Abstract
The eukaryotic translation initiation factor 5A (eIF5A) is essential for growth and cell viability. eIF5A is the only protein that contains hypusine [Nε-(4-amino-2-hydroxybutyl)lysine], which is required for its activity. Hypusine is formed by post-translational modification of one specific lysine residue (Lys50 for human eIF5A) by deoxyhypusine synthase and deoxyhypusine hydroxylase. eIF5A is highly conserved from yeast to mammals, especially around the hypusine modification site. To investigate the features of eIF5A required for its activity and modification, we conducted structure/function studies using human eIF5A-1 mutants with a single amino acid substitution at each of the highly conserved residues and also truncated proteins. The majority of the 49 human eIF5A mutants tested were capable of supporting the growth of a S. cerevisiae eIF5A null strain. Growth-supporting activity was abolished in only a few mutants, K47D, G49A, K50A, K50D, K50I, K50R, G52A and K55A, all with substitutions at, or in the vicinity of, the modification site, and in truncation mutants with deletions of 21 amino acids from N-or C-terminus. Lys50 mutants, G52A and K55A were defective as substrates for deoxyhypusine synthase, and K47D and H51A for deoxyhypusine hydroxylase. These findings demonstrate the critical importance of the hypusine site loop in eIF5A function and hypusine modification. Selected human eIF5A mutants were tested for their activity in protein synthesis in the UBHY-R strain that harbors an unstable eIF5A fusion protein. A close correlation was observed between their ability to enhance protein synthesis and growth, lending further support for a central role of eIF5A in translation.
eIF5A is a putative translation initiation factor and is the only cellular protein that contains the unique modified lysine, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] (1). Hypusine is formed post-translationally at one specific lysine residue of the eIF5A precursor in two consecutive enzymatic reactions (2). The first enzyme, deoxyhypusine synthase (DHS) (3,4) catalyzes the transfer of the aminobutyl moiety from the polyamine spermidine to form an intermediate, deoxyhypusine [Nε-(4-aminobutyl)lysine] residue, which in turn is hydroxylated by deoxyhypusine hydroxylase (DOHH) (5). The absolute requirement for the eIF5A protein and its posttranslational modification was established from gene disruption studies in S. cerevisiae, in which inactivation of the eIF5A genes (TIF51A and TIF51B) (6,7) or of the deoxyhypusine synthase gene (8,9) caused loss of viability.
In spite of the essential nature of the hypusine/deoxyhypusine modification in eukaryotic cell proliferation (2,10–15), the precise cellular function of eIF5A has remained obscure for decades. eIF5A was initially isolated from the high salt washes of reticulocyte lysate ribosomes with other initiation factors (16). eIF5A stimulates methionyl-puromycin synthesis, a model assay for translation initiation. The requirement for deoxyhypusine/hypusine in this assay is remarkably stringent (17,18): the unmodified eIF5A precursor is inactive. Its role as a general translation initiation factor has been disputed, especially since a rapid depletion of UBR5A (a ubiquitin-Arg-yeIF5A fusion protein) caused only a moderate reduction in protein synthesis (19) and did not alter the polysome profile. Recently, however, a role of eIF5A in translation has been revisited more carefully. Association of eIF5A with actively translating ribosomes (20,21) suggests its specific role in translational control. eIF5A has been proposed to be a specific initiation factor for a subset of mRNAs (19,22) or a RNA-binding protein involved in nuclear transport (23). A number of mRNAs have been reported, from differential display analysis, to be candidate targets of eIF5A (24). Furthermore, an eIF5A homolog in archaea Halobacterium sp., as well as eIF5A, was reported to display specific RNA cleavage activity in vitro (25). S. cerevisiae strains harboring eIF5A temperature-sensitive mutants exhibit diverse cellular changes (26–30), suggesting a direct or indirect role of eIF5A in cell wall integrity, mRNA decay, actin polarization, apoptosis and cell cycle progression (see a recent review (31)). It is not yet clear how depletion or dysfunction of eIF5A leads to the pleiotropic phenotypes of the temperature-sensitive eIF5A mutant strains.
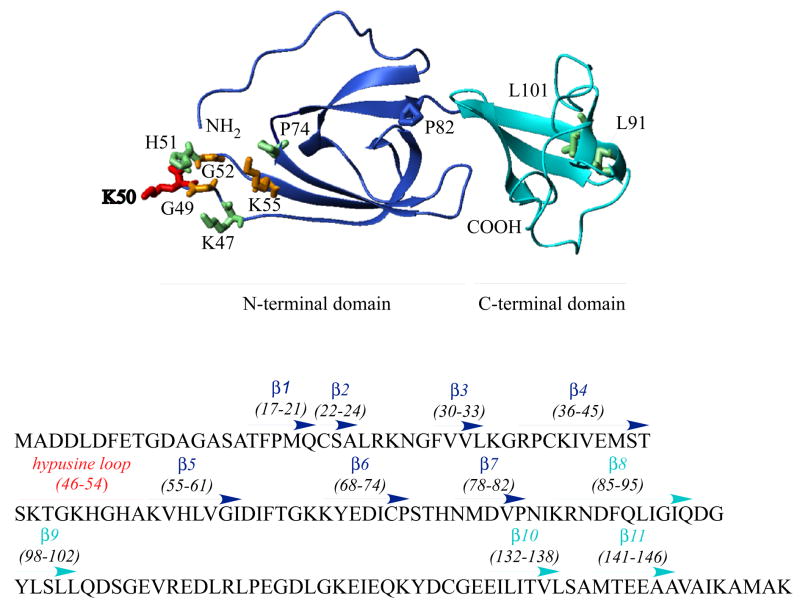
eIF5A is a small acidic protein, highly conserved from yeast to mammals. The archaeal (aIF5A) and bacterial (elongation factor P;EF-P) homologs also share significant sequence identity with eIF5A (32), although the eukaryotic proteins, eIF5As, have longer N- and C-terminal extensions than EF-Ps and aIFs (32). Hypusine modification has evolved in eukaryotes as no hypusine or hypusine modification enzymes have been identified in bacteria. To date, only DHS but no DOHH homologous genes have been identified in archaea (5). Despite the differences in N- and C-terminal sequences and the hypusine modification status, eIF5A and its homologs share significant structural similarities. Crystal structures determined for three aIF5As (PDB 1EIF, 1IZ6, 1BKB) (33–35) and the two Leishmania eIF5As (PDB 1X60, 1XTD) show remarkable overall similarity. The structure model of human eIF5A-1 (PDB 1FH4) based on secondary structure analysis and the structures of the archaeal proteins (36,37) consists of two domains, a basic N-terminal domain and an acidic C-terminal domain connected by a hinge (Scheme 1). The C-terminal domain resembles an oligonucleotide-binding fold of the E. coli cold shock protein and has been implicated in RNA binding. In the basic N-terminal domain, the lysine that undergoes hypusine modification is located at the tip of an exposed loop (Scheme 1, aa46–54). The amino acid sequence surrounding this modification site (STSKTGK50HGHAK) is very basic and hydrophilic. Addition of the 4-amino-2 hydroxybutyl moiety to the ε-amino group of Lys 50 creates a long basic side chain in this loop. The strict conservation of the hypusine loop sequence suggests that it, together with the hypusine residue, serves an essential basic function that has been preserved throughout eukaryotic evolution. The exposed hypusine loop may be the critical focal point of interaction between eIF5A and its downstream effectors, including ribosomes.
Scheme 1. Model structure of human eIF5A with critical amino acid residues.
The structure of human eIF5A-1 is based on the model (PDB 1FH4) constructed by Facchiano et al. (36). Both the N- and C-terminal domains (in blue and aqua-blue) consist of β-sheet core structures and are connected by a hinge at Asn83-Ile84. The hypusine modification site (Lys 50, in red) is located at an exposed loop (hypusine loop aa46–54) in the basic N-terminal domain. No growth is observed upon Ala substitution of the red and orange colored residues (Lys50, Gly49, Gly52 and Lys55) and slow growth upon Ala substitution of the green colored residues (Lys47, His55, Pro74, Leu91 and Leu101).
