Abstract
A new pathway for cysteine biosynthesis has been elucidated in Mycobacterium tuberculosis. This pathway involves a protein bound thiocarboxylate (CysO-SH) as the sulfide donor, similar to thiamin biosynthesis. Cysteine synthase M (CysM) catalyzes the addition of cysteine to the carboxy terminus of the protein bound thiocarboxylate to generate a CysO-cysteine adduct. A protease, Mec+, hydrolyzes the CysO-cysteine adduct to release cysteine and regenerate CysO. Mec+ contains a JAMM motif and this work provides the first functional characterization of the JAMM motif in prokaryotes. MoeZ, a paralog of ThiF, has been shown to transfer sulfur onto CysO.
In the biosynthesis of thiamin and molybdopterin, a small sulfide carrier protein (ThiS 1 and MoaD 4) is converted to a carboxy terminal thiocarboxylate (3, 6) which then functions as the sulfide donor for cofactor biosynthesis.1 Analogous chemistry, involving formation of thioester 9, occurs in the ubiquitin targeting of doomed proteins to the proteasome (Scheme 1).2
Scheme 1.
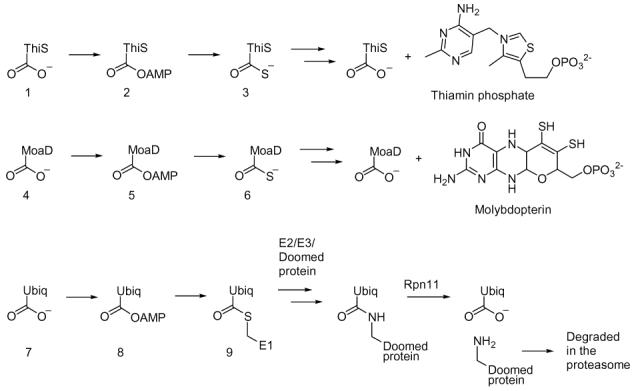
These observations suggested that sulfide carrier proteins may play a role in the biosynthesis of other sulfur-containing natural products. This was supported by the analysis of sequenced genomes for homologs of ThiS and MoaD, which revealed that ThiS-like proteins not only cluster with thiamin and molybdopterin biosynthetic genes, but also with the biosynthetic genes of the sulfur-containing siderophores pyridine-2,6-dithiocarboxylate and quinolobactin.3 In addition, in Mycobacterium tuberculosis and other actinomycetes, a gene similar to thiS and moaD (Rv1335; herein called cysO) was found adjacent to a gene annotated as cysteine synthase (Rv1336; herein called cysM). This raised the possibility that CysO could function as the sulfide carrier 12 for cysteine biosynthesis in M. tuberculosis by displacing the acetate of O-acetylserine (Scheme 2).
Scheme 2.
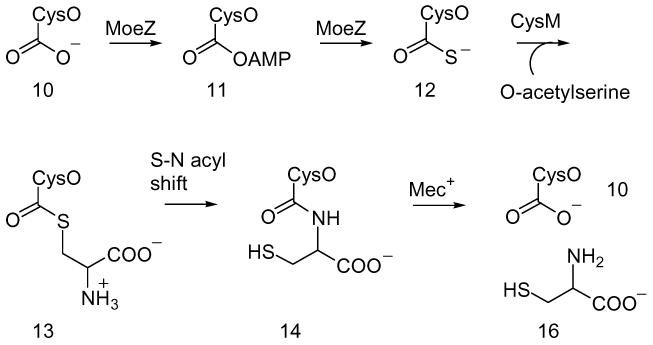
The mec+ gene (Rv1334), which codes for a putative hydrolase (JAMM; Jab1/MPN domain metalloenzyme motif), is also clustered with the cysO and cysM genes in M. tuberculosis.4 This gene was originally identified by its ability to restore a nutritional requirement for cysteine and methionine in a mutant strain of Streptomyces kasugaensis, however, no function was assigned.5 Based on its clustering with CysO and CysM and its predicted hydrolase activity, we proposed that Mec+ might hydrolyze CysO-cysteine 14 generated by an N-S acyl shift of the initially formed adduct 13 (Scheme 2).6 In this communication, we describe the experimental confirmation of this new variation of the cysteine biosynthetic pathway.
