Abstract
Embryos express several unique differentiation characteristics, including the accumulation of a number of metabolites that are generally considered to be unique to seeds. PICKLE (PKL) codes for a CHD3-chromatin remodeling factor that is necessary for repression of embryonic traits in seedlings of Arabidopsis thaliana. In pkl mutants, primary roots are capable of expressing many embryonic traits after germination and are referred to as “pickle roots”. In an attempt to examine the breadth of PKL-dependent repression of embryo-specific differentiation pathways, we determined the extent to which a variety of embryo-specific compounds accumulate in pickle roots. We found that pickle roots accumulate triacylglycerol (TAG) with a fatty acid composition that is similar to that found in seeds. The major seed storage proteins are also present in pickle roots. In addition to these two well-characterized seed storage compounds, we observed that pickle roots accumulate phytate, a form of stored phosphate that is preferentially accumulated in seeds. Seeds of members of the Brassicaceae also accumulate a variety of unique secondary metabolites, including sinapate esters and glucosinolates. Surprisingly, the levels of secondary metabolites in pickle roots were not suggestive of an embryonic differentiation state, but did reveal that a mutation in PKL results in substantial changes in root secondary metabolism. Taken together, these data suggest that PKL is responsible for regulating some but not all aspects of the embryonic program as it relates to the accumulation of embryo-specific metabolites.
Keywords: developmental transition, CHD3, embryo, lipid, protein, phytate, secondary metabolism
INTRODUCTION
The seeds of plants accumulate nutrients reserves during the maturation stage of development, and many of these compounds are found only in seeds. During germination, an imbibed seed undergoes a programmed developmental transformation toward vegetative growth and mobilizes its storage reserves (Mansfield and Briarty, 1992, 1996; Bewley, 1997; Holdsworth et al., 1999). The signals to initiate or to stop accumulating storage reserves remain poorly characterized. Similarly, unknown mechanisms restrict the accumulation of many of these same nutrient reserves to seeds. A number of mutations have been identified in Arabidopsis that generally affect the accumulation of storage reserves. For example, mutants that are defective in abscisic acid biosynthesis or response display dramatic reductions in seed storage reserves (Koornneef et al., 1989; McCarty, 1995; Parcy et al., 1997). Other mutations affect the accumulation of specific reserve materials present in mature embryos. In both triacylglycerol biosynthesis defect1 (tag1) and wrinkled1 (wri1) mutants, for example, triacylglycerol accumulation is specifically reduced (Katavic et al., 1995; Focks and Benning, 1998; Jako et al., 2001). Mobilization of these lipid reserves has been shown to play a key role in germination in Arabidopsis, and as a result, wri seeds exhibit a germination defect (Focks and Benning, 1998). In the comatose (cts) mutant, seed storage lipids accumulate normally but fail to be mobilized during germination resulting in a defect in the transition to vegetative growth (Russell et al., 2000; Footitt et al., 2002).
PICKLE (PKL) codes for a CHD3-chromatin remodeling factor that is necessary for repressing embryonic traits in germinating seedlings, including accumulation of storage reserves (Ogas et al., 1997; Ogas et al., 1999). CHD3 proteins are found throughout eukaryotes and act as negative regulators of transcription that play specific roles in development (Ahringer, 2000). Current data indicates that the ability of PKL to repress embryonic traits in Arabidopsis is also mediated through transcriptional regulation. PKL is necessary for the repression of the LEC class of master regulators of embryonic identity: LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON2 (LEC2) and FUSCA3 (FUS3) (Ogas et al., 1999; Rider et al., 2003). Null mutations in the LEC genes result in cotyledons that display leaf-like characteristics, as well as various defects in seed development including reduced accumulation of storage reserves (Meinke, 1992; Keith et al., 1994; Meinke et al., 1994; West et al., 1994; Parcy et al., 1997). Furthermore, overexpression of LEC1 or LEC2 results in the inappropriate expression of embryonic characteristics in vegetative tissues, including the production of somatic embryos from vegetative tissues of germinating seeds (Lotan et al., 1998; Stone et al., 2001). In germinating pkl seedlings, the transcript level of LEC1 and LEC2 is increased substantially over that found in wild-type seedlings (Ogas et al., 1999; Rider et al., 2003). In pkl primary roots that continue to express embryonic traits after germination (so-called “pickle roots”), LEC1, LEC2 and FUS3 are all expressed at levels more than 100-fold higher than found in wild-type roots (Rider et al. 2003). Thus, PKL is necessary for the repression of genes that play central roles in promoting embryonic identity.
Previous analyses suggested that pickle roots were likely to accumulate storage reserves in an embryo-specific manner. Pickle roots accumulate neutral lipids, as determined by Fat Red 7B staining, and transcripts for an oleosin and the seed storage protein 2S1 are abundant in pickle root tissue. Furthermore, electron micrographs of pickle root cells reveal the presence of structures that strongly resemble the oil bodies found in seeds. These observations indicate that in the absence of PKL-dependent repression, primary roots are capable of producing and storing compounds that are normally found in embryos.
To further investigate the extent to which embryo-specific differentiation traits are expressed in pickle roots, we examined pickle root tissue for the presence of several compounds known to accumulate in embryos. We found that pickle root tissue accumulates several compounds specifically found in seeds, including triacylglycerol containing long chain fatty acids, seed storage proteins, and phytate. In contrast, we found that pickle roots do not accumulate secondary metabolites that are typically found in seeds. Instead, we observed that the roots of pkl plants contain a number of secondary metabolites that are not normally found in embryos or in roots. Thus although PKL acts as a master regulator of the LEC genes, not all embryo-specific traits are derepressed in pickle roots. In addition, we have identified a new role for PKL in the regulation of secondary metabolite production in roots.
MATERIALS AND METHODS
Plant material
Seeds and tissues from the Arabidopsis pkl mutant (in a Columbia ecotype background) and wild-type Columbia were used for all investigations. Plants were grown essentially as described previously (Ogas et al., 1997). Seeds were surface sterilized and plated onto synthetic solid media (Murashige & Skoog media supplemented with Gamborg's vitamins, 1 μg/ml glycine, 100 μg/ml myo-inositol and 1% sucrose at a pH of 5.6). Seeding density was 144 seeds per 4 cm × 4 cm square plate. Seeds were stratified at 4° C in the dark for 4 days and transferred to a CU36L incubator (Percival Scientific Inc., Perry, IA) under constant illumination (85−100 μE m−2 s−1). Seedlings were grown for 10−14 days prior to harvest.
