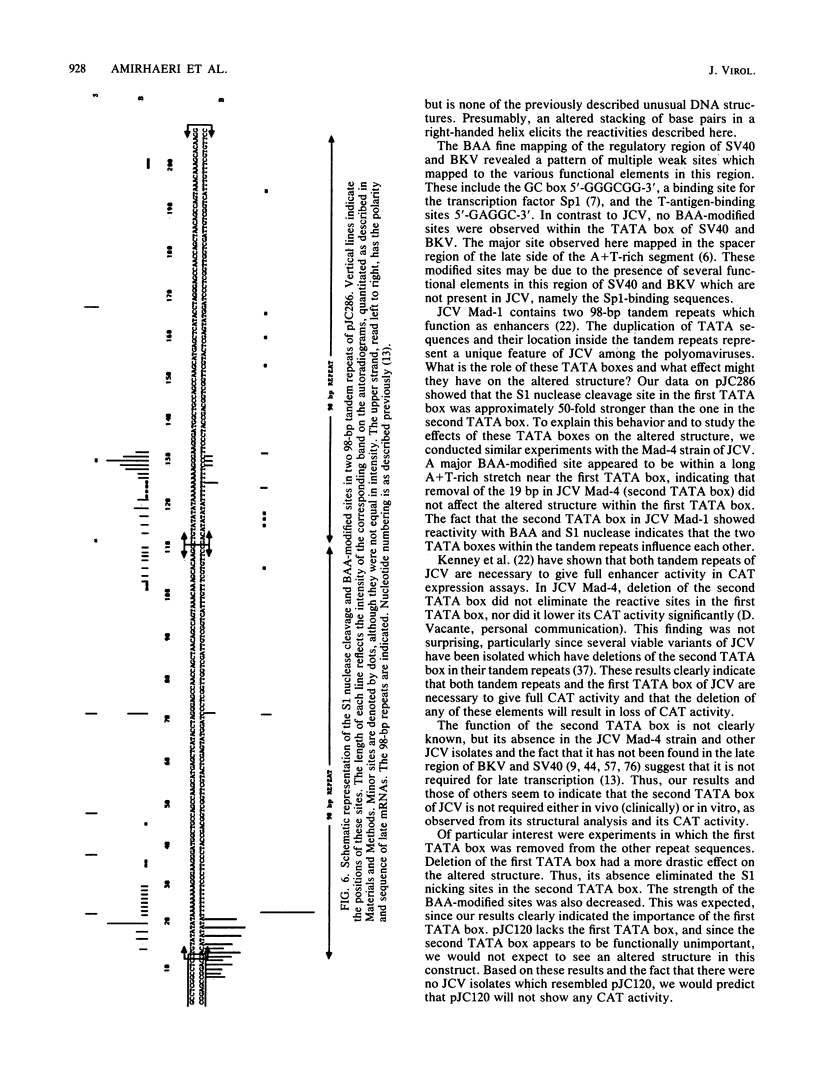
Abstract
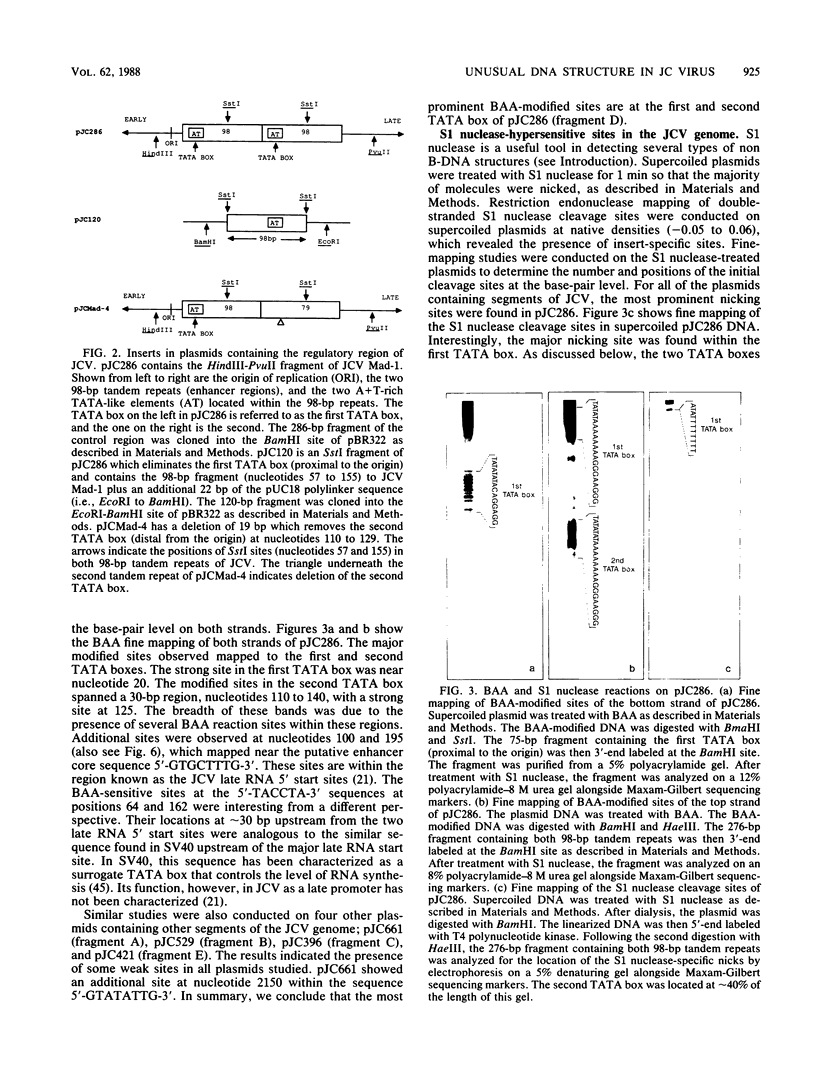
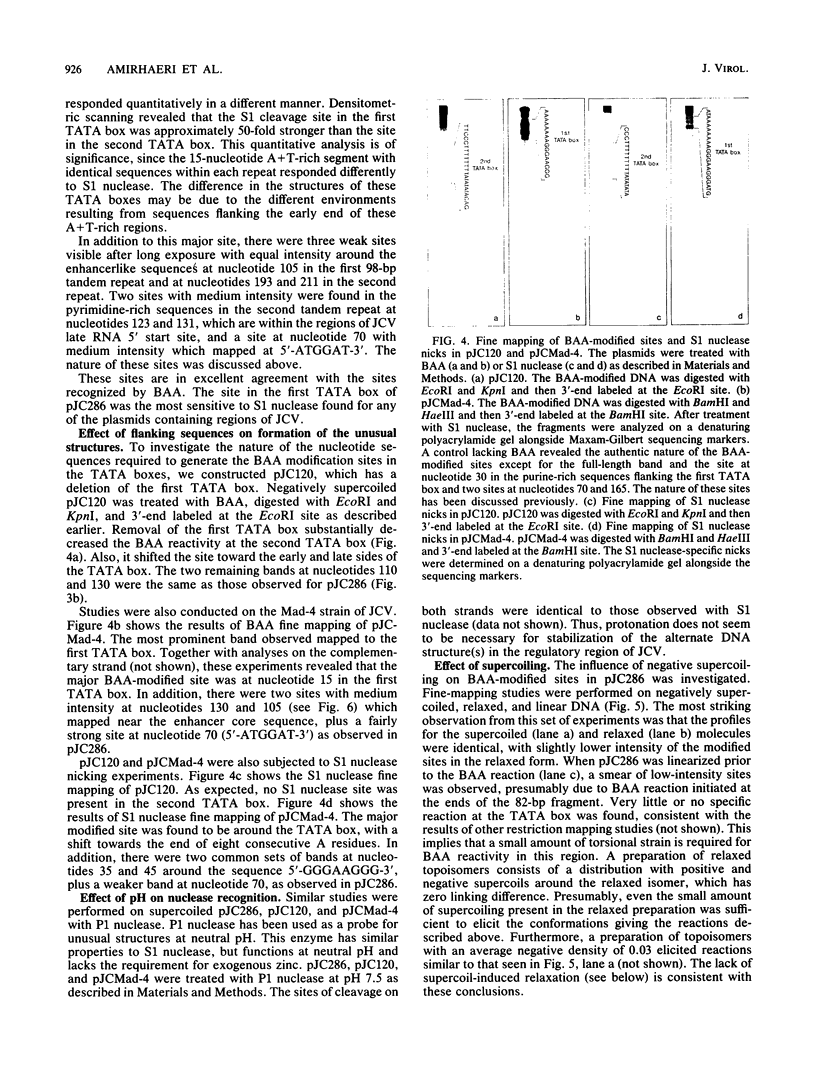
The human papovavirus JC virus (JCV) was analyzed for the presence of unusual DNA conformations. Recombinant plasmids containing 60% of the JCV prototype Mad-1 strain DNA were constructed and analyzed with both enzymatic and chemical probes. Fine-mapping studies revealed that the most prominent S1 nuclease-sensitive and bromoacetaldehyde-modified sites were located within the TATA boxes of each 98-base-pair tandem repeat. Further studies revealed that the S1 nuclease-sensitive site in the first TATA box (proximal to the origin) was approximately 50-fold stronger than the site in the second TATA box (distal from the origin). Deletion of the first TATA box drastically reduced the extent of bromoacetaldehyde modification in the second TATA box, whereas deletion of the second TATA box had little or no effect on the reactivity at the first TATA box. Hence, the biological and conformational role of the second TATA box remains unclear. No supercoil-induced relaxation was found, and reactions with the probes were not pH dependent. Also, fragments containing this regulatory region did not appear to be bent, although the A+T-rich segment contained a tract of eight consecutive A's. We conclude that the regulatory region of JCV contains non-B, but right-handed, DNA conformations which account for this behavior.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C., Walker J. K., Wang M. DNA secondary structures: helices, wrinkles, and junctions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):53–65. doi: 10.1101/sqb.1983.047.01.008. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Koff A., Tsui S., Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986 Dec;6(12):4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Evans T., Schon E., Gora-Maslak G., Patterson J., Efstratiadis A. S1-hypersensitive sites in eukaryotic promoter regions. Nucleic Acids Res. 1984 Nov 12;12(21):8043–8058. doi: 10.1093/nar/12.21.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Fowler R. F., Skinner D. M. Eukaryotic DNA diverges at a long and complex pyrimidine.purine tract that can adopt altered conformations. J Biol Chem. 1986 Jul 5;261(19):8994–9001. [PubMed] [Google Scholar]