To better understand the molecular mechanisms of eIF5A action, it is critical to identify downstream effector molecules and to characterize the structural elements of eIF5A that are involved in these interactions. Several proteins have been reported as binding partners of eIF5A, including DHS (20,38,39) DOHH (yeast Lia1) (39), HIV-1 posttranscriptional activator REV (40), ribosomal protein L5 (41), nuclear actin (42), transglutaminase 2 (43), exportin 4 (23), ribosomes, ribosomal component proteins and ribosome associated proteins, Clu1, Ssb2p, Zuotin, eEF1A, eEF2, P0 and L11(21) (20). Molecular interactions have been extensively characterized for eIF5A binding to DHS (44), and DOHH in vitro (45), but not for other candidate binding partners, some of which may bind to eIF5A indirectly through bridging molecules like RNA. As such, identification of amino acid residues critical for eIF5A activity will provide new insights into molecular interactions and mechanisms by which eIF5A exerts its biological function. Our goal was to assemble an informative set of mutants to address three general questions: i) how stringent is the sequence requirement for eIF5A as substrates for DHS and DOHH? ii) are the N-and C-terminal extensions in the eIF5A functionally required? iii) which structural features of the hypusine loop, other than hypusine residue itself, contribute to eIF5A activity? To this end, we generated a number of human eIF5A mutant proteins through site-directed mutagenesis of each conserved amino acid and by truncation, and tested them as substrates for DHS and DOHH and assessed their activities in supporting growth and protein synthesis under an yeIF5A null background (19). Our data demonstrate that both N-and C-terminal domain β-sheet core structures are required for eIF5A activity and underscore the importance of the conserved hypusine site loop as a focal point for its post-translational modification and its biological action via downstream effectors. They also provide further evidence that eIF5A stimulates protein synthesis in vivo.
Experimental Procedures
Materials
[1,8-3H]Spermidine.HCl (15–25 Ci/mmol) was purchased from PerkinElmer/NEN. Precast Tris-glycine and NuPAGE (Bis-Tris) gels, electrophoresis buffers and Simply Blue staining solution were from Invitrogen, ECL Plus Western Blotting Detection system from GE Healthcare, the Quick Change Site-Directed Mutagenesis Kit from Stratagene. The protease inhibitor cocktail was purchased from Pierce and 5-fluoroorotic acid (5-FOA) from Sigma. A monoclonal antibody raised against recombinant human eIF5A (aa58–154) was from BD Biosciences. Rabbit polyclonal antibodies were produced using purified S. cerevisiae eIF5A (Tif51a). The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. The human and yeast recombinant eIF5A (46), recombinant DHS (47) and DOHH (5) proteins were purified as described previously.
Methods
Yeast strains and methods
HHY13a strain (Table I) with inactivation of both S. cerevisiae eIF5A genes (TIF51A and TIF51B) and whose growth is sustained by plasmid-born TIF51A under the GAL1 promoter (pBM-TIF51A, URA3) (48) was transformed with p414GAL1(TRP1) vectors encoding human eIF5A-1 wild type or mutant proteins. Trp+ transformant colonies harboring both yeast and human eIF5A proteins were purified on selection plates (SGal, -His,-Leu,-Trp,-Ura) to derive HHY212d strains and streaked on SGal medium containing 5-FOA (SGal,-His,-Leu,-Trp, plus 5-FOA) to select the strains (HHY212s) that lost the pBM-TIF51A plasmid and thereby express only human eIF5A proteins. UBHY-R strain (Table I) is a derivative of HHY13a with inactivation of both of S. cerevisiae eIF5A genes (TIF51A and TIF51B) and its growth is supported by a plasmid-born yeast eIF5A fusion protein containing the N-terminal ubiquitin-Arg destabilizing element (UBR5A) under the control of GAL promoter (19). This strain was transformed with p414GAL1(TRP1) plasmids encoding human wild type and mutant eIF5A and Trp+ transformants were selected on SGal, -His,-Leu,-Trp, -Ura medium to derive UBHY-R212d strains (Table I).
Table I.
Strains and plasmids
| Strain | Ref | |
|---|---|---|
| W303-1A | MATα leu2-3, 112his3-11,15ade2-1 ura3-1 trp1-1 can1-100 | (48) |
| HHY13 | MATα leu2 his3 ura3 trp1 can1 tif51A:: LEU2 tif51B::HIS3 (pBM-TIF51A) | (48) |
| UBHY-R | MATα leu2 his3 ura3 trp1 can1 tif51A:: LEU2 tif51B::HIS3 (YCpUB-R5A) | (19) |
| HHY212d | MATα leu2 his3 ura3 trp1 can1 tif51A:: LEU2 tif51B::HIS3 (pBM-TIF51A) (p414GAL1-heIF5A-1m) | This work |
| HHY212S | MATα leu2 his3 ura3 trp1 can1 tif51A:: LEU2 tif51B::HIS3 (p414GAL1-heIF5A-1m) | This work |
| UBHY-R 212d | MATα leu2 his3 ura3 trp1 can1 tif51A:: LEU2 tif51B::HIS3 (YCpUB-R5A) (p414GAL1-heIF5A-1m) | This work |
| Plasmid | ||
| pBM-TIF51A | CEN4, ARS1, AmpR, URA3, GAL10, TIF51A | (48) |
| YCpUB-R5A | CEN11, ARS1, AmpR, URA3, GAL10, UBR5A | (19) |
| P414GAL1-heIF5A-1m | CEN6, ARSH4, AmpR, TRP1, GAL1, heIF5A-1 mutants | This work |
Generation of recombinant plasmids encoding human eIF5A site-directed mutants and truncated proteins
The yeast and bacterial expression vectors (p414GAL1 and pET11a) encoding a number of human eIF5A mutants, including D3A, D4A, L5A, D6A, F7W, D11A, G13A, S15A, T17A, P19A, C22A, P37A, C38A, M43A, K47A, K47D, K47R, G49A, K50A, K50D, K50I, K50R, H51A, G52A, K55A, F64A, P74A, H77A, M79A, P82A, I84A, R86A, L91A, L101A, P115A, E116A, L119A, E144A, K150A, M43A/M79A, C73A/M79A were generated using the QuickChange Site-directed Mutagenesis Kit and employing p414GAL1/heIF5A-1 and pET11a/heIF5A-1 as templates. The vectors encoding truncated eIF5A were generated by subcloning the PCR-amplified ORFs into the BamHI-EcoRI sites of p414GAL1 and NdeI-BamHI sites of pET11a. The entire open reading frame of the site-mutated and truncated eIF5A was sequenced for confirmation of the intended mutation or truncation.
Determination of growth rates
Yeast strains were grown in YPGal medium (1% yeast extract, 2% bactotryptone, 2% Galactose) at 30 °C with shaking at 200 rpm starting with a small inoculum so that the culture would still be in exponential stage after overnight incubation. The overnight cultures were diluted in fresh medium to the density indicated and growth was monitored at 600 nm.
Detection and determination of yeIF5A and heIF5A proteins by western blots
Yeast cells were grown at 30 °C in rich or minimal medium containing galactose or glucose as indicated. Cells were harvested by centrifugation and cell pellets were resuspended in cold Tris buffer (50 mM Tris-HCl, pH 7.5, 1 mM DTT containing 2X yeast protease inhibitor cocktail). Yeast extracts were generated by breaking the cells with glass beads using BioSpec bead beater. Cycles of 10 seconds of agitation and 1 minute cooling on ice were repeated three times. Cell lysates were clarified by centrifugation at 14,000 rpm for 20 min at 4 °C. The total protein concentration of the lysate was determined using the Bio-Rad Protein Assay solution. ~ 20 μg of cellular protein was separated by SDS-PAGE (4–12% gel) in MES buffer. Proteins were transferred to 0.2 μm nitrocellulose membrane for immuno-detection using rabbit polyclonal antibody (1:5,000 dilution) against yeast eIF5A and mouse monoclonal antibody (1:10,000 dilution) against the human eIF5A-1 peptide, aa58–154. The signal was developed with an ECL Plus chemiluminescence reagent.