Lipids
For triacylglycerol identification, 30 mg of tissue was homogenized in 2 mL of chloroform:methanol (1:1). This mixture was then heated at 60°C for 1 minute and then extracted twice with 2 mL of 1 M NaCl/0.1 M HCl. The chloroform (bottom) phase was then dried down under nitrogen and the remaining material was then redissolved in 100 μL of chloroform. Twenty μL of this sample was then separated on a silica TLC plate developed with hexane:diethylether:formic acid (80:20:2). Four standards were run in one lane as a control and included 20 μg each of cetyl alcohol, TAG (18:1/16:0/18:1), 16:0 aldehyde, and palmitic palmityl ester. Lipids were visualized by staining with iodine.
To isolate the TAG fraction from pickle roots for fatty acid methyl ester (FAME) analysis, the TAG spot was scraped off of the plate and vortexed into 2 ml of chloroform:methanol:acetic acid (50:50:1). Debris was removed by centrifugation, the supernatant was dried down under nitrogen, and the remaining material was then dissolved in 1 mL of 1 N methanolic HCl. Preparation of methyl esters from this sample and others and the subsequent gas chromatographic analysis of the resulting extracts was performed using established procedures (Browse et al., 1986) with a Hewlett-Packard 5800 gas chromatograph equipped with a Supelco SP2330 glass capillary column (0.75 mm × 20 m).
Proteins
An analysis of seed or pickle root protein was made using denaturing polyacrylamide gel electrophoresis (Laemmli, 1970). Crude protein extraction was performed using a modified form of the EZ protein extraction method (Martinez-Garcia et al., 1999). Protein from seeds or roots was extracted by homogenizing tissue in buffer E (125 mM Tris pH 8.8, 1% (w/v) SDS, 10% (v/v) glycerol, 50 mM Na2S2O5) containing 1% protease inhibitor cocktail (Sigma, MO). Debris was removed by centrifugation at 10,000 × g for 10 min, followed by the addition of 1/10 volume of buffer Z (125 mM Tris pH 6.8, 12% (w/v) SDS, 10% (v/v) glycerol, 22% β-mercaptoethanol, 0.001% (w/v) bromophenol blue). The samples were then heated at 95°C for 5 min. Protein was quantified using the RC DC assay from BioRad (Hercules, CA). Twenty μg of each sample was electrophoresed in a 15% SDS-polyacrylamide gel containing 0.1% SDS and subsequently stained with coomassie brilliant blue R250.
Phytate
The concentration of free and phytate-bound phosphate present in seeds or roots was measured by spectrometry as described by (Heinonen and Lahti, 1981). Samples were homogenized in 100 μL of 10 mM Tris buffer, pH 8.0, mixed with 90 μL of deionized water, incubated at 37°C in the presence of phytase enzyme (40U Natuphos® in 10 μL; BASF Inc., AG Ludwigshafen, Germany) and stopped at 0 or 30 minutes by the addition of 100 μl of 30% TCA. For color development, the resulting solutions were mixed with 900 μl of acetone-acid molybdate (2.5 mM ammonium molybdate, 50% acetone, 25% sulfuric acid) and 100 μl of 1 M citric acid. Wild-type and pkl seeds were examined as well as root tissues of wild-type and pkl plants. Triplicate biological samples were examined for each tissue for each time point. Phosphorous levels were deduced using absorbance readings at 355nm of samples and the equation of the best-fit line derived from a standard curve. Phytate derived phosphorous was determined as the difference in average detectable phosphorous between samples from the two time points.
Transmission Electron Microscopy
Transmission electron microscopy was performed as described previously (Kolosova et al., 2001) using pickle roots from ten day-old pkl plants grown in the presence of uniconazole-P.
Sinapate Ester and Glucosinolate Analysis
For analysis of soluble secondary metabolites, sinapate esters and glucosinolates were analyzed and quantified as described previously (Hemm et al., 2003).
Quantitative RT-PCR
For analysis of cruciferin (12s seed storage protein) transcript levels in pickle and wild type roots, quantitative RT-PCR was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously (Hemm et al., 2003). Relative abundance of transcript was determined using 18S as a normalization control. PCR primers for 18S were GGTCTGTGATGCCCTTAGATGTT, and GGCAAGGTGTGAACTCGTTGA. Primer sequences for cruciferin (At1g03880) were GTGCCAGCTCGATCAACTCA, and CGACCACCCTCGCTCTTG.
RESULTS
Lipid
Pickle roots stain intensely with Fat Red 7B, a dye that binds neutral lipids such as triacylglycerols (Ogas et al., 1997). To determine whether the staining of pickle roots was indeed the result of triacylglycerol accumulation, lipids were extracted from wild-type and pickle roots and analyzed by TLC (Figure 1A). Lipid extracts from pickle roots (lane 2) clearly contained a fraction that co-migrates with the triacylglycerol standard (lane 3), whereas this fraction was not detected in lipid samples from an equivalent amount of wild-type roots (lane 1).
Figure 1.
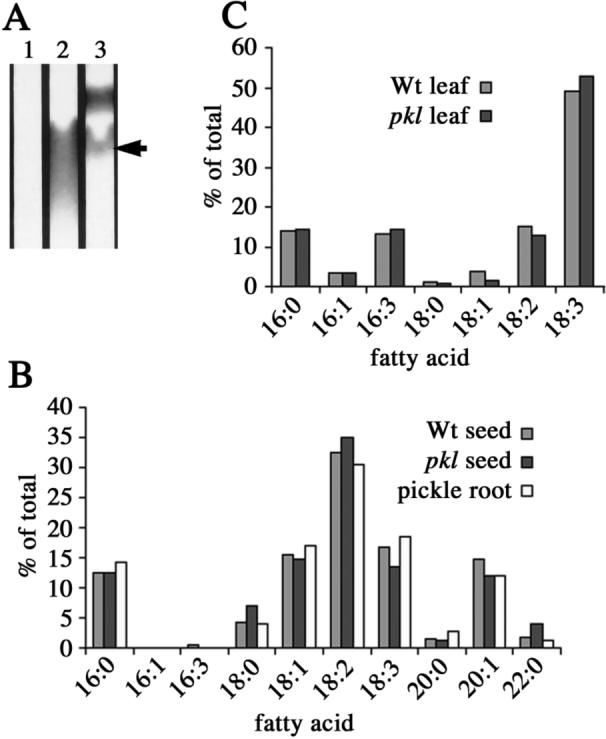
Pickle roots contain triacylglycerols with a fatty acid composition similar to that of seeds. A) Lipid extracts were made from equivalent amounts of wild-type (lane 1) and pickle roots (lane 2) and analyzed by TLC. A portion of the TLC plate is shown. The position of the triacylglycerol standard (lane 3) is indicated by an arrow. B) The fatty acid composition of wild-type seeds, pkl seeds, and the triacylglycerol fraction from pickle roots as determined by GC. C) The fatty acid composition of wild-type and pkl leaves as determined by GC.