- Frisque R. J., Bream G. L., Cannella M. T. Human polyomavirus JC virus genome. J Virol. 1984 Aug;51(2):458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque R. J. Nucleotide sequence of the region encompassing the JC virus origin of DNA replication. J Virol. 1983 Apr;46(1):170–176. doi: 10.1128/jvi.46.1.170-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque R. J. Regulatory sequences and virus-cell interactions of JC virus. Prog Clin Biol Res. 1983;105:41–59. [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. S., Wells R. D. B-Z DNA junctions contain few, if any, nonpaired bases at physiological superhelical densities. J Biol Chem. 1985 Jun 25;260(12):7783–7790. [PubMed] [Google Scholar]
- Kenney S., Natarajan V., Salzman N. P. Mapping 5' termini of JC virus late RNA. J Virol. 1986 Apr;58(1):216–219. doi: 10.1128/jvi.58.1.216-219.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney S., Natarajan V., Strike D., Khoury G., Salzman N. P. JC virus enhancer-promoter active in human brain cells. Science. 1984 Dec 14;226(4680):1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kilpatrick M. W., Klysik J., Singleton C. K., Zarling D. A., Jovin T. M., Hanau L. H., Erlanger B. F., Wells R. D. Intervening sequences in human fetal globin genes adopt left-handed Z helices. J Biol Chem. 1984 Jun 10;259(11):7268–7274. [PubMed] [Google Scholar]
- Kilpatrick M. W., Torri A., Kang D. S., Engler J. A., Wells R. D. Unusual DNA structures in the adenovirus genome. J Biol Chem. 1986 Aug 25;261(24):11350–11354. [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. F., Martin J. D., Takemoto K. K., Howley P. M. The colinear alignment of the genomes of papovaviruses JC, BK, and SV40. Virology. 1979 Jul 30;96(2):576–587. doi: 10.1016/0042-6822(79)90113-2. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Structural perturbation in supercoiled DNA: hypersensitivity to modification by a single-strand-selective chemical reagent conferred by inverted repeat sequences. Nucleic Acids Res. 1983 May 25;11(10):3097–3112. doi: 10.1093/nar/11.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London W. T., Houff S. A., Madden D. L., Fuccillo D. A., Gravell M., Wallen W. C., Palmer A. E., Sever J. L., Padgett B. L., Walker D. L. Brain tumors in owl monkeys inoculated with a human polyomavirus (JC virus). Science. 1978 Sep 29;201(4362):1246–1249. doi: 10.1126/science.211583. [DOI] [PubMed] [Google Scholar]
