Abstract
Predisposition to psoriasis is known to be affected by genetic variation in HLA-C, IL12B and IL23R, but other genetic risk factors also exist. We recently reported three psoriasis-associated single nucleotide polymorphisms (SNPs) in the 5q31 locus, a region of high linkage disequilibrium laden with inflammatory pathway genes. The aim of this study was to assess whether other variants in the 5q31 region are causal to these SNPs or make independent contributions to psoriasis risk by genotyping a comprehensive set of tagging SNPs in a 725 kb region bounded by IL3 and IL4 and testing for disease association. Ninety SNPs, capturing 86.4% of the genetic diversity, were tested in one case–control sample set (467 cases/460 controls) and significant markers (Pallelic < 0.05) (n = 9) were then tested in two other sample sets (981 cases/925 controls). All nine SNPs were significant in a meta-analysis of the combined sample sets. Pair-wise conditional association tests showed rs1800925, an intergenic SNP located just upstream of IL13 (Mantel–Haenszel Pcombined = 1.5 × 10−4, OR = 0.77 [0.67–0.88]), could account for observed significant association of all but one other SNP, rs11568506 in SLC22A4 [Mantel–Haenszel Pcombined = 0.043, OR = 0.68 (0.47–0.99)]. Haplotype analysis of these two SNPs showed increased significance for the two common haplotypes (rs11568506–rs1800925: GC, Pcombined = 5.67 × 10−6, OR = 1.37; GT, Pcombined = 6.01 × 10−5, OR = 0.75; global haplotype P = 8.93 × 10−5). Several 5q31-region SNPs strongly associated with Crohn's disease (CD) in the recent WTCCC study were not significant in the psoriasis sample sets tested here. These results identify the most significant 5q31 risk variants for psoriasis and suggest that distinct 5q31 variants contribute to CD and psoriasis risk.
INTRODUCTION
Genome wide association (GWA) studies have proven powerful in identifying single nucleotide polymorphisms (SNPs) that are significantly associated with common, multi-factorial diseases (1–3). However, given the nature of these scans, the identified markers may not be causal but rather in linkage disequilibrium (LD) with the true causative variants located elsewhere, possibly in a neighboring, unrelated gene. Furthermore, there may be multiple variants in a region that, independently or together, contribute to disease risk (4,5). Consequently, comprehensive follow-up fine mapping of significant GWA markers, which incorporates the rich genetic information revealed by the HapMap project (6), is required to narrow the region of interest and identify true causal variant(s).
Psoriasis is a common, chronic inflammatory skin disease that can affect individuals of all ages with the majority manifesting clinical symptoms before the age of 40 years (7). Susceptibility to psoriasis is strongly influenced by genetic factors, independently or interacting with each other, and may be initiated or exacerbated by environmental factors such as infection. Haplotypes carrying the HLA-Cw*0602 allele confer the strongest genetic effect in whites, particularly among early onset cases (8,9). More recently, association of psoriasis with SNPs in IL12B and IL23R has been identified (2,10) and independently confirmed in other case–control studies (11–13). Thus, a total of three psoriasis risk loci have been established with convincing statistical evidence.
These genetic variations, however, do not explain the totality of psoriasis risk, suggesting other genetic risk factors exist. Indeed, linkage studies have provided evidence suggesting other genomic loci are significantly associated with this disease (14). In addition, numerous putative risk factors have been reported, often with substantial appeal as strong biological and/or positional candidates (reviewed in 15,16). Recent additions to the growing list of potential genetic risk factors for psoriasis include variation in genomic copy number of ß-defensin (17) and SNPs and/or haplotypes in ADAM33 (18), IL15 (19) and CDKAL1 (20).
We recently reported that three SNPs (rs1800925, rs20541 and rs848) in the IL13-region at the 3′-end of the 5q31 cytokine gene cluster were associated with psoriasis in a large, multi-tiered, genetic association study (21). The 5q31 genomic region contains a cluster of cytokine and immune-related genes, including interleukin (IL) genes IL3, IL4, IL5 and IL13; interferon regulatory factor-1 (IRF-1); colony-stimulating factor-2 (CSF2) and T-cell transcription factor-7 (TCF7), all excellent biological candidate genes. This region shows extended marker–marker correlation; for example, examination of the CEU HapMap dataset (www.hapmap.org) reveals that a number of markers within the 100 kb interval immediately upstream of IL13 are in relatively strong LD (r2 > 0.5) with one of the significant IL13 SNPs, rs20541. Numerous other markers extending as far as 1 Mb upstream of IL13 also show moderate LD with rs20541 (r2 > 0.2). Therefore, to more precisely define the causal variants in the 5q31 region, we carried out a comprehensive analysis of the genetic variation in this region using three large, independent sample sets, totaling 1448 cases and 1385 controls, and report findings from association tests of the markers with psoriasis risk.