The fatty acid composition of seed triacylglycerols is distinct from that of leaf-derived lipids (Ohlrogge et al., 1991). We analyzed the fatty acid composition of the TLC-purified pickle root triacylglycerol fraction by gas chromatography to determine if the profile of fatty acids was similar to that of seed-derived lipids. We analyzed the fatty acid composition of wild-type seeds, pkl seeds, and triacylglycerols extracted from pickle roots (Figure 1B). The fatty acid content of pickle root triacylglycerols was remarkably similar to that of seeds. In particular, 16:1 and 16:3 fatty acids were not detected in the pickle root triacylglycerols, 18:2 was the most abundant fatty acid rather than 18:3, and 20- and 22-carbon fatty acids were found in the pickle root triacylglycerols in proportions that were very similar to that of seed-derived lipids.
The presence of triacylglycerol in pickle roots with a fatty acid composition that is characteristic of seeds is unlikely to reflect a general alteration in lipid metabolism in pkl plants. The fatty acid composition of pkl seeds is indistinguishable from that of wild-type seeds (Figure 1B). To determine if a mutation in PKL altered the fatty acid composition of non-embryonic tissues, we analyzed the fatty acid composition of wild-type leaves and pkl leaves (Figure 1C). In contrast to the dramatic effect on lipid composition in pickle roots, we observed that the pkl mutation does not result in a substantive alteration in the fatty acid content of leaves. In particular, we did not observe the presence of long chain fatty acids typical of those found in seeds. This observation is consistent with previous observations that inappropriate expression of embryonic traits in pkl seedlings is only observed in those organs that are produced during embryogenesis (Ogas et al., 1997; Henderson et al., 2004).
Protein
In Arabidopsis, cells of the mature embryo contain protein bodies (Mansfield and Briarty, 1992). Seed storage proteins are rich in nitrogen and are presumed to represent a form of nitrogen storage for use by developing seedlings (Muntz, 1998). A majority of the soluble protein present in Arabidopsis seed is represented by a limited number of seed storage proteins that can be visualized as distinct bands on polyacrylamide gels (Finkelstein and Somerville, 1990; Fujiwara et al., 2002).
Transcripts for the gene encoding the 2S1 seed storage protein are present in pickle root tissue but not in wild-type roots (Ogas et al., 1997). To determine if this level of gene expression results in the accumulation of seed storage proteins, we examined the content of crude protein extracts from pickle root tissue by denaturing polyacrylamide gel electrophoresis. The major seed storage proteins are easily visualized in protein extracts from wild-type and pkl seeds (lanes 1−2). Consistent with the analysis of transcript levels, we observed that extracts from pickle roots clearly gave rise to bands that co-migrated with the 2S seed storage proteins (lane 4), whereas extracts from wild-type roots did not (lane 3). In addition, extracts from pickle roots appeared to give rise to bands that co-migrated with 12S seed storage proteins, although the presence of multiple bands in extracts from wild-type roots that migrate at similar molecular weights makes this observation less obvious than the 2S seed storage proteins. In agreement with this observation, examination of pickle roots by transmission electron microscopy revealed the presence of structures that resemble the protein bodies of seeds (Figure 4). These data thus indicate that pickle roots do accumulate seed storage proteins. Finally, we also examined the relative transcript level of cruciferin, a 12S seed storage protein, by quantitative RT-PCR in wild-type and pickle roots. We found that12S transcript levels were several thousand times higher in pickle root tissues than in wild-type root tissues (>20,452 fold; average of 3 replicates).
Figure 4.
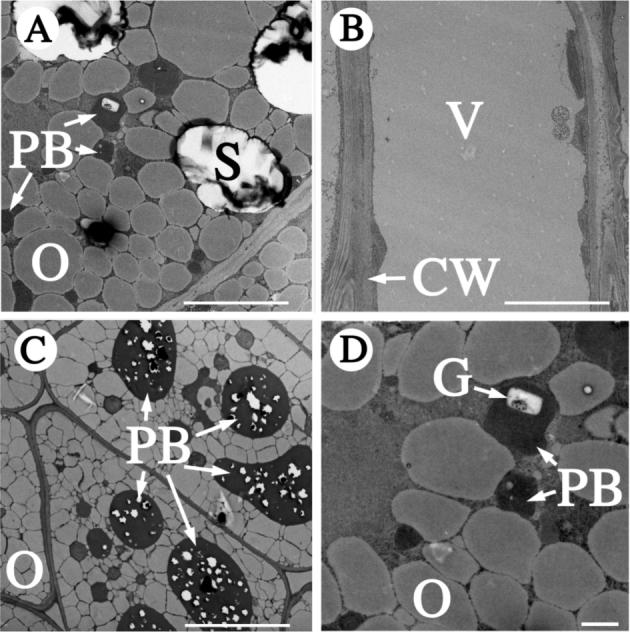
Pickle roots contain protein bodies and phytin globoids. The subcellular organization of a pickle root cell (panels A and D) was examined by transmission electron microscopy. Images from a wild-type root (B) and wild-type seed (C) are included for comparison. The larger protein bodies of seeds (C) contain numerous phytin globoids (white inclusions) that are not specifically marked as such. CW, cell wall; G, phytin globoid; O, oil body; PB, protein body; S, starch granule; V, vacuole. The white scale bars represent 10 μm in A, B and C or 1 μm in panel D.
Although our analyses indicate that pickle roots accumulate seed storage proteins, it is worth noting that our data also reveal that the overall protein composition of pickle roots is significantly more complex than that of seeds as indicated by the number of bands observed in the crude extracts. This observation is consistent with other observations indicating that pickle roots express both embryo and seedling differentiation traits (Ogas et al., 1997).
Phytate
In addition to triacylglycerol and storage proteins, plant embryos accumulate various other compounds and mineral nutrients (Bewley and Black, 1994). Phosphorous is an essential nutrient for plants and is stored within seeds as an organic compound called phytate (myo-inositol hexakisphosphate). Phytate is sequestered in the protein bodies of Arabidopsis seeds as phytin globoids - a complex mixture of proteins, various cations, and phytate (Mansfield and Briarty, 1992). The mineral composition of globoids in Arabidopsis seed protein bodies differs between tissues, but contains relatively high levels of P, Ca, K, and Mg with lower amounts of Fe, Mn and Zn (Lott and West, 2001). Given that seed storage proteins are present in pickle roots (Figure 2), we attempted to determine if phytate was also present in pickle roots.
