Abstract
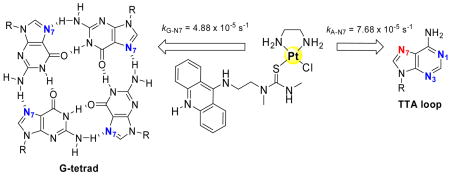
The interactions of PT-ACRAMTU, a cytotoxic platinum–acridine conjugate, with the human telomeric G-quadruplex have been studied using in-line high-performance liquid chromatography–mass spectrometry and footprinting assays. The conjugate reacts significantly faster with quadruplex DNA (t1/2= 1.2 h) than with double-stranded DNA, and A-N7, and not G-N7, is the kinetically preferred target, an unprecedented reactivity feature in platinum–DNA interactions. Unlike the clinical platinum drug cisplatin, which targets the human telomeric sequence nonspecifically, the platinum–intercalator technology has the potential to produce telomere-specific anticancer agents via a mechanism that kinetically discriminates between G and A in the two DNA secondary structures.
Human telomeres consist of noncoding repeats of the guanine (G)-rich sequence 5′-TTAGGG, which protect chromosomes from degradation and end fusion, two factors limiting a normal cell’s life span.1 In most tumors, the telomeres are relatively short compared to normal cells, but are efficiently maintained by up-regulated telomerase, rendering cancer cells immortal.2 It has been proposed that telomere-targeted agents capable of disrupting indefinite cell proliferation might have applications as anticancer therapeutics.2 Telomeres contain 3′ single-stranded overhangs, which have the potential to fold into G-quadruplex structures containing three π-stacked G tetrads (G4). In a G tetrad, the four guanine bases are linked by Hoogsteen H-bonding.3 In this configuration, N7 of guanine is protected from DNA-targeted electrophilic agents, such as platinum-containing drugs, which preferentially bind to this site.
Our previous discovery of the unusual high frequency of monofunctional adenine (A) adducts in double-stranded DNA formed by PT-ACRAMTU (ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea) (Figure 1A), a platinating–intercalating cytotoxic hybrid agent,4 prompted us to study its interactions with the sequence 5′-TTAGGG in a quadruplex secondary structural context. On the basis of high-resolution5 and biophysical6 data available for human telomere sequences, we anticipated that specific adenine bases in the flexible TTA loop regions of the quadruplex might be targeted by PT-ACRAMTU. The current study demonstrates that telomeric A is not only highly susceptible to platination by this agent, but binding to A-N7 is kinetically favored over adduct formation with G-N7. This is an unprecedented reactivity feature in platinum–DNA interactions and suggests a new strategy for telomere-targeted chemotherapeutic intervention.
Figure 1.
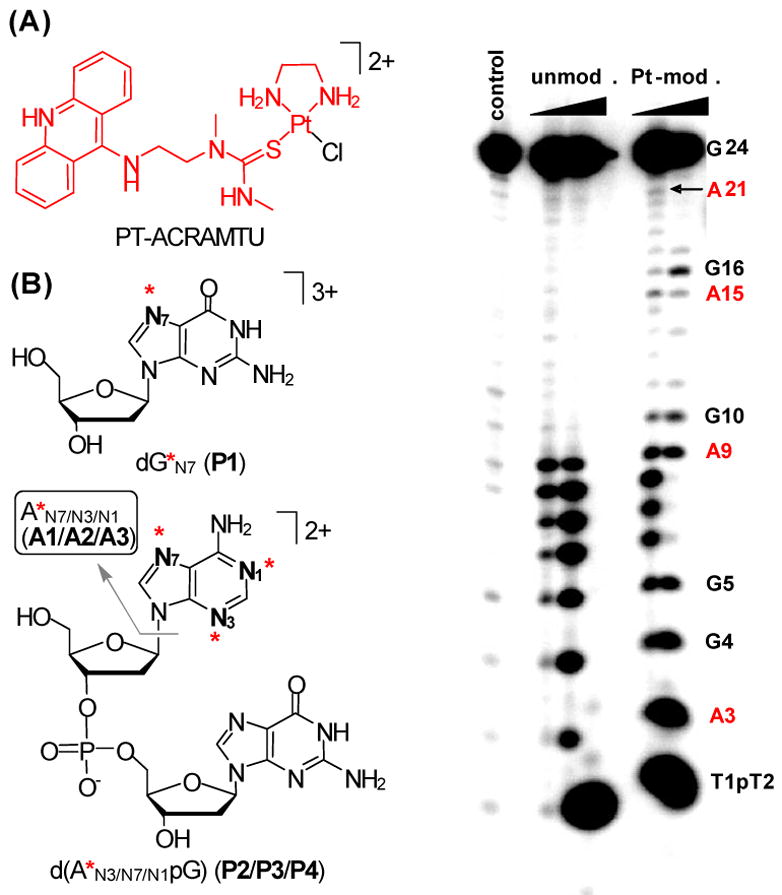
Structures of PT-ACRAMTU (A) and the platinum-modified DNA fragments (B) detected in enzymatic and acidic digests. The asterisks indicate the fragment [Pt(en)(ACRAMTU)]3+, which is highlighted in (A). (C) Denaturing polyacrylamide gel analysis of platinum-modified G4-24 digested with exonuclease I.
