between the enhancer and the promoter supports this model. Strong evidence for this idea is also provided by our H19DMD-9CG mutation. The establishment of the insulator on the paternal H19DMD-9CG allele seems to act as a roadblock for the enhancers, markedly diminishing their interactions with chromatin lying upstream.
The presence of CTCF at the paternal DMD of the H19DMD-9CG allele does not completely block the interaction of the enhancers with the upstream chromatin, although Igf2 levels are decreased (10). This indicates that the insulator established on the methylated DMD is not completely effective in restricting the motility of the enhancers, possibly because of the residual methylation. It also suggests that insulator activity is not all or none, and that there could be varying degrees of enhancer-blocking.
It is striking that in the absence of a blocking activity on the maternal chromosome, the enhancers can both engage the H19 promoter and also advance beyond it to activate the distal Igf2 promoter, although whether contacts with both H19 and Igf2 occur simultaneously within a single cell is not known. This suggests that, by default, the enhancers are equally capable of activating both promoters. It also implicates that the lack of detectable interactions at H19 on the wild-type paternal allele is a result of presence of DNA methylation. In fact, when the H19DMD-ΔR mutation is transmitted maternally, the H19 region is hypermethylated and contact with the enhancers is lost. Furthermore, loss of methylation on the paternal H19 gene in the H19DMD-9CG mutation allows the enhancers to access the promoter, again suggesting that the determinant for H19 silencing and the exclusion of the enhancers is DNA methylation. Thus, our results underscore two roles for paternal DNA methylation as a regulatory switch at this locus. Methylation at the DMD inactivates the insulator by inhibiting CTCF binding. In addition, methylation of the H19 promoter might neutralize it as a target for enhancer interaction, promoting its bypass in favor of Igf2. These results contrast with a previous report that detected enhancer contacts within the H19 region paternally and concluded that the paternal enhancers have equal access to the whole H19 and Igf2 region (17). The discrepancy possibly arises from the differences in choice of restriction enzyme. Our restriction digests clearly separate the DMD, the H19 promoter and the endodermal enhancers into distinct fragments.
The imprinting control region of the H19/Igf2 region is the only vertebrate insulator proven to block enhancer–promoter interactions at the endogenous locus. Two prevailing mechanisms of insulator action have been proposed (28–30). Topological models propose that insulators are vital for establishing discrete structural loops that restrict the possibility of enhancer–promoter interaction to those elements within the domain. Interaction models assign a regulatory role to insulators, proposing that they compete with the promoter for the enhancer. Previous reports applying 3C technology to the H19/Igf2 locus have led to different models of insulator activity. Kurukuti et al. propose a topological model in which the insulator associates with a matrix attachment region and a differentially methylated region at the Igf2 locus. This would establish a transcriptionally inert loop encompassing the Igf2 gene. Yoon et al. favor a model in which the insulator regulates gene activity by acting as a decoy for the enhancers and the inactive Igf2 promoter. It is not clear in this model how the enhancers would be available to activate H19 expression.
Our results support a model in which the enhancers track along the chromosome and engage available promoters. The maternal DMD poses a restriction to this scanning activity, and as a result the enhancers are only present at the H19 promoter. We propose that rather than acting as a decoy for Igf2, CTCF binding to the DMD accomplishes this restriction by acting as a transcriptional facilitator for H19. Two lines of evidence support this idea. First, in the absence of CTCF binding sites (H19DMD-ΔR), initiation of H19 expression is delayed, clearly implicating CTCF binding in transcriptional activity (12). Second, the presence of CTCF binding in the paternal H19DMD-9CG correlates with activation of H19 transcription (10).