RESULTS
Single marker analysis identifies multiple SNPs associated with psoriasis
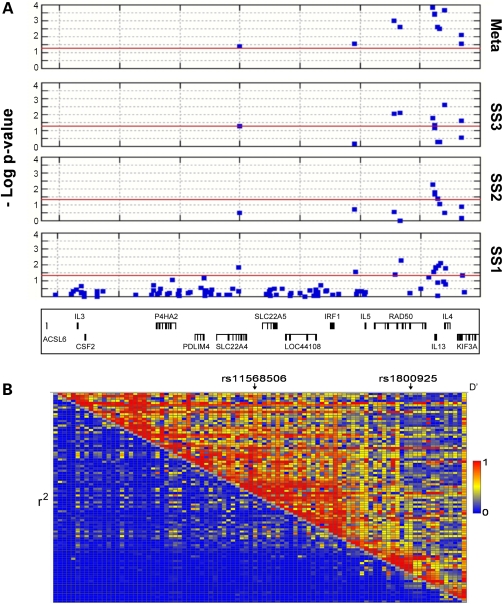
Examination of the 5q31 LD structure in the CEU HapMap Phase II dataset led us to focus our fine mapping on a 725 kb region that delimits the entire cytokine cluster, extending from ∼50 kb upstream of IL3 to ∼50 kb downstream of IL4. Ninety fine mapping SNPs, primarily selected as tagging markers (see Materials and methods), were genotyped in 467 cases and 460 controls (sample set 1) (Fig. 1A). In addition to the three original IL13-region SNPs (21), allelic association tests of the 90 new SNPs identified nine additional markers significantly associated with psoriasis (Pallelic < 0.05, Fig. 1A and Supplementary Material, Table S1) in a 370 kb region extending from SLC22A4 to KIF3A and including IRF1, IL5, IL13 and IL4. These nine SNPs were then genotyped in two other case–control sample sets (sample set 2: 498 cases/498 controls; sample set 3: 483 cases/427 controls) and although the significance of these nine markers varied in the two follow-up sample sets (Fig. 1A and Supplementary Material, Table S2), all nine were significant in a combined analysis of the three sample sets (Mantel–Haenszel Pcombined < 0.05, Table 1 and Supplementary Material, Table S2). Breslow–Day tests provided no evidence for heterogeneity of effect across sample sets except for one marker, rs11568506 [odds ratio (OR) homogeneity: Prs11568506 = 0.032; P > 0.05 for all other markers]. Based on the Pcomb-values, none of the nine new markers were more significant than rs1800925 (Mantel–Haenszel Pcombined = 0.00015), the most significant marker identified in our original report (21), although some were comparable (e.g. P = 0.00022 for the IL4 marker rs2227282). ORs conferred by these variants were all modest (Table 1 and Supplementary Material, Table S2).
Figure 1.
(A) P-values of SNPs tested in the three sample sets individually (SS1, SS2 and SS3) and combined (Meta). A total of 93 SNPs were tested in sample set 1; 12 SNPs were tested in sample sets 2 and 3. rs1800925, rs20541 and rs848, which we previously reported to be associated with psoriasis (21), were tested in all three sample sets. A gene map is shown at the bottom, based on the NCBI b36 genome assembly. (B) Inter-marker LD of the 93 markers in sample set 1. D' values are shown in the top right triangle, and r2 values are in the bottom triangle. The D' and r2 values were calculated using both cases and controls from sample set 1.
Table 1.
Association analysis of the two independent, psoriasis-associated SNPs in the three sample sets combineda
| SNP ID | Gene | Allele/Genotype | Cases, n (%) | Controls, n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| rs11568506 | SLC22A4 | A | 50 (0.017) | 69 (0.025) | 0.68 (0.47–0.99) | 0.043 |
| G | 2820 (0.983) | 2677 (0.975) | 1.00 (reference) | |||
| AA | 2 (0.0014) | 0 (0) | NCb | 0.022 | ||
| AG | 46 (0.032) | 69 (0.050) | 0.63 (0.43–0.92) | |||
| GG | 1387 (0.0967) | 1304 (0.950) | 1.00 (reference) | |||
| rs1800925 | IL13 | T | 453 (0.158) | 540 (0.196) | 0.77 (0.67–0.88) | 0.00015 |
| C | 2417 (0.842) | 2210 (0.804) | 1.00 (reference) | |||
| TT | 36 (0.025) | 56 (0.041) | 0.56 (0.37–0.86) | 0.00081 | ||
| TC | 381 (0.266) | 428 (0.311) | 0.78 (0.66–0.92) | |||
| CC | 1018 (0.709) | 891 (0.648) | 1.00 (reference) |
aData for the 10 other significant SNPs are presented in Supplementary Material, Table S2.
bNot calculated.