- Major E. O., Miller A. E., Mourrain P., Traub R. G., de Widt E., Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl C., Walker D. L., Frisque R. J. Derivation and characterization of POJ cells, transformed human fetal glial cells that retain their permissivity for JC virus. J Virol. 1987 Mar;61(3):755–763. doi: 10.1128/jvi.61.3.755-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. D., King D. M., Slauch J. M., Frisque R. J. Differences in regulatory sequences of naturally occurring JC virus variants. J Virol. 1985 Jan;53(1):306–311. doi: 10.1128/jvi.53.1.306-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. D., Padgett B. L., Walker D. L. Characterization of tissue culture-induced heterogeneity in DNAs of independent isolates of JC virus. J Gen Virol. 1983 Oct;64(Pt 10):2271–2280. doi: 10.1099/0022-1317-64-10-2271. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKeon C., Schmidt A., de Crombrugghe B. A sequence conserved in both the chicken and mouse alpha 2(I) collagen promoter contains sites sensitive to S1 nuclease. J Biol Chem. 1984 May 25;259(10):6636–6640. [PubMed] [Google Scholar]
- McLean M. J., Blaho J. A., Kilpatrick M. W., Wells R. D. Consecutive A X T pairs can adopt a left-handed DNA structure. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5884–5888. doi: 10.1073/pnas.83.16.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Larson J. E., Wohlrab F., Wells R. D. Reaction conditions affect the specificity of bromoacetaldehyde as a probe for DNA cruciforms and B-Z junctions. Nucleic Acids Res. 1987 Sep 11;15(17):6917–6935. doi: 10.1093/nar/15.17.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N. R., Major E. O., Wallen W. C. Transfection of human fetal glial cells with molecularly cloned JCV DNA. Prog Clin Biol Res. 1983;105:29–40. [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B. C., Raphael A. L., Barton J. K. Evidence for altered DNA conformations in the simian virus 40 genome: site-specific DNA cleavage by the chiral complex lambda-tris(4,7-diphenyl-1,10-phenanthroline)cobalt(III). Proc Natl Acad Sci U S A. 1987 Apr;84(7):1764–1768. doi: 10.1073/pnas.84.