CysO-thiocarboxylate was prepared by expressing a CysO-Intein fusion containing a chitin binding domain using the pTYB1 vector. Binding of the fusion protein to a chitin column, followed by soaking in ammonium sulfide (30mM; 40hrs.) resulted in the release of pure CysO-thiocarboxylate.7 His-tagged CysM was overexpressed using the pET16b vector and purified by Ni-Nta chromatography. MS analysis of a reaction mixture containing CysO-thiocarboxylate, CysM, and O-acetylserine demonstrated the formation of a new CysO derived species with a mass increase of 88 Da consistent with the expected mass of CysO-cysteine 14 (Figure 1). MS/MS analysis localized the adduct to the carboxy terminus of CysO. Neither ThiS-thiocarboxylate 3 from Bacillus subtilis nor CysK (Rv2334), the other cysteine synthase from M. tuberculosis, were functional in the reaction mixture demonstrating that adduct formation is specific for CysO/CysM.
Figure 1.
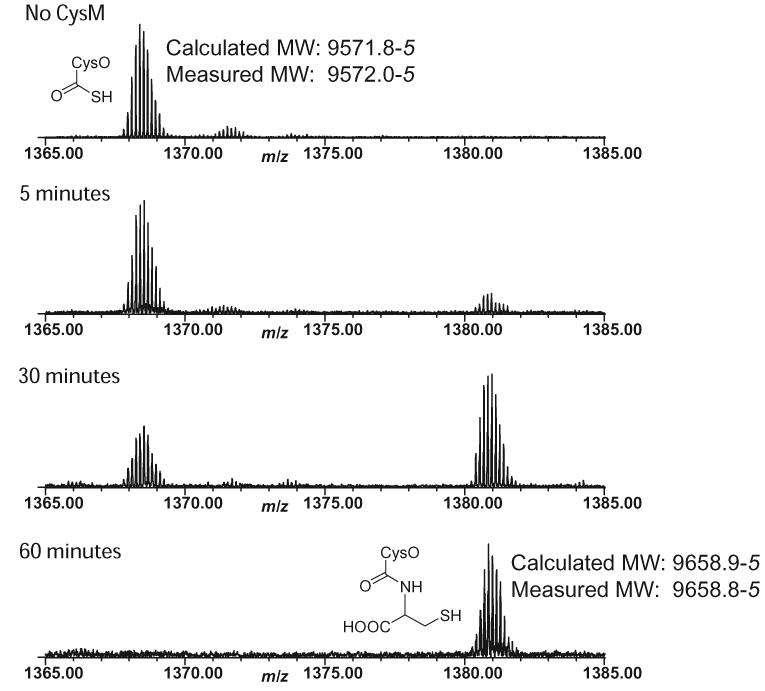
ESI-FTMS analysis of the reaction between CysM and CysO-thiocarboxylate in the presence of O-acetylserine (+7 charge state).
His-tagged Mec+, fused to the maltose binding protein to improve solubility, was overexpressed using a modified pET vector and purified by Ni-Nta chromatography. MS analysis of a reaction mixture containing CysO-thiocarboxylate 12, CysM, O-acetylserine and Mec+ demonstrated the reformation of CysO 10. In addition, the formation of cysteine 16 in the reaction mixture could be detected with ninhydrin.8 When 2.6 nmoles of CysO-thiocarboxylate was added to CysM and Mec+ in the presence of O-acetylserine, 2.7±0.16 nmoles of cysteine were produced, indicating full conversion to cysteine. When Mec+ is pre-incubated with EDTA, no hydrolysis is observed. Addition of Zn2+ to the gel filtered, EDTA containing reaction restored the hydrolytic activity. This suggests that Mec+ is a Zn2+ dependent CysO-cysteine carboxypeptidase.9 To provide additional support for Mec+ hydrolysis of amide 14 rather than thioester 13, His-tagged CysO with an additional cysteine or alanine at the carboxy terminus (His10-CysO-cys; His10-CysO-ala) was overexpressed. Upon incubation of His10-CysO-cys with Mec+, we observed the production of cysteine by the ninhydrin assay and hydrolysis by ESI-FTMS (Figure 2). His10-CysO-ala was also hydrolyzed, but at a reduced rate.