Conditional analysis reveals two independent SNPs
To tease apart association signals from patterns of LD and determine which of these 12 significant SNPs were independently associated with psoriasis risk, we performed pair-wise genotype-conditioned analyses. Using the combined datasets, we calculated a summary statistic for each pair of SNPs using the genotypes of one marker conditioned on another marker, and evaluated the significance of each result using a permutation test (10,000 permutations) (Table 2). This approach is similar to the haplotype method (22) and has been described previously (2). Not surprisingly, the significant association of various SNPs with psoriasis disappeared upon conditioning on other markers. Most notably, the significance of all other markers except rs11568506 (in SLC22A4; P = 0.0105) was abolished upon conditioning on rs1800925. Conversely, association of rs1800925 remained strong (P = 0.0002) upon conditioning on rs11568506. Together these data suggest that rs11568506 and rs1800925 make independent contributions to psoriasis risk. The genotype correlation between rs11568506 and rs1800925 is very low (r2 = 0.006), whereas that between rs1800925 and the other 10 SNPs varied from 0.016 to 0.801 (Supplementary Material, Table S3).
Table 2.
Conditional association tests for 5q31 markers: P-values for marker 1 conditional on marker 2
| Marker 1 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11568506 | rs4143832 | rs2897443 | rs6884762 | rs1800925 | rs20541 | rs848 | rs2243211 | rs762534 | rs2227282 | rs12186803 | rs10069772 | ||
| Marker 2 | rs11568506 | 0.0138 | 0.0012 | 0.0023 | 0.0002 | 0.0012 | 0.0012 | 0.0023 | 0.0094 | 0.0093 | 0.3754 | 0.0390 | |
| rs4143832 | 0.0022 | 0.2503 | 0.0067 | 0.0046 | 0.0591 | 0.0353 | 0.0791 | 0.1072 | 0.0531 | 0.3720 | 0.0873 | ||
| rs2897443 | 0.0071 | 0.9922 | 0.0736 | 0.0423 | 0.1103 | 0.1386 | 0.1807 | 0.4214 | 0.0552 | 0.2559 | 0.1870 | ||
| rs6884762 | 0.0114 | 0.0915 | 0.0740 | 0.0118 | 0.0129 | 0.0099 | 0.0648 | 0.0608 | 0.0148 | 0.0239 | 0.2777 | ||
| rs1800925 | 0.0105 | 0.2493 | 0.2168 | 0.0648 | 0.1538 | 0.1006 | 0.1136 | 0.3292 | 0.0888 | 0.4419 | 0.1153 | ||
| rs20541 | 0.0195 | 0.8541 | 0.2509 | 0.0432 | 0.1123 | 0.8374 | 0.1031 | 0.4035 | 0.1524 | 0.3309 | 0.5399 | ||
| rs848 | 0.0197 | 0.6773 | 0.3075 | 0.0361 | 0.0676 | 0.7771 | 0.2661 | 0.3943 | 0.2287 | 0.5391 | 0.5746 | ||
| rs2243211 | 0.0145 | 0.5871 | 0.2605 | 0.0553 | 0.0330 | 0.0578 | 0.1443 | 0.2897 | 0.0048 | 0.0992 | 0.1132 | ||
| rs762534 | 0.0202 | 0.3471 | 0.2286 | 0.0302 | 0.0378 | 0.0822 | 0.0742 | 0.0999 | 0.0034 | 0.0879 | 0.0365 | ||
| rs2227282 | 0.1132 | 0.6841 | 0.1164 | 0.0588 | 0.0563 | 0.1368 | 0.1943 | 0.0238 | 0.0276 | 0.8115 | 0.8433 | ||
| rs12186803 | 0.1977 | 0.4103 | 0.0326 | 0.0005 | 0.0151 | 0.0153 | 0.0342 | 0.0040 | 0.0079 | 0.0557 | 0.0116 | ||
| rs10069772 | 0.0281 | 0.1877 | 0.0327 | 0.0691 | 0.0054 | 0.0642 | 0.0556 | 0.0372 | 0.0167 | 0.0878 | 0.0289 | ||
P < 0.05 in boldface.
Next we assessed whether SNPs in high LD with rs1800925 could be more significantly associated with disease. Genotype data were retrieved from the HapMap CEU dataset (release no. 22, phase II April 07), covering 1 Mb on each side of the IL13 gene, and the correlation (as determined by r2 values) between rs1800925 and all 1559 SNPs was calculated. A total of 32 markers, including five SNPs in putative transcription factor binding sites but no cSNPs, were highly correlated with rs1800925 (r2 > 0.8) (Supplementary Material, Table S4), 30 of which were in absolute LD (r2 = 1) with one another. One of these 30 markers, rs2897443 (r2 with rs1800925 = 0.897) had been genotyped in all three psoriasis sample sets (Supplementary Material, Table S2) and was nearly 10-fold less significant than rs1800925 (Prs2897443 = 0.001 versus Prs1800925 = 0.00015). In addition, rs2897443 was no longer significant after conditioning on rs1800925 (P = 0.2168), whereas rs1800925 remained significant after conditioning on rs2897443 (P = 0.0423). The remaining two markers (rs2706370 and rs12187537) were not genotyped in our study; however, they were more highly correlated with rs2897443 (r2 = 0.94) than with rs1800925 (r2 = 0.829 and 0.817, respectively) suggesting their association with psoriasis is most likely less significant than rs1800925. All together, these data indicate that there is no evidence that any of the HapMap SNPs are more significantly associated with psoriasis than rs1800925.