Figure 2.
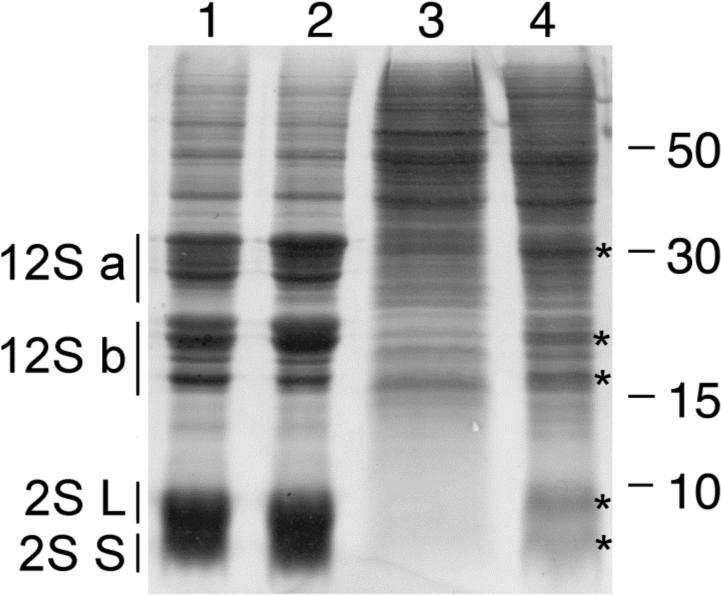
Proteins that co-migrate with seed storage proteins are present in pickle roots. Proteins were extracted from wild-type seed (lane 1), pkl seed (lane 2), wild-type roots (lane 3), and pickle roots (lane 4) and subjected to SDS-PAGE. Positions of the acidic (12S-A) and basic (12S-B) subunits of 12S globulin and the large (L) and small (S) subunits of 2S albumin from seeds are indicated to the left. Positions of proteins isolated from pickle roots that co-migrate with seed storage proteins are marked with an asterisk (*) to the right of the band. 20 μg of protein were loaded per lane. The size (in kDa) of protein molecular weight standards is indicated to the right of the gel.
To this end, we examined seeds and roots for phytate-bound phosphorous using phytase to enzymatically remove phosphate from phytate. We then quantified the inorganic phosphate using a colorimetric assay. Using this assay, we found that the level of phosphate that can be enzymatically removed from phytate by phytase accounted for approximately 60% of the detectable phosphate present in our pkl and wild-type seed extracts (Figure 3A). We also found that the relative abundance of phosphate that can be liberated from pickle root extracts via phytase is similar to that found in seeds (53%). Similar analysis of wild-type root extracts did not reveal the presence of phytate, indicating that phytate-bound phosphate was not abundant in wild-type root tissues (Figure 3A). This observation suggested that pickle roots are accumulating phosphorous as phytate, similar to embryos. Similarly to seed storage proteins, the amount of phytate-derived phosphate in pickle root tissue (as represented by ng/mg fresh weight) is substantially less than that found in seeds (Figure 3B).
Figure 3.
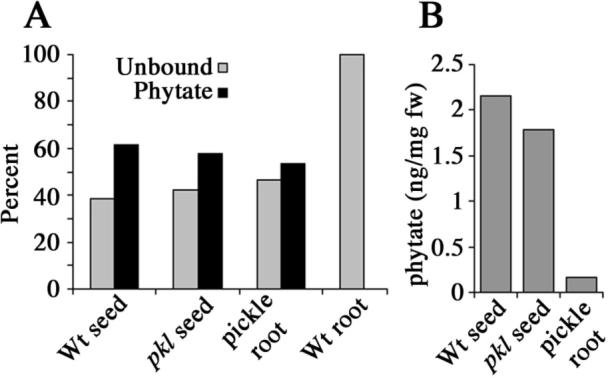
Pickle roots contain phosphorous bound in the form of phytate. A) The relative molar abundance of free and phytate-bound phosphate as determined by enzymatic analysis for seeds and roots from wild-type and pkl plants. B) Mean percent fresh weight of phytate found in wild-type seed, pkl seed, and pickle roots.
To determine whether phytin globoids, storage structures for phytate, were formed in pickle roots, we examined pickle root cortical cells by transmission electron microscopy. This analysis revealed the presence of phytin globoids within protein bodies of pickle root cortical cells (Figure 4). Considering that pickle roots also accumulate lipid in oil bodies (Ogas et al., 1997), these data demonstrate that in pickle roots, multiple seed storage materials accumulate in an organized fashion in a manner analogous to that of seeds.
Secondary metabolites
Seeds of Arabidopsis accumulate a number of characteristic secondary metabolites including specific sinapate esters and glucosinolates (Matthaus, 1998; Kliebenstein et al., 2001). Sinapoylcholine, in particular, is present in seeds and is believed to represent a stored form of choline that is subsequently used by young developing seedlings (Ruegger and Chapple, 2001). The function of seed glucosinolates is unknown, but it is likely that they are involved in plant defense and protection (Rask et al., 2000). Although representatives of these classes of secondary metabolites are present in seeds, other sinapate esters and glucosinolates are present in other tissues (Kliebenstein et al., 2001), thus their presence provides a biochemical fingerprint for the differentiation state of the tissue analyzed. Genotype and environmental conditions may also influence the accumulation of these compounds in various tissues (Kliebenstein et al., 2001; Brown et al., 2003).
To test the hypothesis that pickle roots may accumulate sinapoylcholine or other embryo-specific sinapate esters, methanolic extracts from pickle roots were analyzed by HPLC (Chapple et al., 1992; Lorenzen et al., 1996; Lehfeldt et al., 2000; Hemm et al., 2003). pkl plants were grown on synthetic media containing uniconazole-P to increase pickle root penetrance (Ogas et al., 1997) in order to provide sufficient material for these biochemical analyses. Because these growth conditions may be stressful to the plant and lead to alterations in phenylpropanoid metabolism that are not truly specific to pickle roots, the phenylpropanoid content of wild type roots grown on plates with and without uniconazole-P was examined in parallel. In addition, we examined the phenylpropanoid content of secondary roots of pkl plants grown in the presence or absence of uniconazole-P. Secondary roots of pkl plants (referred to here as pkl secondary roots to distinguish them from embryogenic “pickle roots”) do not express embryonic traits and were examined to determine if mutating PKL had an effect on secondary metabolism in the absence of expression of embryonic traits.