Measurement of Protein synthesis in cells
UBHY-R cells were grown to mid log phase (OD600 <1.0) and quickly washed in sterile room temperature water. For measurement of protein synthesis at 0 time, washed cell pellets (~1 OD unit) were resuspended in 0.2 ml of YPD medium containing 20 μCi of [3H]leucine for incubation. The rest of cells were diluted in YPD medium to ~0.125 (OD, 600 nm) and growth was monitored. To measure protein synthesis at later time points after medium shift (1, 3 and 5 h), cells (~1 OD unit) were centrifuged at room temperature and resuspended in 0.2 ml of YPD labeling medium (containing 20 μCi of [3H]leucine). After incubation of the cells with shaking at 30 °C for 20 min, protein synthesis was stopped by addition of 1 ml of ice cold stop solution containing 0.3 mg/ml of cycloheximide and 1 mg/ml of unlabeled leucine. After incubation on ice for 1 min, cells were harvested by centrifugation and cell pellets were frozen on dry ice. When all samples were collected for all the time points, 1 ml of 15% TCA solution was added to the cell pellets and the suspended samples were heated at 100 °C for 15 min. The samples were cooled on ice and the TCA precipitates were collected by centrifugation at 15,000 xg at 4 °C for 4 min. The precipitates were washed twice with 10 % TCA, dissolved in 0.1 ml of 0.2 N NaOH. 50 μl aliquots were used for measurement of radioactivity in a Beckman Scintillation Counter and 10 μl aliquots for determination of protein amounts by Bio-Rad protein assay. The rate of protein synthesis was calculated for each sample as dpm/μg/20 min.
Combined Deoxyhypusine synthase (DHS)/Deoxyhypusine hydroxylase (DOHH) assays
Human recombinant eIF5A wild type and mutant proteins were tested as substrates for DHS and DOHH in vitro as described previously (45). eIF5A protein expression was induced in E. coli. BL21(DE3) cells transformed with pET11a/heIF5A vectors by 1 mM isopropyl-β-D-thiogalactoside (IPTG) for 3 h. The cell pellets from 5 ml cultures were sonicated using Ultrasonic Processor in 0.2 ml of ice-cold Tris buffer (50 mM Tris-HCl, pH 7.5, 1 mM DTT) containing protease inhibitor cocktail. Cell debris was removed by centrifugation at 15,000 xg at 4 C for 20 min. Aliquots of the clarified lysates (1–10 μl) were used for SDS-PAGE to determine the level of eIF5A protein expression. Aliquots (1–10 μl) containing 2–5 μg of recombinant eIF5A proteins were used for the combined DHS/DOHH assays (45). The reaction mixture contained in 20 μl, 0.125 M Tris. HCl, pH 8.5, 6 mM DTT, 1 mM NAD, 25 μg BSA, 3 μCi [3H]spermidine, 0.1 μg of DHS and 1.0 μg of DOHH and clarified BL21(DE3) lysates containing human eIF5A proteins (2–5 μg). After incubation for 2 h at 37 °C, an aliquot of the reaction mixture was used for SDS-PAGE for fluorographic detection of the radiolabeled peptides. To the rest, 500 μg of carrier BSA was added and the proteins were precipitated with 10% TCA containing polyamines (putrescine, spermidine and spermine, 1 mM each). After removal of [3H]spermidine by repeated washing with 10 % TCA containing polyamines, the TCA precipitated proteins were hydrolyzed in 6 N HCl at 110 °C overnight and the radiolabeled hypusine and deoxyhypusine were measured after ion exchange chromatographic separation as described (49).
Results
Identification of amino acid residues of human eIF5A vital for its activity in supporting yeast growth
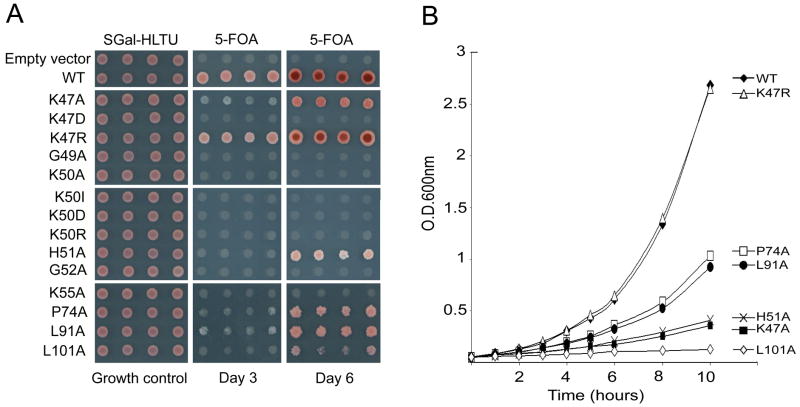
S. cerevisiae contains two eIF5A genes TIF51A and TIF51B that encode two isoforms with highly similar amino acid sequences (92 % amino acid sequence identity). These two genes are reciprocally regulated by oxygen, TIF51A being expressed under aerobic condition and TIF51B only under anaerobic conditions (50). However, the two yeast proteins appear to be functionally identical in S. cerevisiae, since either can support the growth under aerobic as well as anaerobic conditions (48). Furthermore, expression of either of the two human eIF5A isoforms can substitute for the yeast eIF5A (48,51), suggesting functional conservation of eIF5A from yeast to human. In order to identify structural elements of eIF5A required for its activity, we targeted all the highly conserved amino acid residues (including ~30 strictly conserved ones) of human eIF5A-1 by site-directed mutagenesis and generated 41 mutant proteins, including D3A, D4A, L5A, D6A, F7W, D11A, G13A, S15A, T17A, P19A, C22A, P37A, C38A, M43A, K47A, K47D, K47R, G49A, K50A, K50D, K50I, K50R, H51A, G52A, K55A, F64A, P74A, H77A, M79A, P82A, I84A, R86A, L91A, L101A, P115A, E116A, L119A, E144A, K150A, M43A/M79A, C73A/M79A. The ability of each mutant to complement growth of HHY13a haploid strain (Table I) was evaluated by the plasmid shuffle technique (52). In this strain, both yeast eIF5A genes (TIF51A and TIF51B) are inactivated and growth is supported by yeast eIF5A expressed from pBM-TIF51A (URA3). HHY13a was transformed with p414GAL1 (TRP1) recombinant vectors encoding human eIF5A proteins and Trp+ transformants expressing both yeast and human eIF5A proteins were selected on SGal,-His,-Leu,-Trp,-Ura plates (Table 1, HHY212d strains). Purified colonies were resuspended in water and spotted on selection plates (Fig. 1A, left side panels) and on plates containing 5-fluoroorotic acid (SGal,-His,-Leu,-Trp plus 5-FOA) to select those strains that lost the pBM-TIF51A plasmid and thereby express only human eIF5A proteins (Table 1, HHY212s strains). Surprisingly, the majority of human mutant proteins could substitute for yeast eIF5A in supporting growth. Although we characterized each of the 41 mutants listed above by the plasmid shuffle technique (52), data are shown only for mutations at specific sites, i.e. the conserved hypusine region, and those that displayed growth defects.
Fig. 1. Growth analysis of S. cerevisiae strains expressing human eIF5A wild type and mutant proteins.