Given that two markers, rs11568506 and rs1800925, show independent association with psoriasis, we carried out haplotype analyses using both markers in each of the three individual sample sets and in a combined analysis of all three sample sets. Although it is difficult to determine whether the two independent markers reside in the same LD block (Fig. 1B), the number of double heterozygotes was small (7 cases and 16 controls out of 2800 samples; Supplementary Material, Table S5); therefore, phase could be unambiguously established in 99.2% of all individuals. Three haplotypes were observed, with the fourth being relatively rare (<0.001 in cases or controls) due to the low frequency of rs11568506. The two common haplotypes were significantly associated with psoriasis in two of the three sample sets; although they were not significant in sample set 2, the ORs were in the same direction as in the other two sample sets (Table 3). In a meta-analysis, both common haplotypes were significantly associated with psoriasis, and significance of the most common haplotype, GC, was markedly higher than any single SNP (P = 5.67 × 10−6 for the haplotype versus 1.5 × 10−4 for rs1800925). The observed effect sizes in the combined analysis of all sample sets were 1.37 for the predisposing GC (rs11568506–rs1800925) haplotype (frequency: 0.827 in cases versus 0.777 in controls) and 0.75 for the protective GT haplotype (frequency: 0.156 in cases 0.198 in controls).
Table 3.
Haplotype association tests
| Sample set | Haplotypea (rs11568506/rs1800925) | Cases, n (%) | Controls, n (%) | Phaplotype | Global P | OR |
|---|---|---|---|---|---|---|
| SS1 | GC | 771 (0.844) | 713 (0.787) | 0.0019 | 0.0028 | 1.46 |
| GT | 131 (0.143) | 167 (0.184) | 0.018 | 0.74 | ||
| AC | 12 (0.013) | 26 (0.029) | 0.019 | 0.45 | ||
| SS2 | GC | 796 (0.807) | 765 (0.781) | 0.13 | 0.11 | 1.18 |
| GT | 168 (0.170) | 199 (0.203) | 0.052 | 0.81 | ||
| AC | 22 (0.022) | 15 (0.015) | 0.36 | 1.48 | ||
| SS3 | GC | 799 (0.830) | 651 (0.764) | 0.00091 | 0.0028 | 1.51 |
| GT | 147 (0.153) | 175 (0.205) | 0.0069 | 0.70 | ||
| AC | 13 (0.014) | 26 (0.031) | 0.031 | 0.45 | ||
| Allb | GC | 2366 (0.827) | 2128 (0.777) | 5.67E−06 | 8.93E−05 | 1.37 |
| GT | 446 (0.156) | 542 (0.198) | 6.01E−05 | 0.75 | ||
| AC | 47 (0.017) | 68 (0.025) | 0.053 | 0.66 |
aFrequency of AT haplotype is <0.001 in both cases or controls.
bHaplotype counts were estimated and may vary slightly from the sum of the three individual sample sets.
Distinct SNPs are associated with psoriasis and Crohn's disease (CD)
Previous studies have identified linkage of the 5q31 locus and/or association of specific variants in this region with several other diseases, including CD and asthma/allergic disorders (23–25). Because psoriasis and CD may share a common genetic etiology [psoriasis is approximately five times more common in CD patients than in controls (26) and the same IL23R missense SNP is associated with both diseases (1,2,11–13)], we tested our psoriasis sample sets for 5q31 SNPs showing strong association with CD (3). Of the 90 fine-mapping markers described above, five, strongly associated with CD in the WTCCC study (rs2285673, rs4540166, rs2522057, rs6596075 and rs10077785; all with P < 5 × 10−5 in ∼2000 cases and ∼3000 controls (3)], were genotyped in our three psoriasis sample sets. None were significant in the individual sample sets (all P > 0.05) (data not shown), although one SNP, rs2522057, was significantly associated with psoriasis in a meta-analyses of all three sample sets combined (P = 0.022) (Table 4). The pair-wise conditional association test, however, showed that this significant association could be accounted for by the IL13 marker, rs1800925 (P = 0.60 for rs2522057 conditioned upon rs1800925, whereas rs1800925 remained significant when conditioned upon rs2522057, P = 0.017).
Table 4.