The results of these analyses reveal that pickle roots uniquely accumulate sinapic acid and sinapoylglucose, a sinapate ester found in both leaves and seeds but not in normal roots (Figure 5). In concentrated samples, compounds with the same retention time and spectra as sinapoylmalate and sinapoylcholine were detected in pickle roots but were absent in all other samples assayed (data not shown). These results indicate that phenylpropanoid biosynthesis is altered in pickle roots, although not in a manner that is consistent with an embryonic state of phenylpropanoid metabolism. Furthermore, pkl secondary roots hyperaccumulate other phenylpropanoid-derived compounds in comparison to wild-type roots (Figure 5). Thus, the loss of PKL results in changes in phenylpropanoid metabolism in roots in a manner that is distinct from the failure to repress embryonic traits.
Figure 5.
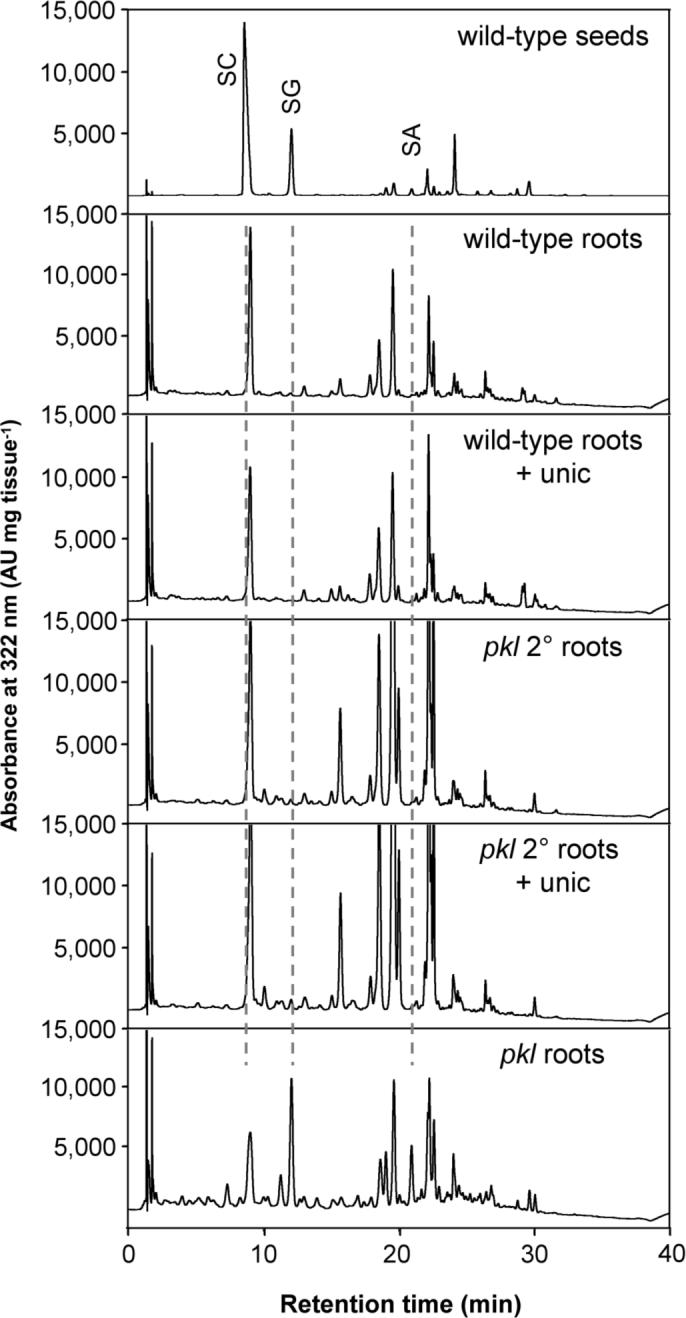
Secondary metabolism is altered in roots of pkl plants. HPLC analysis of soluble secondary metabolites that accumulate in the roots of wild-type and pkl plants grown in the presence or absence of uniconazole-P as compared to wild-type seeds. The elution of UV-absorbing compounds was monitored at 322 nm. Phenylpropanoids are identified as follows: SA, sinapic acid; SG sinapoylglucose; SC sinapoylcholine.
To determine if pickle roots accumulate embryo-specific glucosinolates, methanolic extracts of pickle roots, wild-type roots, and pkl secondary roots (grown as described above) were fractionated by anion exchange chromatography and analyzed by HPLC (Hemm et al., 2003). The most abundant glucosinolates found in wild-type roots were 4-methylthiobutyl (4MTB), indol-3-ylmethyl (I3M), 1-methoxyindol-3-ylmethyl (1MOI3M) and 4-methyoxyindol-3-ylmethyl (4MOI3M) glucosinolates (Table 1). Wild-type roots grown on uniconazole-P showed similar levels of these glucosinolates as well as a slight accumulation of the methionine-derived 8-methylsulfinyloctyl glucosinolate (8MSOO) (Table 1). In contrast, pkl secondary roots accumulated a significant amount of 8MSOO and greater levels of I3M while showing decreased levels of 4MOI3M. Growth on uniconazole-P did not significantly affect glucosinolate levels in pkl secondary roots, with the exception of decreased I3M. In comparison to these tissues, the glucosinolate content of pickle roots was most similar to that of pkl secondary roots grown on uniconazole-P, although pickle roots accumulated significantly less 4MOI3M and 1MOI3M (Table 1). Accumulation of embryo-specific glucosinolates such as 3-benzoyloxypropyl glucosinolate (3BZO) and 4-benzoyloxybutyl glucosinolate (4BZO) was not observed in pickle roots. Thus glucosinolate biosynthesis is altered in the roots of pkl plants, although pickle roots do not accumulate a profile of glucosinolates similar to that observed in embryos.
Table 1.