(A) The haploid strain HHY13a was transformed with recombinant p414GAL1 plasmid encoding human eIF5A-1 wild type or mutant proteins with single amino acid substitutions, indicated on the left side. Trp+ transformants were selected on minimal galactose plates (SGal,-His, -Leu, -Trp, -Ura). Four individual transformant colonies (HHY212d) were resuspended in water and spotted in parallel on the same selection plates (left panels) and on the 5-FOA containing plates (SGal, -His,- Leu, -Trp, plus 5-FOA) (middle and right panels) to derive HHY212s that lost pBM-TIF51A. The plates were photographed after incubation at 30 C for 2 days without 5-FOA (left panels), or 3 and 5 days with 5-FOA (middle and right panels).
(B), Growth curves of HHY212s strains harboring only the human eIF5A proteins. The experiments were repeated two to three times with virtually the same results: a typical experiment is shown.
Only eight mutations at five sites, K47D, G49A, K50A, K50I, K50D, K50R, G52A, and K55A, caused total inactivation of eIF5A in supporting yeast growth, judging from the lack of growth on 5-FOA plates (Fig. 1A, middle and right panels). It is interesting that the residues identified to be critical for eIF5A function, namely Gly49, Lys50, Gly52 and Lys55, are all clustered around the hypusine modification site (Scheme 1). At no other site tested did alanine substitution abolish the growth supporting activity of eIF5A. The inability of the yeast eIF5A mutant protein K51R (Lys51 is the hypusine modification site of yeast eIF5A) to rescue the eIF5A null strain was previously reported (6). That no other amino acid, including Asp, Ala and Ile, can substitute for the Lys at residue 50 confirms the absolute requirement for deoxyhypusine/hypusine for eIF5A activity in cell proliferation.
Five other mutations (K47A, H51A, P74A, L91A and L101A) also seemed to impair eIF5A function, since strains expressing these mutant eIF5A grew at much slower rates than that expressing the wild type protein (Fig. 1). Upon selection on 5-FOA, there was only minimal growth on day 3, but growth was apparent by day 5 for the strains carrying these mutations (Fig. 1A). Moreover, varying growth rates were observed among these mutant strains. The slow growth phenotype was confirmed in liquid culture containing YPGal (Fig. 1B). Doubling times were estimated to be 234.4± 5.2, 113.8±10.8, 238.7±11.3, 144.5±14.2, 150.1±12.5 and 444.0±32.6 minutes for K47A, K47R, H51A, P74A, L91A and L101A, respectively, while the growth rate for the strain expressing wild type human eIF5A was 112.3±6.8 min. It is noteworthy that three different substitutions at Lys47 resulted in distinct growth phenotypes: K47R displayed normal growth, K47A, reduced growth, whereas no growth was observed with the K47D mutation.
Expression and stability of human eIF5A mutant proteins in yeast
The deficiency of certain human eIF5A mutant constructs in supporting growth may be due to inactivity or instability of the mutant proteins. Therefore, we examined the expression levels of these human eIF5A mutant proteins and yeast eIF5A before (Fig. 2A, HHY212d strains) and after 5-FOA selection (Fig. 2B, HHY212s strains) using specific antibodies. Although human eIF5A-1 is functionally similar to yeast eIF5A in complementing growth of HHY13a null strain, there is no cross-reactivity between anti-human and anti-yeast eIF5A antibodies. In addition to yeIF5A, most human eIF5A mutant proteins were readily detectable in HHY212d strains harboring both recombinant plasmids pBM-TIF51A and p414GAL1-heIF5A (Fig. 2A). Only two mutant proteins, L91A and L101A, were markedly reduced. The L91A protein level was low, and L101A was undetectable, suggesting instability of these two mutated proteins. In HHY212s strains harboring wild type, K47A, K47R, H51A, P74A, L91A and L101A, only human eIF5A proteins, but no yeast eIF5A protein, were detected (Fig. 2B), suggesting that these human eIF5A proteins can either fully or partially substitute for yeast eIF5A in supporting S. cerevisiae growth. Interestingly, the levels of the two unstable mutant proteins, L91A and L101A, were notably increased after 5-FOA selection (Fig. 2B vs. Fig. 2A). This may reflect stabilization of the mutant proteins by their association with eIF5A partners and engagement in eIF5A function when present as the sole eIF5A source. Although the L101A signal was consistently enhanced and clearly visible after 5-FOA selection (compare Fig. 2B with 2A), its level was much lower than other heIF5A proteins (Fig. 2B). The slow growth of HHY212s strain bearing L101A (Fig. 1B) may be caused by its reduced level, as well as its reduced activity. On the other hand, the slow growth rate of the strains expressing the apparently stable human eIF5A mutants, K47A, H51A, and P74A (Fig. 1A and 1B) may be largely due to compromised activity of these mutant proteins.
Fig. 2. Expression and stability of human eIF5A wild type and mutant proteins in S. cerevisiae strains.
Western blot analyses of proteins of HHY212d strains harboring both recombinant plasmids, pBM-TIF51A and p414GAL1-heIF5A (A) and of HHY212s strains expressing only the human eIF5A proteins (B). All the strains in A were cultured in SGal,-His,-Leu,-Trp,-Ura and those in B, cultured in SGal,-His,-Leu,-Trp. The experiments were repeated two times with virtually the same results: a typical experiment is shown.
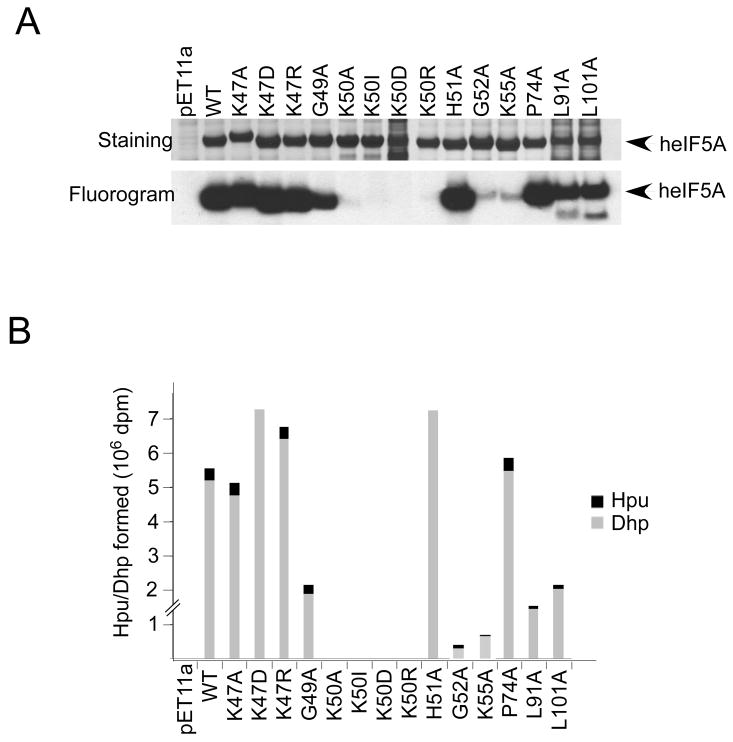
Human eIF5A mutant proteins as substrates for deoxyhypusine synthase and deoxyhypusine hydroxylase
The impaired eIF5A activity of human eIF5A mutants could be due to their deficiency as substrates for deoxyhypusine/hypusine modification. We, therefore, tested recombinant heIF5A mutant proteins as substrates for S. cerevisiae DHS and DOHH in a combined in vitro assay. While all the mutant proteins were overexpressed in BL21(DE3) E. coli cells, the levels of K50D, L91A, and L101A mutant proteins were lower than others in the bacterial lysates, presumably due to their instability. Upon reaction of the lysates with DHS and DOHH in the presence of [3H]spermidine, strong radiolabeling of eIF5A proteins was observed for the wild type and the mutant proteins K47A, K47D, K47R, G49A, H51A, P74A, L91A and L101A (Fig. 3A), indicating that these mutant proteins are effective substrates for DHS. Thus slow or no growth of the HHY212s strains carrying these mutant proteins (K47A, K47D, G49A, H51A, P74A, L91A and L101A) (Fig. 1) cannot be attributed to impaired deoxyhypusine modification. As expected, no labeling was observed for any of the substitution mutants at Lys50 (the hypusine modification site) (Fig. 3A). Only very weak radiolabeling was observed for G52A and K55A (less than 4 % of wild type), indicating poor substrate property of these two mutant proteins toward DHS. The radiolabeled proteins were hydrolyzed and analyzed for their content of deoxyhypusine and hypusine (Fig. 3B). Radiolabeled hypusine was formed in K47A, K47R, G49A, P74A, L91A and L101A mutant proteins indicating that they acted as substrates for DOHH as well as for DHS. In contrast, only radioactive deoxyhypusine was detected in the mutants K47D and H51A. The fact that H51A mutant, which does not undergo hydroxylation, can support yeast growth at all is consistent with the viability of the DOHH null S. cerevisiae strain (5). The basic charges of Lys47 and His51 may be important for the interaction of the eIF5A substrate with DOHH. It is evident that all substitution mutants at Lys50 are totally inactive in supporting yeast growth, underscoring the notion that the deoxyhypusine/hypusine modification is absolutely required for eIF5A activity. Two other eIF5A mutants, G52A and K55A, also failed to support yeast growth, partly because they are poor substrates for DHS (Fig. 3) and probably because Gly52 and Lys55 contribute to the proper configuration of the hypusine loop in eIF5A binding to its downstream effectors. In contrast to G52A and K55A, the K47D and G49A mutant proteins were effective substrates for DHS, yet no growth was observed in the strains expressing these two mutant proteins, pointing to important roles of Lys 47 and Gly52 in downstream biological events of eIF5A action. Another mutant, P74A, also showed impaired eIF5A activity (Fig. 1), without apparent loss of its stability (Fig. 2) or its modification by DHS (Fig. 3), suggesting a potential role of Pro74 in eIF5A/partner interactions.