Association of putative CD SNPs with psoriasis
| Marker | Chr | Position (bp) | Gene | Psoriasis (SS1+SS2+SS3) |
CD (WTCCC)a |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele frequency |
Allele frequency |
||||||||||
| Case | Control | Pallelic | OR (95% CI) | Case | Control | Pallelic | OR (95% CI) | ||||
| rs6596075 | 5 | 131 770 127 | 0.162 | 0.164 | 0.82 | 0.98 (0.85–1.14) | 0.127 | 0.166 | 5.40E−07 | 0.73 (0.65–0.83) | |
| rs2285673 | 5 | 131 783 868 | LOC441108 | 0.226 | 0.235 | 0.43 | 0.95 (0.84–1.08) | 0.212 | 0.252 | 1.50E−05 | 0.80 (0.73–0.89) |
| rs4540166 | 5 | 131 807 756 | LOC441108 | 0.205 | 0.212 | 0.52 | 0.96 (0.84–1.09) | 0.189 | 0.228 | 8.98E−06 | 0.79 (0.71–0.88) |
| rs10077785 | 5 | 131 829 057 | 0.209 | 0.218 | 0.45 | 0.95 (0.84–1.09) | 0.192 | 0.234 | 1.81E−06 | 0.78 (0.70–0.86) | |
| rs2522057 | 5 | 131 829 846 | 0.444 | 0.414 | 0.022 | 1.13 (1.02–1.26) | 0.478 | 0.422 | 1.01E−07 | 1.29 (1.20–1.33) | |
aData are from WTCCC (3).
Assuming a type I error rate of 0.05 and the population allele frequency observed in the WTCCC study, our combined analysis had 90% power to detect an allelic effect equal to an OR of 1.25, similar to the reported OR of 1.37 for rs6596075, which tags the 5q31 CD risk haplotype. Thus, variants contributing to CD risk in this genomic region, which map 5′ of the psoriasis-associated SNPs near the LOC441108-region, do not make an independent contribution to the etiology of psoriasis in our sample sets. In addition, although neither of the two independent psoriasis markers, rs1800925 and rs11568506, was tested in the WTCCC's CD sample set, four other markers in high LD with rs1800925 (r2 = 0.9 for all) were tested and were not significant in their study. Therefore, association with CD and psoriasis in this region appears to be explained by different variants.
DISCUSSION
Our analysis of the 5q31 cytokine cluster reveals that multiple SNPs are significantly associated with psoriasis risk; however, incorporating LD into our analysis revealed two independent, psoriasis-associated SNPs: rs1800925 located 5′ of IL13 and rs11568506, an intronic SNP in SLC22A4, also known as organic cation transporter 1 (OCTN1). The observed significance was only marginal for the SLC22A4 marker (Mantel–Haenszel Pcombined = 0.043). However, combining the genotype information for this marker with the more frequent IL13 marker, which is also more significant (Mantel–Haenszel Pcombined = 0.00015), resulted in two common psoriasis-associated haplotypes that were both highly significant. In particular, the major risk haplotype, with a control allele frequency of 0.777 increasing to 0.827 in cases, showed more pronounced association with psoriasis (Pcombined = 5.67 × 10−6, OR = 1.37) than any of the individual markers.
The biological interpretation of this observation (i.e. variants in two linked genes) is not straight-forward; however, both SLC22A4 and IL13 are putative genetic risk factors for other autoimmune or autoinflammatory disorders. Specifically, two-locus functional polymorphisms (haplotypes) in SLC22A4 and the adjacent SLC22A5 genes have been reported to be associated with CD (27), although definitive causal variants remain to be determined (28,29). Variation in IL13, which encodes an immunoregulatory cytokine produced by activated T helper 2-type (Th2) cells, has been most prominently associated with asthma (23), as well as atopic dermatitis (30). Thus, both genes have strong a priori plausibility as risk factors of psoriasis, particularly as the evidence for shared risk variants across multiple autoimmune disorders increases (31,32). Alternatively, rs11568506 might be in high LD with another, untested SNP closer to IL13 so that the ‘independent’ effect could be explained by another variant within or near IL13. Further experimentation should help determine whether one or both of these genes play a role in the disease process.
The two identified SNPs are located in an intron of SLC22A4 and near the 5′-end of IL13, respectively, with no predictive functional significance based on current SNP annotation, although rs11568506 is 43 bp from a POU2F1 transcription factor binding site, which exhibits a high degree of conservation across multiple placental mammals. In the absence of a validated role for these SNPs in gene regulation, and the presence of several other excellent biological candidate genes within this region as well as critical regulatory elements (33), it is tempting to speculate that untested, causal genetic variant(s) may lie on the haplotype defined by rs11568506 and rs1800925. Although our comprehensive, HapMap-based analysis of the variation in this region provided no evidence for such a marker, we were unable to develop assays for tagging SNPS covering 13.6% of the variation. In addition, SNPs not available in the HapMap, such as our most significant marker, rs1800925, may explain these results. Similarly, other genetic variants (insertions, deletions, etc.) which have been implicated in the etiology of various complex diseases, including psoriasis (17,34), may be responsible for modulating disease susceptibility.