Glucosinolate content of wild-type and pkl roots. (n=3)
| Sample | Glucosinolate content (nmol per mg tissue ± S.E.) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4MTB |
8MSOO |
I3M |
4MOI3M |
1MOI3M |
4OHB |
4MSOB |
7MSOH |
7MTH |
3BZO |
4BZ0 |
8MTO |
|
| wt roots | 0.81 ± 0.13 | 0.11 ± 0.03 | 0.12 ± 0.02 | 0.40 ± 0.06 | 0.22 ± 0.05 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| wt roots + Unic | 0.76 ± 0.04 | 0.12 ± 0.01 | 0.14 ± 0.01 | 0.34 ± 0.01 | 0.25 ± 0.02 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| pkl 2° roots | 0.75 ± 0.09 | 0.53 ± 0.03 | 0.53 ± 0.01 | 0.13 ± 0.01 | 0.21 ± 0.03 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| pkl 2° roots + Unic | 0.62 ± 0.11 | 0.44 ± 0.03 | 0.28 ± 0.02 | 0.10 ± 0.01 | 0.22 ± 0.02 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| pickle roots + Unic | 0.46 ± 0.07 | 0.26 ± 0.01 | 0.22 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Col seeds | 178.90 ± 9.43 | 54.67 ± 1.77 | 22.08 ± 0.89 | n.d. | n.d. | 80.28 ± 2.98 | 14.10 ± 3.09 | 13.36 ± 0.45 | 61.58 ± 1.95 | 29.06 ± 1.52 | 79.23 ± 4.42 | 47.83 ± 1.99 |
Glucosinolates are identified as follows: 4MTB, 4-methylthiobutyl; 8MSOO, 8-methylsulfinyloctyl; I3M, indol-3-ylmethyl; 4MOI3M 4-methoxyindol-3-ylmethyl; 1MOI3M 1-methoxyindol-3-ylmethyl; 4OHB, 4-hydroxybutyl; 4MSOB, 4-methylsulfinylbutyl; 7MSOH, 7-methylsulfinylheptyl; 7MTH, 7-methylthioheptyl; 3BZO, 3-benzoyloxypropyl; 4BZO, 4-benzoyloxybutyl; 8MTO, 8-methylthiooctyl
DISCUSSION
The Arabidopsis PICKLE gene (PKL) encodes a member of the CHD3 class of chromatin remodeling complexes. In Xenopus and mammalian cell lines, a CHD3 protein is a component of a major histone deacetylation complex (Mi-2/NURD) that is presumed to generally function as a negative regulator of transcription (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998). A growing body of work indicates that CHD3 proteins play specific roles in the development of plant and animal systems and act as repressors of key regulators of developmental identity (Ahringer, 2000). In Drosophila melanogaster, a CHD3 homolog is required for the repression of homeotic genes during embryogenesis (Kehle et al., 1998), whereas in C. elegans a CHD3 homolog is necessary for the repression of germ-line specific genes in somatic cells (Unhavaithaya et al., 2002). Furthermore, a human CHD3 protein has been demonstrated to play a role in epithelial cell differentiation as an estrogen-dependent repressor of the master regulator Snail (Fujita et al., 2003). Similar to its animal CHD3 counterparts, PKL plays a developmental role in plants (Ogas et al., 1997) and is required for the repression of regulators of embryonic identity in vegetative tissues (Ogas et al., 1999; Rider et al., 2003).
Previous analyses have revealed that the pickle roots of pkl seedlings display several embryonic characteristics (Ogas et al., 1997). Although pickle roots can undergo somatic embryogenesis when excised from the plant (Ogas et al., 1997) and possess greatly elevated transcript levels for the LEC class of master regulators (Rider et al., 2003), the extent to which embryonic traits are derepressed in pickle roots has not been systematically examined. Here we report an analysis of the biochemical composition of pickle root tissue with an emphasis on the characterization of compounds known to accumulate in embryos.
Pickle roots accumulate embryonic storage materials
Although lipids usually comprise less than 1% of the weight of tissues such as roots or leaves (Broun et al., 1999), previous histological analyses of pickle roots indicated the presence of large quantities of neutral lipids that are organized into oil bodies that are morphologically similar to those from seeds (Ogas et al., 1997). We found that pickle roots specifically accumulate triacylglycerol with a fatty acid composition that is indistinguishable from that of seed lipid (Figure 1A, B). Thus many of the genes involved in synthesis of seed lipid are likely to be expressed in pickle roots. Furthermore, although long chain fatty acids that are typical of seed lipids are present in pickle roots (Figure 1B), we found that these long chain fatty acids were not detectable in lipids from pkl leaf tissue (Figure 1C). These data support previous observations that derepression of embryonic traits in pkl plants is restricted to organs produced during embryogenesis (Ogas et al., 1997).
More than half of the protein in mature seeds is represented by the 12S seed storage proteins and the remaining protein is comprised mostly of the 2S seed storage proteins (Finkelstein and Somerville, 1990; Fujiwara et al., 2002). Several different types of analyses strongly suggest that seed storage proteins are abundant in pickle roots as well. Transcripts for the AT2S1 seed storage protein gene have previously been reported to be present in pickle roots but not wild-type roots (Ogas et al., 1997), and we report here that the transcript for a 12S seed storage protein is elevated more than 20,000-fold in pickle roots than in wild-type roots. Analysis of the protein content of pickle roots on a Coomassie-stained gel reveals the presence of bands that co-migrate with the 12S and 2S seed storage proteins from seeds (Figure 2). Finally, analysis of pickle roots by transmission electron microscopy reveals the presence of structures that resemble the protein bodies of seeds. The presence of seed storage proteins in pickle roots is consistent with the elevated expression of LEC1, LEC2, and FUS3 in pickle roots (Rider et al., 2003), all of which have been implicated in promoting expression of storage proteins in embryos (Meinke et al., 1994; Parcy et al., 1997; Harada, 2001). The pattern of proteins that accumulate in pickle roots is considerably more complex than that observed in seeds (Figure 2), again supporting previous observations that pickle roots express the differentiation characteristics of both embryonic and non-embryonic tissue (Ogas et al., 1997).
In addition to accumulating storage reserves of lipid and protein, seeds accumulate phosphorous which is stored in seed as an organic compound called phytate (Mansfield and Briarty, 1992). We found that phytate is also present in pickle roots (Figure 3A), revealing that pickle roots also accumulate embryonic storage forms of minerals. The proportion of phosphorous that is accumulated as phytate in pickle roots is equivalent to that observed in seeds when adjusted for the total level of phosphate that is found in each sample. In addition, pickle roots possess phytin globoids (Figure 4), thus demonstrating that pickle roots are storing phytate in a manner analogous to that employed by the seed. In fact, all three seed storage compounds investigated – lipids, proteins, and phytate – accumulate in pickle roots in forms that are indistinguishable from those observed in seeds (Ogas et al., 1997). Thus in addition to expressing those factors necessary for accumulation of seed storage compounds, pickle roots also express those factors necessary for those compounds to be sequestered properly within the cell.