In conclusion, we present strong evidence that at the H19/Igf2 locus, transcriptional activity is correlated with co-localization of promoters and enhancers. Our data provide new evidence suggesting that the enhancer reaches the target promoter by tracking along the intervening DNA and then looping it out. Our analyses also indicate that the DMD blocks the enhancer from tracking further upstream. One possibility is that the region upstream of the DMD is not accessible to the enhancers because the DMD is in contact with the inactive Igf2 promoter, as suggested by Yoon et al. (18) or with the Igf2 DMR, as reported by Murrell et al. (16). Either of these interactions leads to the formation of a loop that would exclude the presence of the enhancers. The analyses of our mutations lend support to a different model, in which the enhancers are blocked because CTCF is acting as a transcriptional factor for H19. It will be interesting to determine if there are additional proteins involved in insulator activity, as has been reported for cohesins (31). Furthermore, it will be exciting to see what factors are involved in the tracking motions of cis-acting sequences. For instance, a recent report found that some loci can be recruited to specific nuclear positions in an actin-dependent manner (32), and it is tempting to speculate that actin could be involved in enhancer tracking. Ultimately, conformational studies at the human H19/Igf2 locus will be important to gain insight into how loss of imprinting affects the region in disease processes.
MATERIALS AND METHODS
Chromosome conformation capture (3C)
Livers from newborn mice were homogenized and filtered through a 100 µm nylon cell strainer into 10% FBS/DMEM. Cross-linking with 2% formaldehyde was carried out for 10 min at room temperature and the reaction was quenched with glycine to 0.125 m. Cells were lysed on ice in lysis buffer (10 mm Tris pH 8.0, 10 mm NaCl, 0.2% NP-40 and protease inhibitors). Nuclei were resuspended in restriction buffer containing 0.3% SDS and incubated for 1 h at 37°C. Triton X-100 was added to 1.8% and samples were incubated at 37°C for an additional hour. Digestion with PsuI was carried out overnight at 37°C. The reaction was stopped by adding SDS to 2% and incubating at 65°C. An aliquot of the reaction was removed and analyzed and compared with a digest of naked DNA to demonstrate that the digest was complete. DNA was diluted to 2.5 ng/µl with ligase buffer and ligation was carried out on one half of the sample with T4 ligase at 15°C for 4 h. Proteinase K was added to and the samples incubated at 65°C overnight. DNA was purified by phenol extraction and ethanol precipitation.
To create a pool of all possible interacting products, control templates of defined molar amounts were prepared. Control template was prepared from a BAC encompassing the H19/Igf2 region (RP23–50N22, CHORI). The BAC was digested with the appropriate enzymes and ligated. Ligated fragments of interest were amplified by PCR, gel purified and their concentration determined with a Nanodrop spectrophotometer. A mix of equimolar amounts of all the fragments was prepared and used in each reaction. The linear range of amplification was determined for each 3C sample. An amount of DNA within the linear range was used for amplification. All PCR reactions were performed with RubyTaq™ (USB) as follows: 67° 20′′, 72° 20′′ for 15 cycles, with a decrease in the annealing temperature of 2° every 3 cycles, followed by 25 cycles of 55° 20′′, 72° 20′′ and a final extension step of 72° 1′30′′. For all 3C assays, duplicate PCR analyses of two independent 3C preparations were performed. Primer pairs are described in Table 1 and their location depicted in Figure 2. PCR products were digested with either NlaIII or HaeIII to distinguish the C57BL/6J and B6(CAST7) alleles and run on 12% acrylamide gels.
Table 1.
Primers used in the 3C assay and the sizes (in base pairs) of the undigested and digested products for each strain
| H19 primers | 3C product NlaIII | C57BL/6J | B6(CAST7) | |
|---|---|---|---|---|
| 1 | GTCTCTGTAGGCCATCACTAAAGAGTATTA | 281 | 136 | 281 |
| 2 | CATTAGAAGAGAACATTTAGACTCAGACAT | 251 | 136, 115 | 251 |
| 3 | GTTGCTTTCTGTTTTAGTCAGTGTTCTATT | 283 | 136, 147 | 234 |
| 4 | CTAGTCCTCAATGTCACGTACTATTACAA | 305 | 136, 135 | 170 |
| 5 | GTACAATACTACATATTGCTCGGCAGAC | 294 | 136, 158 | 294 |
| 6 | TTTTCACAGAGTAAAAATGAAGGATCACTA | 222 | 136 | 207 |
| 7 | ACATAGAAAGGCAGGATAGTTAGCAAAG | 350 | 136, 214 | 290 |
| 8 | TATGTTTCAGTGACAAGTTAAGGTTGGACAAAG | 339 | 136, 158 | 181, 158 |
| 9 | GGCAGTAGACCTGACACAGCTTTTCTTC | 293 | 128 | 275 |
| EE/NlaIII | GTACAGGAGGCTCTACCCCCACCCTCCGTGTG | |||
| Igf2 primers | HaeIII | |||
| 10 | CAGTGATAACTTTAGATTGTGGATTGTAA | 268 | 224 | 185 |
| 11 | CCTGTTTTTATATCCTGTACCTCCTAACTA | 202 | 158 | 119 |
| 12 | CCTCTGCCACCAAGGCCCGAGCCGAGGCCTC | 250 | 206 | 167 |
| 13 | TGGCTAGAAGGCGAAAGAACGAAAAATGAAG | 275 | 231 | 192 |
| 14 | CTCGGATTCCAGAAAATGGA | 417 | 201, 216 | 193 |
| EE/Hae III | AAAAGGGACTTCAGACCTTATGCCCCCCACCTACCAG |
FUNDING
National Cancer Institute grant (CA115906 to N.E.); National Institutes of Health (GM51279 to M.S.B.).