Figure 2.
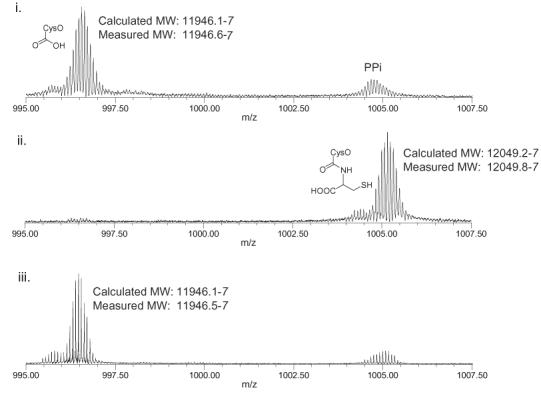
ESI-FTMS analysis of the Mec+ catalyzed hydrolysis of His10-CysO-cys (+12 charge state). i. His10-CysO-cys treated with Mec+; ii. His10-CysO-cys, treated with Mec+ that was pre-incubated with EDTA; iii. Reaction ii + Zn2+.
The reaction catalyzed by Mec+ resembles the deubiquitination reaction of the 26S proteasomal subunit Rpn11, which sends doomed proteins into the proteasome for degradation and regenerates ubiquitin (Scheme 1). Both Rpn11 and Mec+ contain the JAMM motif, which is found in bacteria, archaea, and eukaryotes.10 Due to the complexity of the proteasome, it has been difficult to study Rpn11 activity on its own. Our demonstration of Mec+ activity represents the first functional assignment for a JAMM motif protein in prokaryotes and may be a useful model for Rpn11 and the other deubiquitinating enzymes.
We have not yet identified the sulfur donor for CysO. In the formation of ThiS-thiocarboxylate 3 in thiamin biosynthesis, ThiS is activated as an acyl adenylate 2 in a reaction catalyzed by ThiF. The resulting ThiS-COOAMP then reacts with IscS persulfide. Since cysteine is the sulfur donor for IscS persulfide, it is unlikely that this species is the sulfur donor for CysO-thiocarboxylate formation.11 We have identified and cloned a ThiF paralog in M. tuberculosis (moeZ, Rv3206) that may catalyze the CysO adenylation and sulfur transfer reaction. MoeZ was expressed from a pET16b vector and purified by Ni-Nta chromatography. When MoeZ was combined with pure CysO, we were unable to observe thiocarboxylate formation on CysO using various sulfur sources (sulfide, thiosulfate). However, when CysO and MoeZ overexperssion strains were co-lysed and co-purified from E. coli crude cell lysate, we observed CysO-thiocarboxylate (Figure 3). This indicates that MoeZ is able to catalyze sulfur transfer from an unidentified sulfur source onto CysO.
Figure 3.
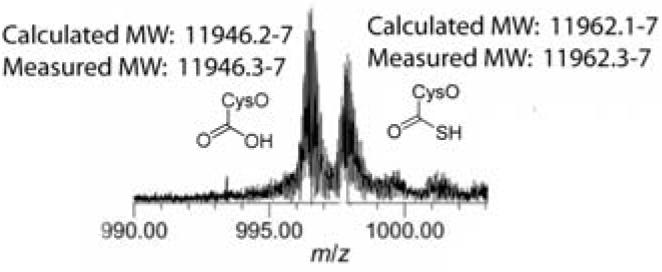
ESI-FTMS analysis of the MoeZ catalyzed sulfur transfer onto CysO (+12 charge state).