The recent identification of IL23R variants associated with both psoriasis and CD solidifies the notion that genetic etiology may overlap between these two diseases (1,2). However, even though 5q31 linkage signals have been observed for both diseases (35–37), our data suggest that distinct markers within this region contribute to CD and psoriasis risk. Several previously reported CD-associated markers in the 5q31 region, including one that tags a risk haplotype (rs6596075), were not significant in our psoriasis sample sets despite having sufficient power to detect an effect size similar to that observed in the CD study (3). In addition, the modest association of rs2522057 with psoriasis in the combined analysis of our sample sets could be explained by LD with the most significant marker, rs1800925. This observation is consistent with a recent report that failed to detect a significant association between psoriasis and three other CD-associated SNPs in SLC22A4 and SLC22A5 (38). It is possible that the lack of association may be partly confounded by the fact that the actual causal CD variants have not been tested. However, judging from the significance level observed in the WTCCC study (3), and unless there are significant differences in the LD structure between their sample sets and ours (which is unlikely), we favor the interpretation that susceptibility to psoriasis and CD are affected by distinct genetic variants at this locus or the same variants but with different effect sizes.
Finally, it is possible that we may not have the power to detect independent effects in our sample sets. Thus, although the significance of all other markers except rs11568506 can be accounted for by rs1800925, we cannot exclude the possibility that other markers, particularly the stronger ones in IL13 and IL4, make additional, independent contributions to disease risk. In addition, the possibility that our current finding is false positive cannot be completely discounted, since the observed significance will not survive multiple testing corrections if all markers tested in our previous genome-wide scan and in this study are considered—which is generally deemed conservative. However, in an unpublished GWA study of ∼1400 white North American cases and ∼1400 white North American controls, two of our original IL13-associated SNPs, rs20541 and rs848, exhibit very strong evidence for association (Abecasis, Bowcock, Krueger and Elder, personal communication). These data suggest markers in the 5q31 region are true psoriasis risk markers; validation in other sample sets of similar characteristics is warranted to bring further definition to the causal variant(s).
MATERIALS AND METHODS
Clinical samples
Three sample sets of psoriasis cases and unaffected controls were assembled for this study. Detailed demographics and diagnostic/enrollment criteria have been described in a previous publication (2). Briefly, all individuals are North American whites of European descent and were 18 years or older when they contributed their samples (sample set 1: 467 cases/460 controls; sample set 2: 498 cases/498 controls; sample set 3: 483 cases/427 controls). Informed written consent was obtained from all participants, and all protocols were approved by national and/or local institutional review boards.
Selection of tagging SNPs and overall study design
To comprehensively yet efficiently investigate the 5q31 region, we ran the MIT tagger program (http://www.broad.mit.edu/mpg/tagger/server.html) (39) with the CEU HapMap phase II dataset [the landmarks were set at chr5:131373131…132097638 bp (NCBI Build 36); minimal allele frequency was set at 0.02; r2 threshold was set at 0.8; and tagging mode was set at pair-wise]. A total of 103 tagging SNPs (including the previously tested IL13 marker rs20541) are required to capture the SNP diversity (610 polymorphic sites) within this 725 kb region. Assay designs were attempted on the default tagging markers, and when not feasible, on an alternative marker within the tagging group. We referenced the WTCCC study to preferentially select markers that were significantly associated with CD as tagging markers. We were able to develop assays for 90 of these tagging SNPs (excluding the assay already available for rs20541). In addition, assays for two SNPs in predicted transcription factor binding sites (rs381870 and rs4540166) were developed.
The above 92 SNPs were genotyped in one of the three psoriasis case–control sample sets (sample set 1; 467 cases/460 controls). Genotyping data for 90 markers, including 88 tagging markers (equivalent to 86.4% coverage for all tagging markers), passed quality control and were subjected to further analysis. Hardy–Weinberg equilibrium tests identified six markers in controls and two others in cases with P < 0.05 (Supplementary Material, Table S1). However, analysis of the data showed no apparent error in genotype assignment for these markers, thus they were included in the association tests. As predicted, inter-marker LD between the genotyped markers was low in our samples (Fig. 1B). Significant SNPs (P < 0.05) were then genotyped in two other case–control sample sets [sample set 2 (498 cases/498 controls) and sample set 3 (483 cases/424 controls)].
Genotyping
Genotyping was carried out by the method of real-time, allele-specific PCR with primers designed and validated in-house (40) (primer information is available upon request). Genotype calls of cases and controls, randomly arrayed onto genotyping plates, were made using an automated algorithm and were subjected to manual examination without knowledge of case–control status, prior to statistical analysis. Genotyping accuracy has been consistently better than 99% (2,41).
Statistical analysis
Hardy–Weinberg equilibrium was examined by an exact test. Allelic association of SNPs with psoriasis risk was determined by the χ2 test in individual sample sets and by meta-analysis using fixed effects of Mantel–Haenszel methods to combine ORs across the sample sets. Marker conditional association tests were done using an approach similar to the haplotype method (22) as previously described (2). In brief, cases and controls were partitioned on the basis of genotypes at one SNP, and counts at an interrogated SNP were subjected to analysis, where statistical significance was assessed by a permutation method (10 000 permutations). The marker–marker LD measurements, D' and r2 were calculated using LDMax (www.sph.umich.edu/csg/abecasis/GOLD/docs/ldmax.html). Haplotypes were estimated and tested for association with disease status using a score test with haplotypes coded in an additive fashion (42). Power calculations were done using the PS:Power and Sample Size Calculation Program, obtained from the Department of Biostatistics, Vanderbilt University website (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize).