Fig. 3. Human eIF5A mutant proteins as substrates for deoxyhypusine synthase and deoxyhypusine hydroxylase in vitro.
Human recombinant eIF5A proteins were expressed in E. coli, BL21(DE3) and cell lysates were used as substrates for DHS and DOHH in a combined assay. Coomassie-Blue staining of BL21(DE3) lysates expressing human mutant proteins (A, top panel) and fluorogram of the SDS gel of DHS/DOHH reaction mixture showing labeling of several mutant proteins (A, bottom panel). Portions of DHS/DOHH reaction mixtures were analyzed for radioactive deoxyhypusine and hypusine content in the products by ion exchange chromatographic separation (B) as described previously (49). The experiments were repeated two to three times with virtually the same results: a typical experiment is shown.
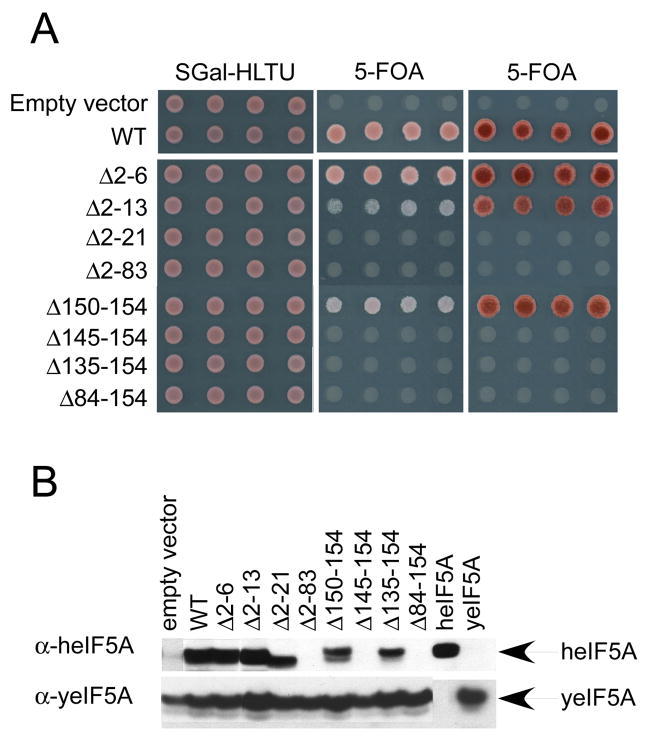
Effects of N- or C-terminal truncation of eIF5A on its stability and activity
eIF5A has longer N- and C-terminal extensions than the bacterial (EF-P) and archaeal (aIF5A) homologs (32). In order to determine whether these terminal extensions are functionally significant and whether intact N- or C-terminal domains of eIF5A are required for eIF5A activity, we generated truncated eIF5A mutants. eIF5A deletions of six or thirteen amino acids from the N-terminus or five amino acids from the C-terminus supported growth, whereas no growth was observed with further truncations (Fig. 4A). When the expression levels of truncated human eIF5A proteins in HHY212d strains were examined (Fig. 4B), truncated human eIF5A proteins Δ2-6, Δ2–13, Δ 2–21, Δ150–154 and Δ135–154 were clearly detectable with a commercial monoclonal antibody generated against human recombinant eIF5A-1 peptide (aa58–154). Three other peptides, the N-terminal domain (Δ84–154), C-terminal domain (Δ2–83) and Δ145–154, were not detected (either due to their inability to be recognized by the monoclonal antibody or due to their instability) and no direct conclusion can be drawn on their activity. That Δ2–13 and Δ150–154 proteins are functional suggests that the N- and C-terminal extensions outside of β-sheet cores are not required for eIF5A activity in yeast. However, the finding that Δ2–21 and Δ135–154 are inactive, while being stably expressed, indicates that eIF5A cannot afford to lose more than 20 amino acids from either terminus and that both N- and C-terminal core domains are required for eIF5A activity. In our previous study, we determined the structural requirements of eIF5A as substrates for DHS and DOHH (45,46). A large polypeptide (>aa20–90), encompassing almost the entire N-terminal domain, is required to be an effective substrate for both enzymes. Therefore, whereas only the N-terminal core domain is sufficient for modification by DHS and DOHH, both the N-terminal domain β-sheet core structure (aa17–82) and the C-terminal domain β-sheet core structure (aa85–146) (Scheme 1), seem to be required for the biological activity of eIF5A in cells.
Fig. 4. Growth analysis of S. cerevisiae strains expressing truncated human eIF5A and expression and stability of truncated proteins.
Growth analysis was performed as described in Fig. 1 (A) and western blots of proteins of HHY212d strains harboring both recombinant plasmids, pBM-TIF51A and p414GAL1-heIF5A (B) are shown. The strains were cultured in minimal galactose medium (SGal,-His,-Leu,-Trp,-Ura). The experiments were repeated two to three times with virtually the same results: a typical experiment is shown.