Analysis of the M. tuberculosis genome suggests that there are three routes to cysteine in this microorganism: the sulfide dependent pathway, the cystathionine pathway and the CysO-thiocarboxylate pathway.12 The relative importance of these pathways is currently unknown. Two observations, however, may point to a function for the CysO dependent pathway. First, transcriptional profile analysis of M. tuberculosis demonstrates that mec+, cysO, cysM and moeZ are all upregulated under oxidative stress conditions.13 Second, thiocarboxylates are much more resistant to oxidation than thiols. These observations suggest that when M. tuberculosis is in the highly oxidizing environment of the macrophage, CysO-thiocarboxylate may be used as an oxidation-resistant form of sulfide for cysteine biosynthesis.
Herein we show that a sulfide carrier protein from M. tuberculosis, CysO (Rv1335), in its thiocarboxylate form 12, is alkylated by O-acetylserine to give 13. This reaction is catalyzed by CysM. Following an S-N acyl rearrangement to give CysO-cys 14, Mec+ (Rv1334) hydrolyzes CysO-cys 14, releasing cysteine and regenerating CysO 10. We have also demonstrated that MoeZ (Rv3206), in crude cell lysates, can catalyze the conversion of CysO 10 to CysO-thiocarboxylate 12 (Scheme 2) using an unidentified sulfur source. We propose that this novel pathway, involving a protein bound thiocarboxylate instead of sulfide, could be necessary for cysteine biosynthesis in M. tuberculosis in the oxidizing environment of the macrophage.
Supplementary Material
Acknowledgement
This paper is dedicated to Peter Dervan on the occasion of his 60th birthday. We would like to thank Cynthia Kinsland of Cornell University Protein Facility for cloning the CysO, CysM, Mec+ cluster and Joanne Widom of Cornell University for the plasmid pMBT-HT. This research was supported by grants from NIH (DK44083 to TPB and GM16609 to FWM).
Footnotes
Supporting Information Available: Cloning, expression, and purification of CysO-SH, His10-CysO-cys, CysO, CysM, and Mec+, MS protocol. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a. Park J-H, Dorrestein P, Zhai H, Kinsland CL, McLafferty FW, Begley TP. Biochemistry. 2003;42:12430–12438. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]; b. Wang C, Xi J, Begley TP, Nicholson LK. Nat. Struct. Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]; c. Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Nat. Struct. Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.a. Lewis TA, Cortese MS, Sebat JL, Green TL, Lee C-H, Crawford RL. Env. Microbio. 2000;2:407–416. doi: 10.1046/j.1462-2920.2000.00122.x. [DOI] [PubMed] [Google Scholar]; b. Matthijs S, Baysse C, Koedam N, Tehrani KA, Verheyden L, Budzikiewicz H, Schaefer M, Hoorelbeke B, Meyer J, De Greve H, Cornelis P. Mol. Microbio. 2004;52:371–384. doi: 10.1111/j.1365-2958.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- 4.Yao T, Cohen RE. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 5.Hirasawa K, Ichihara M, Okanishi M. J. Antibiotics. 1985;38:1795–1798. doi: 10.7164/antibiotics.38.1795. [DOI] [PubMed] [Google Scholar]
- 6.Shao Y, Paulus HJ. Peptide Res. 1997;50:193–198. doi: 10.1111/j.1399-3011.1997.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 7.Kinsland CL, Taylor SV, Kelleher NL, McLafferty FW, Begley TP. Prot. Science. 1998;7:1839–1842. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaitonde MK. Med. Res. Council. 1967;104:627–633. [Google Scholar]
- 9.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 10.Cope GA, Suh GSB, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 11.Lauhon CT, Kambampati R. J. Biol. Chem. 2000;275:20096–20103. doi: 10.1074/jbc.M002680200. [DOI] [PubMed] [Google Scholar]
- 12.Wooff E, Michell S. Ll., Gordon SV, Chambers MA, Bardarov S, Jacobs WR, Jr., Hewinson RG, Wheeler PR. Mol. Microbio. 2002;43:653–663. doi: 10.1046/j.1365-2958.2002.02771.x. [DOI] [PubMed] [Google Scholar]
- 13.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Mol. Microbio. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