SUPPLEMENTARY MATERIAL
FUNDING
Financial support was in part provided by a Public Health Services research grant to the Huntsman General Clinical Research Center at the University of Utah, by National Center for Research Resources grant M01-RR00064, and by generous gifts from the W.M. Keck Foundation and from the George S. and Delores Dore Eccles Foundation. Additional support was provided by Celera.
Supplementary Material
ACKNOWLEDGEMENTS
Contribution of clinical samples by the patients and other individuals made this project possible, and the technical support from our Celera colleagues, particularly C. Rowland and V. Garcia for statistical analyses and D. Ross and D. Wolfson for assay design, was a critical part of the study. All are greatly appreciated. We also thank J. Sninsky and T. White for guidance.
Conflict of Interest statement. Authors employed by Celera declare their financial interest in the company.
REFERENCES
- 1.Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cargill M., Schrodi S.J., Chang M., Garcia V.E., Brandon R., Callis K.P., Matsunami N., Ardlie K.G., Civello D., Catanese J.J., et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 2007;80:273–390. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham R.R., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M., Atmaca-Sonmez P., Othman M., Branham K.E., Khanna R., Wade M.S., Li Y., Liang L., Zareparsi S., Swaroop A., et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat. Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths C.E., Barker J.N. Pathogenesis and clinical features of psoriasis. Lancet. 2007;370:263–271. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 8.Henseler T., Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J. Am. Acad. Dermatol. 1985;13:450–456. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 9.Nair R.P., Stuart P.E., Nistor I., Hiremagalore R., Chia N.V., Jenisch S., Weichenthal M., Abecasis G.R., Lim H.W., Christophers E., et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsunemi Y., Saeki H., Nakamura K., Sekiya T., Hirai K., Fujita H., Asano N., Kishimoto M., Tanida Y., Kakinuma T., et al. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibilty to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 2002;30:161–166. doi: 10.1016/s0923-1811(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 11.Capon F., Di Meglio P., Szaub J., Prescott N.J., Dunster C., Baumber L., Timms K., Gutin A., Abkevic V., Burden A.D., et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum. Genet. 2007;122:201–206. doi: 10.1007/s00439-007-0397-0. [DOI] [PubMed] [Google Scholar]
- 12.Smith R.L., Warren R.B., Eyre S., Ho P., Ke X., Young H.S., Griffiths C.E., Worthington J. Polymorphisms in the IL-12beta and IL-23R Genes Are Associated with Psoriasis of Early Onset in a UK Cohort. J. Invest. Dermatol. 2007;128:1325–1327. doi: 10.1038/sj.jid.5701140. [DOI] [PubMed] [Google Scholar]
- 13.Nair R.P., Ruether A., Stuart P.E., Jenisch S., Tejasvi T., Hiremagalore R., Schreiber S., Kabelitz D., Lim H.W., Voorhees J.J., et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J. Invest. Dermatol. 2008;128:1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Psoriasis Genetics Consortium. The International Psoriasis Genetics Study: assessing linkage to 14 candidate susceptibility loci in a cohort of 942 affected sib pairs. Am. J. Hum. Genet. 2003;73:430–437. doi: 10.1086/377159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capon F., Trembath R.C., Barker J.N. An update on the genetics of psoriasis. Dermatol. Clin. 2004;22:339–347. doi: 10.1016/S0733-8635(03)00125-6. vii. [DOI] [PubMed] [Google Scholar]
- 16.Bowcock A.M. The genetics of psoriasis and autoimmunity. Annu. Rev. Genomics Hum. Genet. 2005;6:93–122. doi: 10.1146/annurev.genom.6.080604.162324. [DOI] [PubMed] [Google Scholar]
- 17.Hollox E.J., Huffmeier U., Zeeuwen P.L., Palla R., Lascorz J., Rodijk-Olthuis D., van de Kerkhof P.C., Traupe H., de Jongh G., den Heijer M., et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesueur F., Oudot T., Heath S., Foglio M., Lathrop M., Prud'homme J.F., Fischer J. ADAM33, a new candidate for psoriasis susceptibility. PLoS ONE. 2007;2:e906. doi: 10.1371/journal.pone.0000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.J., Yan K.L., Wang Z.M., Yang S., Zhang G.L., Fan X., Xiao F.L., Gao M., Cui Y., Wang P.G., et al. Polymorphisms in interleukin-15 gene on chromosome 4q31.2 are associated with psoriasis vulgaris in Chinese population. J. Invest. Dermatol. 2007;127:2544–2551. doi: 10.1038/sj.jid.5700896. [DOI] [PubMed] [Google Scholar]
- 20.Wolf N., Quaranta M., Prescott N.J., Allen M., Smith R., Burden A.D., Worthington J., Griffiths C.E., Mathew C.G., Barker J.N., et al. Psoriasis is associated with pleiotropic susceptibility loci identified in Type II Diabetes and Crohn disease. J. Med. Genet. 2008;45:114–116. doi: 10.1136/jmg.2007.053595. [DOI] [PubMed] [Google Scholar]