ACKNOWLEDGEMENTS
We thank Chris Vakoc for valuable advice on the 3C assay and Melanie Hullings for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bartolomei M.S., Zemel S., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 2.Brannan C.I., Bartolomei M.S. Mechanisms of genomic imprinting. Curr. Opin. Genet. Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 3.Leighton P.A., Saam J.R., Ingram R.S., Stewart C.L., Tilghman S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 4.Kaffer C.R., Grinberg A., Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaffer C.R., Srivastava M., Park K.-Y., Ives E., Hsieh S., Batlle J., Grinberg A., Huang S.-P., Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 6.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 7.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;2000:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 8.Szabo P.E., Pfeifer G.P., Mann J.R. Parent-of-origin-specific binding of nuclear hormone receptor complexes in the H19-Igf2 imprinting control region. Mol. Cell Biol. 2004;24:4858–4868. doi: 10.1128/MCB.24.11.4858-4868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pant V., Kurukuti S., Pugacheva E., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel N., West A.G., Felsenfeld G., Bartolomei M.S. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat. Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay K.D., Saam J.R., Ingram R.S., Tilghman S.M., Bartolomei M.S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 12.Engel N., Thorvaldsen J.L., Bartolomei M.S. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 13.Tremblay K.D., Duran K.L., Bartolomei M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorvaldsen J.L., Mann M.R., Nwoko O., Duran K.L., Bartolomei M.S. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 2002;22:2450–2462. doi: 10.1128/MCB.22.8.2450-2462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 16.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 17.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon Y.S., Jeong S., Rong Q., Park K.Y., Chung J.H., Pfeifer K. Analysis of the H19ICR Insulator. Mol. Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 20.Moore T., Constancia M., Zubair M., Bailleul B., Feil R., Sasaki H., Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl Acad. Sci. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartolomei M.S., Webber A.L., Brunkow M.E., Tilghman S.M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 22.Delaval K., Govin J., Cerqueira F., Rousseaux S., Khochbin S., Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verona R.I., Thorvaldsen J.L., Reese K.J., Bartolomei M.S. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol. Cell Biol. 2008;28:71–82. doi: 10.1128/MCB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorvaldsen J.L., Duran K.L., Bartolomei M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo P.E., Tang S.-H., Rentsendorj A., Pfeifer G.P., Mann J.R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Current Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 26.Pant V., Mariano P., Kanduri C., Mattsson A., Lobanenkov V., Heuchel R., Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X., Ling J., Zhang L., Pi W., Wu M., Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geyer P.K., Clark I. Protecting against promiscuity: the regulatory role of insulators. Cell Mol. Life Sci. 2002;59:2112–2127. doi: 10.1007/s000180200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West A.G., Gaszner M., Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 30.Engel N., Bartolomei M.S. Mechanisms of insulator function in gene regulation and genomic imprinting. Int. Rev. Cytol. 2003;232:89–127. doi: 10.1016/s0074-7696(03)32003-0. [DOI] [PubMed] [Google Scholar]
- 31.Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., Matera A.G. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]