Effects of eIF5A depletion on growth and protein synthesis in S. cerevisiae
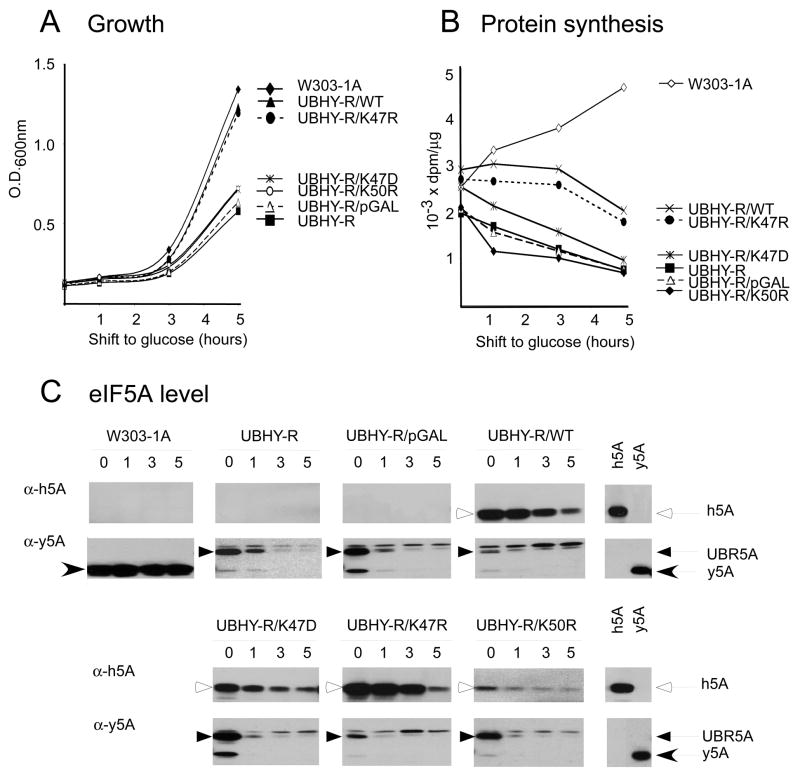
Since its discovery, eIF5A has remained as a putative translation initiation factor for decades, for its mode of action in translational control has yet to be elucidated. The role of eIF5A in translation has re-emerged recently with molecular evidence for its association with ribosomes actively engaged in translation (20,21). Furthermore, careful reevaluation of protein synthesis in the UBHY-R strain revealed a pronounced depression of protein synthesis and a decrease in polysomes upon UBR5A depletion in YPD medium (starting with a high basal level of protein synthesis) (CA Henderson and JWB Hershey, unpublished results), in contrast to modest changes in minimal medium (19). Therefore, we assessed the role of human eIF5A wild type and mutant proteins in growth and protein synthesis in rich medium (Fig. 5A and 5B). Growth of UBHY-R is supported by unstable ubiquitin-Arg-yeIF5A fusion protein (UBR5A) expressed under the control of the GAL promoter. Upon shift from a galactose medium (YPGal) to a glucose medium (YPD), transcription from the GAL promoter is turned off leading to a halt in new synthesis of UBR5A and rapid degradation of preexisting UBR5A (Fig. 5C). The protein synthesis rate in UBHY-R decreased after shift to glucose medium (YPD) to ~50 % at 1 h and to ~30 % after 5 h, while that in the wild type strain W303-1A increased in YPD medium. Cell growth slowed in accordance with inhibition of protein synthesis. Although we observed a much greater inhibition of protein synthesis in rich YPD medium than that previously reported in minimal medium, complete inhibition of protein synthesis was not observed upon depletion of UBR5A. There was a basal level of [3H]leucine incorporation (~30 % of control), even when UBR5A was virtually depleted.
Fig. 5. The effects of human wild type and mutant eIF5A expression on growth and protein synthesis in UBHY-R.
Exponential cultures (in YPGal medium) of W303-1A, UBHY-R, and UBHY-R strains transformed with p414GAL1 vectors encoding human eIF5A-1 wild type or mutant proteins K47A, K47R, and K50R were washed with water, resuspended in YPD medium at the density of ~0.125 (OD 600 nm) and growth was followed for 5 h (A). At 0, 1, 3, and 5 h after shift to the glucose medium, aliquots of cells were used to measure protein synthesis as described under Experimental Procedures and the rate of protein synthesis was calculated for each sample as dpm/μg/20 min (B). The levels of human and yeast eIF5A and yeast UBR5A proteins were determined by western blot analysis (C). The experiments were repeated two to three times with virtually the same results: a typical experiment is shown.
In order to determine the activity of human eIF5A wild type and mutant proteins in translation, we transformed the UBHY-R strain with p414GAL1 empty vector or the recombinant vectors expressing human wild type eIF5A and three mutant eIF5A proteins, K47D, K47R, and K50R. We chose to analyze the early stages of culture after shift to glucose medium in order to avoid indirect effects of eIF5A depletion on protein synthesis, which may be caused by decreased growth rate or other factors. Expression of wild type eIF5A or functional mutant K47R supported growth of UBHY-R in glucose medium in the first 5 h (Fig. 5A) at a rate slightly lower than that in W303-1A. Unlike UBR5A which decays rapidly upon shift to YPD, native human eIF5A is a stable protein with a long half life. Even though new synthesis of the human eIF5A proteins would be halted upon shift to glucose medium due to transcriptional repression, they did not decay rapidly (Fig. 5C, see α-heIF5A signals 0–5 h). The levels of human eIF5A proteins were reduced mainly due to dilution during growth. In the initial period (0 to 5 h), the protein synthesis rate gradually declined, reaching to ~70% of the initial rate at 5 h for both WT and K47R, presumably due to the decreased eIF5A protein level. In contrast, expression of nonfunctional eIF5A mutants K47D and K50R did not cause any significant enhancement in the growth rate or protein synthesis over those in UBHY-R or UBHY-R carrying an empty vector. Parenthetically, the UBR5A level (at 0 time) was markedly reduced in strains UBHY-R/WT and UBHY-R/K47R harboring a high level of functional human eIF5A proteins, compared to those in strains not expressing human protein (UBHY-R, or UBHY-R/pGAL) or expressing nonfunctional human mutant proteins (K47D and K50R) (Fig. 5C). One plausible explanation is that the UBR5A expression plasmid is more likely to be retained when cells are solely dependent on UBR5A for survival, than when they are not absolutely required, in the presence of functional heIF5A expression. Alternatively, it may reflect stabilization of UBR5A protein upon its functional engagement by association with eIF5A binding partners. Conversely, the levels of functional human proteins (WT and K47R) were higher than those of the nonfunctional proteins (K47D and K50R). This may be partly due to differential loss of plasmids encoding active vs. inactive mutants and/or stabilization of active proteins through their functional engagement. Taken together, these results confirm an essential role of eIF5A in cell proliferation and support a primary role of eIF5A in translational control.
Discussion
eIF5A is unique in that it is the only cellular protein activated by hypusine modification. eIF5A consists of two β-sheet core domains, a basic N-terminal domain with an exposed hypusine site loop and an acidic C-terminal domain (Scheme 1). The high sequence and structural conservation of eIF5A may be dictated by structural requirements for its interaction with the hypusine modification enzymes, eIF5A downstream effectors or both. We undertook a comprehensive mutagenesis study of human eIF5A-1 to dissect the structural elements of this protein required for its biological activity and for its hypusine modification and thereby to gain insights into its function. In spite of the high sequence conservation of eIF5A, it was remarkably resilient to individual alanine substitutions and a majority of mutant proteins actively supported S. cerevisiae growth. We have identified only a very few amino acid residues in the exposed hypusine loop as the critical sites for its activity (Scheme 1 and Table II). Our results confirm the absolute requirement for the deoxyhypusine/hypusine modification in eIF5A function and provide new evidence that the hypusine loop is critical for its interaction with downstream effector molecules and that the β-sheet core structures of both the N- and C-terminal domains of eIF5A (Scheme 1, aa17–82 and aa85–146) are essential for eIF5A biological function.
Table II.
Summary of characteristics of heIF5A mutant proteins.
Abbreviations, s.g.= slow growth. n.d. = not determined
| Mutant | Substrate for | Growth | Protein synthesis | |
|---|---|---|---|---|
| DHS | DOHH | |||
| Wild type | +++ | + | + | + |
| K47A | +++ | + | s.g. | n.d. |
| K47D | +++ | − | − | − |
| K47R | +++ | + | + | + |
| G49Aa | +++ | + | − | n.d. |
| K50A | − | − | − | n.d. |
| K50D | − | − | − | n.d. |
| K50I | − | − | − | n.d. |
| K50R | − | − | − | − |
| H51A | +++ | − | s.g. | n.d. |
| G52Aa | + | + | − | n.d. |
| K55A | + | + | − | n.d. |
| P74A | ++ | + | s.g. | n.d. |
| L91Ab | ++ | + | s.g. | n.d. |
| L101Ab | ++ | + | s.g. | n.d. |
In addition to the deoxyhypusine/hypusine at residue 50, these two glycine residues are likely the most critical residues in eIF5A/effector interactions.