To probe the interactions between PT-ACRAMTU and telomeric G4 DNA, we generated the K+-forms of the sequences d[AG3(T2AG3)3] (G4-22) and d[(T2AG3)4] (G4-24) and confirmed their quadruplex structures by variable-temperature UV and CD spectroscopy (Supporting Information) prior to incubation with platinum. G4-22 was incubated with PT-ACRAMTU at a drug-to-nucleotide ratio (rb) of 0.2 at 37 °C for 24 h. The samples were then subjected to enzymatic and acid digestion and analyzed by in-line liquid chromatography–electrospray mass spectrometry (LC–ESMS) according to previously established protocols.4 To test the effects of DNA secondary structure on the adduct profile, the corresponding hybridized duplex, d[AG3(T2AG3)3]/d[(C3TA2)3C3T] (ds-22), and the single-stranded hexanucleotide, d[T2AG3] (ss-6), were also studied. The base and donor-site specificity of platination in G4-22 (as well as in ds-22 and ss-6) was deduced from the LC–MS analysis of fragments detected in the enzymatic and acidic digests. Structural assignments were based on molecular masses and characteristic fragmentation patterns observed in positive- and negative-mode ESMS and tandem MS/MS spectra (Supporting Information).
Four fragments were observed in the enzymatic digests of drug-modified G4-22, all of which could be unambiguously identified: 2′-deoxyguanosine, platinated at N7 (dG*N7, P1, Mr = 847.3), and three isomers of the dideoxyribonucleotide fragment d(A*pG), in which platinum is bound to A-N3, A-N7, and A-N1 (P2–P4, Mr = 1159.3), respectively (Figure 1B). The absence of dA* in the mixtures indicates that the d(ApG) phosphodiester linkage is resistant to endonucleolytic hydrolysis, which has been previously observed specifically for adenine adducts formed by PT-ACRAMTU.4b The platinum binding sites in the linkage isomers and their approximate abundances were further determined by selective depurination of adenine, yielding the corresponding A* fragments (A1–A3, Mr = 715.7) (Figure 1B). All of the adducts identified in G4-22 were also observed in digests of ds-22 and ss-6, albeit at varying abundances characteristic of template secondary structure (vide infra).
To study the sequence preference of adduct formation in telomeric DNA, the platinum-modified (rb = 0.08) template G4-24 was subjected to 3′→5′ exonuclease digestion. In the denaturing gels, multiple damage sites were detected with exonuclease I. The most intense bands are observed at the terminal base, G24, and the first two AG steps, A3G4 and A9G10. Significantly weaker damage is also observed at A15G16 and A21 (Figure 1C). These observations are consistent with previous biophysical6a and footprinting7 studies, which show the first two A bases in the sequence and the outermost of the three stacked G tetrads are more accessible and susceptible to attack by electrophilic agents.
The relative abundances of A and G adducts in enzymatic and acidic digests of G4-22, ds-22, and ss-6 were deduced from reverse-phase HPLC profiles (Figure 2). The enzymatic digests indicate a dramatic increase in A adducts (P2–P4) for G4 DNA compared to ds and ss DNA (Figure 2A). Approximately 50% of the adducts formed by PT-ACRAMTU at rb = 0.2 in G4-22 are formed with A, which has to be attributed to secondary structural effects in this type of DNA. Similarly, higher levels of A adducts are observed in ds-22 compared to ss-6, a consequence of the base-pair step recognition of the conjugate.4a The effects of DNA secondary structure on the damage profiles are also reflected in the frequency of the A-N1, A-N3, and A-N7 linkage isomers (Figure 2B). N1 and N7 are the preferred targets for PT-ACRAMTU in G4-22 with a minor fraction also binding to N3. In contrast, N1 is disfavored in ds-22, because it is involved in Watson–Crick H-bonding, and becomes even less abundant than sterically hindered N3, whose reactivity is enhanced due to PT-ACRAMTU’s unique minor-groove recognition.4a Characteristically, N3 is the least favored A binding site in ss-6.