Secondary metabolism is PKL-dependent in root tissue
Seeds also accumulate unique secondary metabolites including specific sinapate esters and glucosinolates (Matthaus, 1998; Kliebenstein et al., 2001). In contrast to the seed storage compounds analyzed above, we found that these secondary metabolites do not accumulate in an embryo-specific manner in pickle roots (Figure 5, Table 1). Thus inhibition of embryo-specific secondary metabolic pathways in seedlings may be PKL-independent. Alternatively, it is possible that phenylpropanoid metabolism is regulated by PKL whereas the biosynthesis of choline is not. Choline is the substrate for enzyme sinapoylglucose: choline sinapoyltransferase (SCT) and thus required for formation of sinapoylcholine. This model of regulation would explain the accumulation of sinapoylglucose in pickle roots, a phenotype similar to that observed in the SCT-deficient sinapoylglucose accumulator2 (sng2) mutant of Arabidopsis (Shirley et al., 2001). These data further suggest that the LEC genes may not be involved in regulating secondary metabolism. Transcript levels for all three of these master regulators are elevated at least 100-fold in pickle roots in comparison to wild-type roots (Rider et al., 2003) and yet accumulation of secondary metabolites does not adopt an embryonic pattern.
Surprisingly, we found that secondary metabolism is disrupted in the pkl mutant and that pkl roots accumulate abnormal amounts of a number of compounds derived from both the glucosinolate and phenylpropanoid pathways. These changes appear to reflect a previously uncharacterized role for PKL in regulating metabolism. It is currently not clear if metabolic flux through the phenylpropanoid pathway is generally derepressed in roots, or if instead specific branches of the phenylpropanoid pathway are PKL-dependent. Similarly, the specific effect of PKL on expression of genes involved in glucosinolate biosynthesis remains to be determined.
PKL has been proposed to mediate GA-dependent responses (Ogas et al., 1997), but GA levels have not previously been reported to affect accumulation of these secondary metabolites. In the course of carrying out these analyses, we examined the effect of 10−8 M uniconazole-P on accumulation of these compounds in wild-type roots. We observed that accumulation of both phenylpropanoid-derived compounds and glucosinolate-derived compounds in wild-type roots is largely insensitive to treatment with 10−8 M uniconazole-P. These observations indicate that the role of PKL in regulation of secondary metabolism is unlikely to be reflective of its role in mediating GA-dependent responses. Consequently, these data suggest we have uncovered a new role for PKL in growth and development of Arabidopsis that is distinct from previously characterized roles.
The specific role of PKL in regulation of the genes involved in these biosynthetic pathways – lipid, protein, phytate, or secondary metabolites - remains to be determined. Analysis of the transcript levels of the LEC genes and of genes that code for seed storage proteins reveals that the transcript levels of these genes are initially wild-type in imbibed seeds but then subsequently increase during the germination of pkl seeds (Rider et al., 2003) (data not shown). Thus inappropriate expression of these genes appears to be a result of germination-specific derepression of these loci rather than carryover from seed formation itself. Given that the LEC gene products promote seed maturation (Meinke, 1992; Keith et al., 1994; Meinke et al., 1994; West et al., 1994; Parcy et al., 1997) and that expression of LEC genes is elevated during germination of pkl seeds and in pickle roots (Ogas et al., 1999; Rider et al., 2003), perhaps PKL acts as a repressor of LEC genes, which when expressed inappropriately lead to the ability to express compounds in pickle roots associated with seed maturation such as oil, protein, and phytate. It is also possible, however, that PKL is directly involved in regulating one or more of the genes involved in the synthesis of these compounds.
The observation that pkl plants exhibit novel accumulation of secondary metabolites that are not embryo-specific also raises the possibility that PKL is directly involved in regulation of one of more of these genes. In the absence of additional data, however, it is also quite possible that these effects could be indirect. Nonetheless, the observation that secondary metabolites are inappropriate expressed in pkl plants reveals that they may contribute to some aspect of the visible mutant pleiotropies exhibited by pkl plants, including aberrant shoot and root development.
Pickle roots represent a novel context for characterizing biosynthetic pathways of agronomic importance
Identification of the embryo-specific metabolites in the roots of pkl seedlings demonstrates that it is possible for plant roots to accumulate novel compounds when given the appropriate developmental context. The presence of these active metabolic pathways, whether the embryonic pathways in the pickle root or the novel secondary metabolic pathways of all pkl roots, provides a unique context for further genetic and biochemical characterization of these pathways. Such characterization would be of practical value as these pathways generate traits that can be of significant agronomic importance.
Phytate, for example, is a negative agronomic trait in a food crop. It is indigestible by humans and other nonruminant animals and can act as an anti-nutritional compound by sequestering essential trace minerals (Lott et al., 2000; Raboy et al., 2001). As a result, much effort has been placed on the production of low phytate plant seeds (Raboy et al., 2001) or on the production of transgenic animals capable of expressing phytase enzymes to digest phytate (Golovan et al., 2001; Golovan et al., 2001; Hostetler, 2003) . Characterization of phytate accumulation in seeds from crop plants has revealed that accumulation of phytate can be quite variable and is affected by growth conditions and genotype (Lott et al., 2000). Pickle roots represent a new context in which to undertake genetic and biochemical characterization of phytate accumulation. Such studies may facilitate the identification of previously uncharacterized factors and mechanisms that govern phytate accumulation in seeds.
It has previously been noted that production of agronomically significant quantities of triacylglycerols in a root crop would be of significant economic value (Ogas et al., 1997). Our observations extend this concept and suggest that roots represent a potential site for accumulation of valuable secondary compounds. A plethora of secondary compounds are produced in plants and many of them are only found in useable quantities in specific tissues of certain species. Several reviews regarding the commercial production of plant secondary metabolites (especially pharmaceuticals) have emphasized the need for innovation and large-scale, economical options for the production of fine chemicals in plants and plant cells (Verpoorte et al., 1999). One alternative to cell culture that is widely believed to have promise is the use of hairy root cultures to produce specialty chemicals, although hairy root cultures are still not economically feasible in most cases (Shanks and Morgan, 1999). Further characterization of altered secondary metabolism in pkl roots may circumvent current limitations by revealing new factors that will facilitate the accumulation of additional secondary compounds in plant roots.
In summary, we have shown that pickle roots of pkl seedlings accumulate a variety of compounds that are normally preferentially accumulated in embryos. Not all embryo-specific pathways are derepressed in pickle roots, however, and embryo-specific secondary metabolites do not accumulate in pickle roots. Instead, PKL plays a previously undiscovered role in non-embryonic secondary metabolism and is required for normal accumulation of secondary metabolites in roots. It is not clear what properties of the pkl roots make them capable of accumulating the compounds described herein, or if other vegetative tissues could similarly accumulate these compounds given the appropriate developmental context. Nonetheless, it should be possible to further dissect the pathways that lead to lipid, protein, phytate and secondary metabolite accumulation in pickle roots and pkl secondary roots through genetic or biochemical analyses. It may also be possible to further engineer the production of related compounds in economically significant quantities in root crops.