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A., Das G., Salzman N. P. Characterization of a surrogate TATA box promoter that regulates in vitro transcription of the simian virus 40 major late gene. Mol Cell Biol. 1985 Mar;5(3):591–594. doi: 10.1128/mcb.5.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Herrera R. E., Rich A. Binding of anti-Z-DNA antibodies to negatively supercoiled SV40 DNA. Nucleic Acids Res. 1987 Feb 25;15(4):1661–1677. doi: 10.1093/nar/15.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett B. L., Rogers C. M., Walker D. L. JC virus, a human polyomavirus associated with progressive multifocal leukoencephalopathy: additional biological characteristics and antigenic relationships. Infect Immun. 1977 Feb;15(2):656–662. doi: 10.1128/iai.15.2.656-662.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett B. L., Walker D. L., ZuRhein G. M., Hodach A. E., Chou S. M. JC Papovavirus in progressive multifocal leukoencephalopathy. J Infect Dis. 1976 Jun;133(6):686–690. doi: 10.1093/infdis/133.6.686. [DOI] [PubMed] [Google Scholar]
- Padgett B. L., Walker D. L., ZuRhein G. M., Varakis J. N. Differential neurooncogenicity of strains of JC virus, a human polyoma virus, in newborn Syrian hamsters. Cancer Res. 1977 Mar;37(3):718–720. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Reynolds V. L., Molineux I. J., Kaplan D. J., Swenson D. H., Hurley L. H. Reaction of the antitumor antibiotic CC-1065 with DNA. Location of the site of thermally induced strand breakage and analysis of DNA sequence specificity. Biochemistry. 1985 Oct 22;24(22):6228–6237. doi: 10.1021/bi00343a029. [DOI] [PubMed] [Google Scholar]
- Rich A. Right-handed and left-handed DNA: conformational information in genetic material. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):1–12. doi: 10.1101/sqb.1983.047.01.003. [DOI] [PubMed] [Google Scholar]