Figure 2.
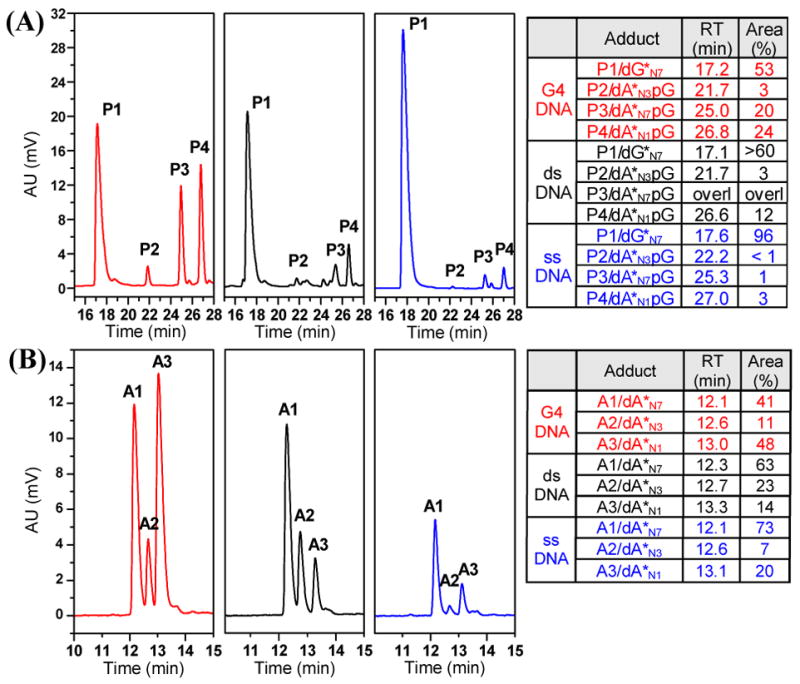
Reverse-phase HPLC profiles and relative adduct abundances for enzymatic (A) and acidic (B) digests of G4-22 (red), ds-22 (black), and ss-6 (blue) DNA treated with PT-ACRAMTU at rb = 0.2.
To establish if a kinetic preference exists for any of the detected binding sites, the reaction between PT-ACRAMTU and G4-22 was quenched at appropriate time intervals with thiourea and its progress monitored by quantitative HPLC analysis. The study was performed at a low rb of 0.1 to enhance the binding selectivity and establish pseudo-first-order reaction conditions. Based on the concentration of adducts P1–P4 in these mixtures, the order of binding preference is: A-N7 > G-N7 > A-N1 > A-N3 (Figure 3). These data confirm that A-N7 is the preferred target of intercalator-driven platination in G-quadruplex DNA.
Figure 3.
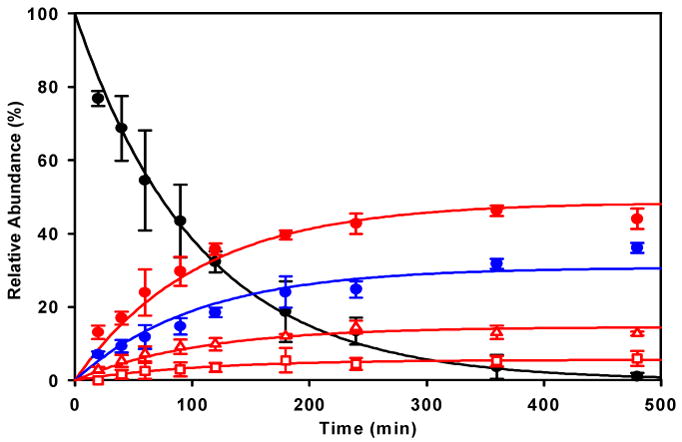
Progress of the reaction of PT-ACRAMTU with G4-22 (37 °C, pH 7.2, rb = 0.1) monitored by quantitative HPLC analysis of enzymatic digests. The solid lines are exponential curve fits assuming 4 parallel reactions and pseudo-first-order conditions. Trace assignments: black circles: quenched, unreacted drug; red open squares: A-N3 adduct P2; red open triangles: A-N1 adduct P4; red full circles: A-N7 adduct P3; blue full circles: G-N7 adduct P1. Error bars indicate ±standard deviations for a set of 3 independent incubations/digestions.