- 21.Chang M., Li Y., Yan C., Callis-Duffin K.P., Matsunami N., Garcia V.E., Cargill M., Civello D., Bui N., Catanese J.J., et al. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun. 2008;9:176–181. doi: 10.1038/sj.gene.6364451. [DOI] [PubMed] [Google Scholar]
- 22.Valdes A.M., Thomson G. Detecting disease-predisposing variants: the haplotype method. Am. J. Hum. Genet. 1997;60:703–716. [PMC free article] [PubMed] [Google Scholar]
- 23.Heinzmann A., Mao X.Q., Akaiwa M., Kreomer R.T., Gao P.S., Ohshima K., Umeshita R., Abe Y., Braun S., Yamashita T., et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum. Mol. Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 24.Howard T.D., Whittaker P.A., Zaiman A.L., Koppelman G.H., Xu J., Hanley M.T., Meyers D.A., Postma D.S., Bleecker E.R. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am. J. Respir. Cell Mol. Biol. 2001;25:377–384. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 25.Postma D.S., Bleecker E.R., Amelung P.J., Holroyd K.J., Xu J., Panhuysen C.I., Meyers D.A., Levitt R.C. Genetic susceptibility to asthma–bronchial hyperresponsiveness coinherited with a major gene for atopy. N. Engl. J. Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 26.Lee F.I., Bellary S.V., Francis C. Increased occurrence of psoriasis in patients with Crohn's disease and their relatives. Am. J. Gastroenterol. 1990;85:962–963. [PubMed] [Google Scholar]
- 27.Peltekova V.D., Wintle R.F., Rubin L.A., Amos C.I., Huang Q., Gu X., Newman B., Van Oene M., Cescon D., Greenberg G., et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat. Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 28.Silverberg M.S. OCTNs: will the real IBD5 gene please stand up? World J. Gastroenterol. 2006;12:3678–3681. doi: 10.3748/wjg.v12.i23.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverberg M.S., Duerr R.H., Brant S.R., Bromfield G., Datta L.W., Jani N., Kane S.V., Rotter J.I., Philip Schumm L., Hillary Steinhart A., et al. Refined genomic localization and ethnic differences observed for the IBD5 association with Crohn's disease. Eur. J. Hum. Genet. 2007;15:328–335. doi: 10.1038/sj.ejhg.5201756. [DOI] [PubMed] [Google Scholar]
- 30.Tsunemi Y., Saeki H., Nakamura K., Sekiya T., Hirai K., Kakinuma T., Fujita H., Asano N., Tanida Y., Wakugawa M., et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J. Dermatol. Sci. 2002;30:100–107. doi: 10.1016/s0923-1811(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 31.Gregersen P.K., Lee H.S., Batliwalla F., Begovich A.B. PTPN22: setting thresholds for autoimmunity. Semin. Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Yamada R., Ymamoto K. Recent findings on genes associated with inflammatory disease. Mutat. Res. 2005;573:136–151. doi: 10.1016/j.mrfmmm.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 33.Mohrs M., Blankespoor C.M., Wang Z., Loots G.G., Afzal V., Hadeiba H., Shinkai K., Rubin E.M., Locksley R.M. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 34.Fanciulli M., Norsworthy P.J., Petretto E., Dong R., Harper L., Kamesh L., Heward J.M., Gough S.C., de Smith A., Blakemore A.I., et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., McLeod R.S., Griffiths A.M., Green T., Brettin T.S., Stone V., Bull S.B., et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am. J. Hum. Genet. 2000;66:1863–1870. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rioux J.D., Daly M.J., Silverberg M.S., Lindblad K., Steinhart H., Cohen Z., Delmonte T., Kocher K., Miller K., Guschwan S., et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat. Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 37.Samuelsson L., Enlund F., Torinsson A., Yhr M., Inerot A., Enerback C., Wahlstrom J., Swanbeck G., Martinsson T. A genome-wide search for genes predisposing to familial psoriasis by using a stratification approach. Hum. Genet. 1999;105:523–529. doi: 10.1007/s004399900182. [DOI] [PubMed] [Google Scholar]
- 38.Friberg C., Bjorck K., Nilsson S., Inerot A., Wahlstrom J., Samuelsson L. Analysis of chromosome 5q31–32 and psoriasis: confirmation of a susceptibility locus but no association with SNPs within SLC22A4 and SLC22A5. J. Invest. Dermatol. 2006;126:998–1002. doi: 10.1038/sj.jid.5700194. [DOI] [PubMed] [Google Scholar]
- 39.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 40.Germer S., Holland M.J., Higuchi R. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 2000;10:258–266. doi: 10.1101/gr.10.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Rowland C., Catanese J., Morris J.C., Lovestone S., O'Donovan M.C., Goate A., Owen M., Williams J., Grupe A. SORL1 variants and risk of late-onset Alzheimer's disease. Neurobiol. Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.