These proteins appear to be unstable and their cellular levels were low.
The loss of eIF5A function in various mutants may result from their defects in effector binding, instability and/or inability to be modified by DHS. Careful analysis of properties of the selected eIF5A mutant proteins, summarized in Table II, revealed distinct sequence requirements for its growth supporting activity and as substrates for DHS and DOHH. We have identified Lys47, Gly49, Dhp/Hpu50, Gly52 and Lys55 as those residues vitally important for the biological activity of eIF5A in supporting yeast growth (Scheme 1). The absolute requirement of the deoxyhypusine/hypusine residue is confirmed by the total lack of activity of Lys50 site mutants, including K50A, K50D, K50I and K50R (regardless of substituting amino acid), when these mutant proteins are stably expressed (Fig. 2A). Beside Lys50, the two neighboring Gly residues, Gly49 and Gly52 are probably most critical in eIF5A activity. No growth support was observed, even though both mutant proteins G49A and G52A are stable in S. cerevisiae (Fig. 2A) and are modified by DHS and DOHH, albeit at reduced efficiency. Being adjacent to Lys50, these two Gly residues are likely to be critical for the β-turn structure of -Gly-X-Y-Gly- motif, the proper orientation of deoxyhypusine/hypusine side chain and the precise configuration of the hypusine loop in its binding to effectors. K55A human mutant is totally inactive in supporting growth, although Lys55 is located adjacent to the hypusine loop (aa46–54, Scheme 1). In an independent yeast eIF5A mutation study, the yeast K56A, a counterpart of human K55A, is modified by yeast DHS and supports S. cerevisiae growth at 25 °C (Dias et. al., Unpublished results), however it does exhibit a temperature sensitive growth phenotype at 37 °C, in spite of its stability. Judging from this temperature sensitive phenotype of yeast K56A and its location at the base of the hypusine loop, Lys55 is not likely to be centrally involved in effector binding. Instead, it may indirectly affect the proper orientation of the hypusine loop. Comparison of the three different heIF5A mutants, K47A, K47D and K47R, provides an interesting clue on the role of Lys47. The three mutant proteins are all effectively modified by DHS, yet widely differ in their growth supporting activity. K47R supports growth as well as the wild type protein, and K47A does at a reduced rate. In contrast, no growth is observed upon expression of the K47D mutant. Therefore, the basic residue at 47 is predicted to be involved in an ionic interaction with an acidic effector adaptor site. Two other mutants, H51A and P74A, supported growth but at much reduced rates (Fig. 1B), suggesting that these proteins are not functioning optimally. Since they are apparently stable and are effectively modified by DHS, alanine substitution of His51 or Pro74 may adversely affect effector binding.
Since the amino acid sequence surrounding the hypusine residue (STSKTGHpu50HGHAKVH) is very basic and hydrophilic, this loop may interact with specific nucleotide sequences of RNA (52), acidic proteins or ribonucleoprotein complexes. The β-sheet structure of the C-terminal domain of eIF5A resembles an oligonucleotide binding fold and has also been implicated in RNA binding. This C-terminal domain contains a stretch of highly conserved hydrophobic amino acids (89-FQLIGIQDGYLSLL-102) that was proposed as a potential effector domain involved in protein-protein interaction (53). Indeed, alanine substitution of Leu91 or Leu101 caused a reduction in growth rate. Analyzing the position of both amino acids in the human eIF5A-1 model, Leu91 and Leu101 are localized at the hydrophobic core of the β-barrel (Scheme 1). Substitution of either of the two leucines by alanine could easily disrupt the tertiary structure. Without a properly folded β-barrel, the mutant proteins are likely more sensitive to to proteolytic degradation as evidenced from their reduced level in HHY212d strains and in BL21(DE3) lysates. Moreover, they may lose important interactions with downstream effectors. The growth defect of L101A may be largely due to instability of the mutant protein, since its level is drastically reduced (Fig. 2). Furthermore, the yeast L102A counterpart exhibits a temperature sensitive phenotype, being unstable only at the non-permissive temperature (29). Since the growth defect is observed even at 25–30 C for the human L101A mutation, the severity of destabilization differs in the two species.
Although DHS and DOHH enzymes are totally specific for eIF5A and likely recognize the β-sheet core structure of eIF5A N-terminal domain (45,46), specific sequence requirements for their substrates were unknown. As for the DHS reaction, in addition to the key residue Lys50, Gly52 and Lys55 seem important for DHS reaction, judging from markedly reduced efficiency of deoxyhypusine synthesis in G52A and K55A mutant proteins. The charge at residue 47 of eIF5A apparently does not matter, since all three mutants, K47A, K47D and K47R are excellent substrates for DHS (Fig. 3). However, in contrast to DHS, DOHH discriminates against K47D, while effectively hydroxylating K47A and K47R. Another residue, His51, also is important for the DOHH, but not for the DHS reaction, since alanine substitution at this site blocks hydroxylation without affecting deoxyhypusine synthesis. Our recent eIF5A/DOHH binding study revealed that the four conserved glutamic acid residues of the DOHH active site are critical for substrate binding (45), anchoring the deoxyhypusine side chain of eIF5A(Dhp) and possibly other basic residues in the vicinity. Thus, neighboring basic residues, such as Lys47 and His51 of the eIF5A substrate, may also contribute to its binding to DOHH through ionic interaction with the enzyme’s active site glutamic acids.
The role of eIF5A and its hypusine modification in translation has been a longstanding mystery. eIF5A enhances methionyl-puromycin synthesis in a deoxyhypusine/hypusine-dependent manner in vitro (17,18). Recently, it was shown that eIF5A binds to actively translating ribosomes and that conditional mutants of eIF5A are super-sensitive to protein synthesis inhibitors (20,21). The aIF5A structure is partially superimposable on its bacterial ortholog, elongation factor P (EF-P) which contains a third domain and resembles the structure of tRNA. Whether there is an actual functional mimicry, over ribosome association, to accompany the tRNA-related structures of eIF5A/aIF5A/EF-P will require further experimentation. Addition of modified eIF5A (eIF5A(Dhp)) to an eIF5A-depleted lysate of UBHY-R strain enhances total protein synthesis by two-fold (CA Henderson and JWB Hershey, unpublished results) in vitro, whereas no enhancement is observed with unmodified eIF5A precursor. Expression of human eIF5A (wild type or K47R) in the UBHY-R strain restores protein synthesis in vivo as well as growth. All these findings are consistent with a role of eIF5A in translation. However, rapid eIF5A depletion did not result in complete inhibition of protein synthesis in vitro or in vivo. While eIF5A is a cellular factor essential for cell proliferation and viability, there seems to be no absolute requirement for it for global protein synthesis. Instead, it may be required for optimal and balanced translation of a large number of endogenous mRNAs, especially ones involved in cell-cycle progression. Recently, genes involved in actin polarization, a process necessary for the G1/S transition in yeast, were isolated as high-copy suppressors of temperature-sensitive eIF5A mutants (28). It remains to be determined whether eIF5A affects the translation of overall mRNAs or a subpopulation of mRNAs that are critical for cell proliferation.
eIF5A activity in vitro and in vivo depends not only on a long and basic deoxyhypusine/hypusine side chain, but also on a specific configuration of the exposed hypusine loop (aa46–54) with Gly-Dhp/Hpu-His-Gly- tip structure properly oriented by neighboring residues such as Lys55 and a basic charge at residue 47. Such a precise structural requirement suggests that this deoxyhypusine/hypusine loop docks into a defined rigid space of its biological effector molecule. It is a great future challenge to identify this binding pocket of eIF5A partner molecules (proteins, RNA or complexes) and to elucidate the mode of eIF5A action.