- Rieth K. G., Di Chiro G., London W. T., Sever J. L., Houff S. A., Kornblith P. L., McKeever P. E., Buonomo C., Padgett B. L., Walker D. L. Experimental glioma in primates: a computed tomography model. J Comput Assist Tomogr. 1980 Jun;4(3):285–290. doi: 10.1097/00004728-198006000-00001. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. The genome of human papovavirus BKV. Cell. 1979 Dec;18(4):963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Wells R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 May;80(9):2447–2451. doi: 10.1073/pnas.80.9.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. Relationship between superhelical density and cruciform formation in plasmid pVH51. J Biol Chem. 1982 Jun 10;257(11):6292–6295. [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Small J. A., Scangos G. A., Cork L., Jay G., Khoury G. The early region of human papovavirus JC induces dysmyelination in transgenic mice. Cell. 1986 Jul 4;46(1):13–18. doi: 10.1016/0092-8674(86)90855-x. [DOI] [PubMed] [Google Scholar]
- Tenen D. G., Haines L. L., Hansen U. M., Martin R. G., Livingston D. M. Formation of a cruciform structure at the simian virus 40 replication origin abolishes T-antigen binding to the origin in vitro. J Virol. 1985 Oct;56(1):293–297. doi: 10.1128/jvi.56.1.293-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Walker D. L., Padgett B. L., ZuRhein G. M., Albert A. E., Marsh R. F. Human papovavirus (JC): induction of brain tumors in hamsters. Science. 1973 Aug 17;181(4100):674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Brennan R., Chapman K. A., Goodman T. C., Hart P. A., Hillen W., Kellogg D. R., Kilpatrick M. W., Klein R. D., Klysik J. Left-handed DNA helices, supercoiling, and the B-Z junction. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):77–84. doi: 10.1101/sqb.1983.047.01.010. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- Wohlrab F., McLean M. J., Wells R. D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987 May 5;262(13):6407–6416. [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Wu R. BK virus DNA: complete nucleotide sequence of a human tumor virus. Science. 1979 Oct 26;206(4417):456–462. doi: 10.1126/science.228391. [DOI] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell. 1986 Jun 6;45(5):743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985 Dec 16;4(13A):3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]