Acknowledgements
The authors wish to thank the Life Science Microscopy Facility (Purdue University, West Lafayette, IN) for processing samples for transmission electron microscopy. We would also like to thank Bill Muir for his support of HAH and Chris Somerville for use of facilities at The Carnegie Institution of Washington, Department of Plant Biology. This work was supported by a grant from the National Institutes of Health (R01GM059770-01A1) to JO, and by a grant from the Division of Energy Biosciences, United States Department of Energy (DE-FG02-94ER20138) to CC. SDR was supported by funds from the BASF Corporation. This is journal paper number 17290 of the Purdue University Agricultural Experiment Station.
REFERENCES
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed Germination and Dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. Ed second Plenum Press; New York, USA: 1994. [Google Scholar]
- Broun P, Gettner S, Somerville C. Genetic engineering of plant lipids. Annu Rev Nutr. 1999;19:197–216. doi: 10.1146/annurev.nutr.19.1.197. [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry. 2003;62:471–481. doi: 10.1016/s0031-9422(02)00549-6. [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt P, Somerville C. A mutant of Arabidopsis deficient in C18−3 and C16−3 leaf lipids. Plant Physiol. 1986;81:859–864. doi: 10.1104/pp.81.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR. 3 classes of abscisic-acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. Embo J. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD Complex Subunit, Regulates an Invasive Growth Pathway in Breast Cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Nambara E, Yamagishi K, Goto DB, Naito S. Storage proteins. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. American Society of Plant Biologists; Rockville, MD: 2002. http://www.aspb.org/publications/arabidopsis/ pp doi/10.1199/tab.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovan SP, Hayes MA, Phillips JP, Forsberg CW. Transgenic mice expressing bacterial phytase as a model for phosphorus pollution control. Nat Biotechnol. 2001;19:429–433. doi: 10.1038/88091. [DOI] [PubMed] [Google Scholar]
- Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, Plante C, Pollard JW, Fan MZ, Hayes MA, Laursen J, Hjorth JP, Hacker RR, Phillips JP, Forsberg CW. Pigs expressing salivary phytase produce low-phosphorus manure. Nat Biotechnol. 2001;19:741–745. doi: 10.1038/90788. [DOI] [PubMed] [Google Scholar]
- Harada JJ. Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J Plant Physiol. 2001;158:405–409. [Google Scholar]
- Heinonen JK, Lahti RJ. A new and convenient colorimetric determination of inorganic ortho-phosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem. 1981;113:313–317. doi: 10.1016/0003-2697(81)90082-8. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell. 2003;15:179–194. doi: 10.1105/tpc.006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li H-C, Rider SD, Mordhorst AP, Romero-Severson J, Cheng J-C, Robey J, Sung ZR, de Vries SC, Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in GA-dependent responses. Plant Physiol. 2004 doi: 10.1104/pp.103.030148. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M, Kurup S, McKibbin R. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci. 1999;4:275–280. [Google Scholar]
- Hostetler HA. Genetic Engineering of IP6 Metabolism (Inositol Hexaphosphate) in the Japanese Medaka. Ph. D. Purdue University; West Lafayette: 2003. [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. Seed-Specific Over-Expression of an Arabidopsis cDNA Encoding a Diacylglycerol Acyltransferase Enhances Seed Oil Content and Seed Weight. Plant Physiol. 2001;126:861–874. doi: 10.1104/pp.126.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L. Alteration of seed fatty acid composition by an ethyl methanesulfonate- induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995;108:399–409. doi: 10.1104/pp.108.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. fusca3: A Heterochronic Mutation Affecting Late Embryo Development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Sherman D, Karlson D, Dudareva N. Cellular and subcellular localization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiol. 2001;126:956–964. doi: 10.1104/pp.126.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HWM, Karssen CM. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants of Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of bacteriophage T. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C. Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell. 2000;12:1295–1306. doi: 10.1105/tpc.12.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen M, Racicot V, Strack D, Chapple C. Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiology. 1996;112:1625–1630. doi: 10.1104/pp.112.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lott JNA, Ockenden I, Raboy V, Batten GD. Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Sci Res. 2000;10:11–33. [Google Scholar]
- Lott JNA, West MM. Elements present in mineral nutrient reserves in dry Arabidopsis thaliana seeds of wild type and pho1, pho2, and man1 mutants. Can J Bot. 2001;79:1292–1296. [Google Scholar]
- Mansfield SG, Briarty LG. Cotyledon cell-development in Arabidopsis thaliana during reserve deposition. Can J Bot. 1992;70:151–164. [Google Scholar]
- Mansfield SG, Briarty LG. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int J Plant Sci. 1996;157:280–295. [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999;20:251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- Matthaus B. Isolation, fractionation and HPLC analysis of neutral phenolic compounds in rapeseeds. NAHRUNG. 1998;42:75–80. [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Ann Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- Meinke DW. A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy Cotyledon mutants of Arabidopsis. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntz K. Deposition of storage proteins. Plant Mol Biol. 1998;38:77–99. [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Browse J, Somerville CR. The genetics of plant lipids. Biochim Biophys Acta. 1991;1082:1–26. doi: 10.1016/0005-2760(91)90294-r. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V, Young KA, Dorsch JA, Cook A. Genetics and breeding of seed phosphorus and phytic acid. J Plant Phys. 2001;158:489–497. [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Rider SD, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313x.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Chapple C. Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics. 2001;159:1741–1749. doi: 10.1093/genetics/159.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, Larner V, Kurup S, Bougourd S, Holdsworth M. The Arabidopsis COMATOSE locus regulates germination potential. Development. 2000;127:3759–3767. doi: 10.1242/dev.127.17.3759. [DOI] [PubMed] [Google Scholar]
- Shanks JV, Morgan J. Plant ’hairy root’ culture. Curr Opin Biotechnol. 1999;10:151–155. doi: 10.1016/s0958-1669(99)80026-3. [DOI] [PubMed] [Google Scholar]
- Shirley AM, McMichael CM, Chapple C. The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J. 2001;28:83–94. doi: 10.1046/j.1365-313x.2001.01123.x. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a Homolog of the NURD Complex Component Mi-2 Act Together to Maintain Germline-Soma Distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Verpoorte R, van der Heijden R, ten Hoopen HJG, Memelink J. Metabolic engineering of plant secondary metabolite pathways for the production of fine chemicals. Biotechnol Lett. 1999;21:467–479. [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]