Acknowledgments
We thank Edith C. Wolff (NIDCR/NIH) for helpful discussion and critical reading of our manuscript.
Abbreviation footnote
- aIF5A
archaeal initiation factor 5A
- EF-P
elongation factor P
- eIF5A
eukaryotic initiation factor 5A
- eIF5A-1
major isoform of eIF5A
- eIF5A(Lys)
eIF5A precursor
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- yeIF5A
yeast eIF5A
- heIF5A
human eIF5A
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- ORF
open reading frame
- 5-FOA
5-fluoroorotic acid
- UBR5A
ubiquitin-arginine-yeast eIF5A fusion protein
- SGal
Synthetic minimal medium containing galactose
- YPGal
rich medium containing galactose
- YPD
rich medium containing glucose
References
- 1.Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Proc Natl Acad Sci U S A. 1983;80(7):1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park MH. J Biochem (Tokyo) 2006;139(2):161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolff EC, Lee YB, Chung SI, Folk JE, Park MH. J Biol Chem. 1995;270(15):8660–8666. doi: 10.1074/jbc.270.15.8660. [DOI] [PubMed] [Google Scholar]
- 4.Tao Y, Chen KY. J Biol Chem. 1995;270(41):23984–23987. doi: 10.1074/jbc.270.41.23984. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Proc Natl Acad Sci U S A. 2006;103(1):51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Mol Cell Biol. 1991;11(6):3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wohl T, Klier H, Ammer H, Lottspeich F, Magdolen V. Mol Gen Genet. 1993;241(3–4):305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki K, Abid MR, Miyazaki M. FEBS Lett. 1996;384(2):151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 9.Park MH, Joe YA, Kang KR. J Biol Chem. 1998;273(3):1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- 10.Byers TL, Ganem B, Pegg AE. Biochem J. 1992;287(Pt 3):717–724. doi: 10.1042/bj2870717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerner EW, Mamont PS, Bernhardt A, Siat M. Biochem J. 1986;239(2):379–386. doi: 10.1042/bj2390379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay MK, Tabor CW, Tabor H. Proc Natl Acad Sci U S A. 2003;100(24):13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen KY, Liu AY. Biol Signals. 1997;6(3):105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K. Biochem J. 2005;385(Pt 3):779–785. doi: 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caraglia M, Marra M, Giuberti G, d’Alessandro AM, Budillon A, del Prete S, Lentini A, Beninati S, Abbruzzese A. Amino Acids. 2001;20(2):91–104. doi: 10.1007/s007260170050. [DOI] [PubMed] [Google Scholar]
- 16.Benne R, Brown-Luedi ML, Hershey JW. J Biol Chem. 1978;253(9):3070–3077. [PubMed] [Google Scholar]
- 17.Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. J Biol Chem. 1989;264(31):18527–18530. [PubMed] [Google Scholar]
- 18.Park MH. J Biol Chem. 1989;264(31):18531–18535. [PubMed] [Google Scholar]
- 19.Kang HA, Hershey JW. J Biol Chem. 1994;269(6):3934–3940. [PubMed] [Google Scholar]
- 20.Jao DL, Chen KY. J Cell Biochem. 2006;97(3):583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- 21.Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. Biochem Biophys Res Commun. 2006;348(4):1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 22.Hanauske-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano-Coico L, Hanauske AR, McCaffrey T, Szabo P. FEBS Lett. 1995;366(2–3):92–98. doi: 10.1016/0014-5793(95)00493-s. [DOI] [PubMed] [Google Scholar]
- 23.Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay UDG. EMBO J. 2000;19(16):4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu A, Jao DL, Chen KY. Biochem J. 2004;384(Pt 3):585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner S, Klug G. J Biol Chem. 2007 doi: 10.1074/jbc.M701166200. [DOI] [PubMed] [Google Scholar]
- 26.Zuk D, Jacobson A. Embo J. 1998;17(10):2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetics. 2002;160(2):393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanelli CF, Valentini SR. Genetics. 2005;171(4):1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee I, Gross SR, Kinzy TG, Chen KY. Mol Genet Genomics. 2006;275(3):264–276. doi: 10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- 30.Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. J Biol Chem. 2006;281(46):35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- 31.Zanelli CF, Valentini SR. Amino Acids. 2007 doi: 10.1007/s00726-007-0533-0. in press. [DOI] [PubMed] [Google Scholar]
- 32.Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Proc Natl Acad Sci USA. 2004;101(26):9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KK, Hung LW, Yokota H, Kim R, Kim SH. Proc Natl Acad Sci U S A. 1998;95:10419–10424. doi: 10.1073/pnas.95.18.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peat TS, Newman J, Waldo GS, Berendzen J, Terwilinger TC. Structure. 1998;6:1207–1214. doi: 10.1016/s0969-2126(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 35.Yao M, Ohsawa A, Kikukawa S, Tanaka I, Kimura M. J Biochem (Tokyo) 2003;133(1):75–81. doi: 10.1093/jb/mvg011. [DOI] [PubMed] [Google Scholar]
- 36.Facchiano AM, Stiuso P, Chiusano ML, Caraglia M, Giuberti G, Marra M, Abbruzzese A, Colonna G. Protein Eng. 2001;14(11):881–890. doi: 10.1093/protein/14.11.881. [DOI] [PubMed] [Google Scholar]
- 37.Costa-Neto CM, Parreiras ESLT, Ruller R, Oliveira EB, Miranda A, Oliveira L, Ward RJ. Biochem Biophys Res Commun. 2006;347(3):634–640. doi: 10.1016/j.bbrc.2006.06.119. [DOI] [PubMed] [Google Scholar]
- 38.Lee YB, Joe YA, Wolff EC, Dimitriadis EK, Park MH. Biochem J. 1999;340(Pt 1):273–281. [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson GM, Cano VS, Valentini SR. FEBS Lett. 2003;555(3):464–468. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- 40.Ruhl M, Himmelspach M, Bahr GM, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington GK, Probst H, Bevec D, et al. J Cell Biol. 1993;123(6 Pt 1):1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schatz O, Oft M, Dascher C, Schebesta M, Rosorius O, Jaksche H, Dobrovnik M, Bevec D, Hauber J. Proc Natl Acad Sci U S A. 1998;95(4):1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann W, Reichart B, Ewald A, Müller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, Dabauvalle MC. J Cell Biol. 2001;152(5):895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh US, Li Q, Cerione R. J Biol Chem. 1998;273(4):1946–1950. doi: 10.1074/jbc.273.4.1946. [DOI] [PubMed] [Google Scholar]
- 44.Lee CH, Um PY, Park MH. Biochem J. 2001;355(Pt 3):841–849. doi: 10.1042/bj3550841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang KR, Kim YS, Wolff EC, Park MH. J Biol Chem. 2007;282(11):8300–8308. doi: 10.1074/jbc.M607495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joe YA, Park MH. J Biol Chem. 1994;269(41):25916–25921. [PubMed] [Google Scholar]
- 47.Joe YA, Wolff EC, Park MH. J Biol Chem. 1995;270(38):22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- 48.Schwelberger HG, Kang HA, Hershey JW. J Biol Chem. 1993;268(19):14018–14025. [PubMed] [Google Scholar]
- 49.Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. Int J Cancer. 2002;100(4):491–498. doi: 10.1002/ijc.10515. [DOI] [PubMed] [Google Scholar]
- 50.Mehta KD, Leung D, Lefebvre L, Smith M. J Biol Chem. 1990;265(15):8802–8807. [PubMed] [Google Scholar]
- 51.Clement PC, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JWB, Park MH, Johansson HE. Eur J Biochem. 2003;147(21):4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- 52.Boeke JD, Trueheart J, Natsoulis G, Fink GR. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 53.Liu YP, Nemeroff M, Yan YP, Chen KY. Biol Signals. 1997;6(3):166–174. doi: 10.1159/000109123. [DOI] [PubMed] [Google Scholar]