In this study we report the highest frequency of adenine adducts detected to date (> 50% of total adducts) for a platinum agent in this G-rich form of DNA. (For comparison, PT-ACRAMTU forms ~20% A adducts in ds calf thymus DNA.4) This is in distinct contrast to the reactivity of the clinical drug cisplatin (cis-diamminedichloroplatinum(II)), which induces > 80% G and < 15% A adducts in telomeric DNA,8 similar to its adduct distribution in ds DNA.9 Most strikingly, PT-ACRAMTU reacts with G4 DNA significantly faster (t1/2 = 1.2 h) than with ds DNA (t1/2 ~ 2–3 h4). The proposed binding mechanism involves →-stacking of acridine with the outermost G tetrads, which can be expected to stabilize the G4 structure and favor reaction of the A-affinic4 platinum moiety in PT-ACRAMTU with the TTA loops. It is noteworthy to mention that a structurally related quinacridine-tethered “classical” platinum triamine complex10 induces major damage only at thermodynamically preferred G in the same sequence, indicating major differences in the binding mechanisms of the two complexes. In contrast, our platinum–intercalator technology has the potential to produce telomere specificity for agents that are able to discriminate between G4 and ds DNA via a mechanism that combines specific recognition of DNA secondary structure and nucleobase nitrogen. While ACRAMTU itself binds to and stabilizes G4 DNA (ΔTm~ 13 °C in G4-22; Supporting Information), it was designed as a groove-specific intercalator for ds DNA.11 Thus, to generate a platinum–intercalator conjugate that is truly selective for telomeric adenine, it will be necessary to replace acridine in PT-ACRAMTU with a highly G4-specific ligand. Synthetic studies of complexes containing suitable extended polyaromatic carrier systems are currently underway to achieve this goal.
Supplementary Material
Experimental details, characterization of the DNA sequences, ESMS and MS/MS data, and details of the modeling and calculation of kinetic data. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by the National Institutes of Health (Grant CA101880).
References
- 1.Maizels N. In: Quadruplex Nucleic Acids. Neidle S, Balasubramanian S, editors. RSC Publishing; Cambridge: 2006. pp. 228–252. [Google Scholar]
- 2.Shay JW, Wright WE. Nature Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson GN. In: Quadruplex Nucleic Acids. Neidle S, Balasubramanian S, editors. RSC Publishing; Cambridge: 2006. pp. 1–30. [Google Scholar]
- 4.(a) Barry CG, Day CS, Bierbach U. J Am Chem Soc. 2005;127:1160–1169. doi: 10.1021/ja0451620. [DOI] [PubMed] [Google Scholar]; (b) Barry CG, Baruah H, Bierbach U. J Am Chem Soc. 2003;125:9629–9637. doi: 10.1021/ja0351443. [DOI] [PubMed] [Google Scholar]
- 5.(a) Dai J, Carver M, Punchiheva C, Jones RA, Yang D. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Parkinson GN, Lee MP, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 6.(a) Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Proc Natl Acad Sci USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) He Y, Neumann RD, Panyutin IG. Nucleic Acid Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Balagurumoorthy P, Brahmachari SK. J Biol Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 8.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fichtinger-Schepman AMJ, van der Veer JL, den Hartog JHJ, Lohman PHM, Reedijk J. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand H, Bombard S, Monchaud D, Teulade-Fichou MP. J Biol Inorg Chem. 2007;12:1003–1014. doi: 10.1007/s00775-007-0273-3. [DOI] [PubMed] [Google Scholar]
- 11.Baruah H, Bierbach U. Nucleic Acids Res. 2003;31:4138–4146. doi: 10.1093/nar/gkg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental details, characterization of the DNA sequences, ESMS and MS/MS data, and details of the modeling and calculation of kinetic data. This material is available free of charge via the Internet at http://pubs.acs.org.


