Abstract
Using one-bead one-compound (OBOC) combinatorial method, four heptapeptide ligands of CD21 receptor, a cell surface marker of malignant B cell lymphoma, were identified with an innovative two-step fluorescence screening method to overcome the limitation caused by autofluorescence of TentaGel resin. The binding affinities of selected peptides, YILIHRN (B1), PTLDPLP (B2), and LVLLTRE (B3), were in the micromolar region as determined by a fluorescence quenching assay. Peptide B1 was conjugated to N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer via spacers of different lengths, composed of one to four repeats of the 8-amino-3,6-dioxaoctanoic acid (A) group. The evaluation of the biorecognizability of HPMA copolymer-B1 conjugates by the CD21 receptor revealed that increasing the number of repeats of A in the spacer from one to three, resulted in continuous improvements in the biorecognition by the CD21 receptor; the increase from three to four repeats showed no significant effect. This work showed the potential of the OBOC combinatorial approach to select peptide ligands as targeting moieties for CD21 specific polymeric drug carriers.
INTRODUCTION
The water-soluble N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer is an extensively studied anticancer drug carrier.1, 2 As other water-soluble macromolecules,1, 3 HPMA copolymer–drug conjugates accumulate passively in solid tumors as a result of enhanced permeation and retention (EPR) effect.4, 5 Active targeting of HPMA copolymer–drug conjugates can be achieved with the incorporation of cancer cell specific ligands, such as carbohydrates,6 lectins,7 antibodies,8, 9 antibody fragments,10, 11 and peptides,12, 13 resulting in enhanced uptake of conjugates by cancer cells through receptor-mediated endocytosis with concomitant improvement of therapeutic efficacy. Among different cancer targeting molecules, peptides are of particular interest, as they may be readily identified with combinatorial peptide libraries.14 Enhanced peptide targeting efficiency could be achieved through multivalent interactions between targets and HPMA copolymer–peptide conjugates containing multiple copies of peptides within a single polymer chain.12, 15
Antigenic targets of lymphoma include receptors CD19, CD20, CD22, and CD37.16 The CD21 receptor (complement receptor 2, CR2) is expressed primarily on mature B lymphocytes, also on epithelial cells, thymocytes and follicular dendritic cells.17 Overexpression of CD21 receptor was found on lymphoblastoid cell lines such as Raji cells,18 consequently, it has been used as an alternative target for lymphomas.12, 19 CD21 receptor is a 145 kDa transmembrane glycoprotein with its extracellular region consisting of 15 – 16 short consensus repeats (SCRs), of which SCR1 and SCR2 are responsible for the interaction with several natural ligands, such as C3d, C3dg and Epstein Barr virus (EBV).17 Gold particles coated with C3dg were internalized into Raji cells (CD21 positive) through a receptor mediated endocytosis by CD21 receptor.20 Therefore, CD21 targeting could not only direct the tumor specific delivery of anticancer drugs, but also mediate their internalization, thus providing a suitable target for a HPMA copolymer-based delivery system for lymphoma chemotherapy. For convenience, a truncated recombinant CD21 receptor, rsCR2.1-4,21 was prepared. rsCR2.1-4 consists of first four SCRs of CD21 receptor and it is fully functional in interactions with ligands of CD21 receptor. Therefore, it was used as a target molecule for the screening of peptide ligands of CD21 receptor with combinatorial peptide libraries in this work.
The emergence of combinatorial peptide library techniques has dramatically expedited the screening and identification of novel peptides with desired properties.22 The methods to prepare combinatorial peptide libraries can be generally divided into two categories: chemically prepared libraries, using methods such as one-bead one-compound (OBOC)23–25 and SPOT synthesis;26, 27 and biologically prepared libraries using methods such as phage display28, 29 and bacterial display.30, 31 Both OBOC and phage display methods have been extensively used for selection and identification of oligopeptide ligands for tumor targeting.14 However, most of peptide ligand identification studies ended at verifying the validity of identified sequences, without evaluating their potentials in drug delivery systems applications.
Previously, five distinctive pentadecapeptide ligands of CD21 receptor were identified with phage display, and the specificity of selected peptides was confirmed by phage ELISA and competitive phage ELISA.15 The dissociation constants of three peptides, RMWPSSTVNLSAGRR (P1), PNLDFSPTCSFRFGC (P2), and GRVPSMFGGHFFFSR (P3), were determined to be within the micromolar range with a fluorescence quenching assay. To verify the applicability of selected peptides for targeting within a polymeric drug delivery system, the binding properties of HPMA copolymer–P1 conjugate was examined with surface bound CD21 receptor. It was found that peptide P1 retained its biorecognizability after it was conjugated with HPMA copolymer, and that the multivalency effect was important for interaction between the conjugate and the receptor.15 Interestingly, varying the length of spacer between peptide P1 and polymer backbone had trivial effect on the conjugate binding, suggesting that peptide P1 was readily accessible for interaction.
Alternatively, peptide ligands of CD21 receptor could also be identified using one-bead one-compound (OBOC) method with a two-step fluorescence screening process. Based on the “split and mix” concept,32, 33 Lam et al. developed an OBOC combinatorial method which consists of library preparation, biological screening, and structure determination of individual hits.23 With OBOC, a peptide library containing millions of different peptides can be synthesized on a resin so that each bead contains only one distinctive peptide. A most widely used solid support is the TentaGel resin, which has good swelling properties in both organic solvent and aqueous solutions.24 However, fluorescence screening, one of the most direct and convenient screening methods, of peptide libraries on TentaGel resin has been greatly limited, because of the autofluorescence of the TentaGel resin.34, 35
In this work, four heptapeptide ligands of CD21 receptor, YILIHRN (B1), PTLDPLP (B2), LVLLTRE (B3), and IVFLLVQ (B4), were identified with OBOC method using a two step fluorescence screening method to avoid the limitation caused by autofluorescence of TentaGel. The binding affinities of selected heptapeptides, except the hydrophobic B4, were determined with a fluorescence quenching assay. Peptide B1 was conjugated to N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer via spacers of different length, composed of one to four repeats of the 8-amino-3,6-dioxaoctanoic acid (A) group (Figure 1). The biorecognizability of these HPMA copolymer-B1 conjugates by the CD21 receptor was evaluated. Finally, the results on the biorecognition of HPMA copolymer conjugates, containing either a heptapeptide selected by OBOC or a pentadecapeptide selected by phage display, were compared.
Figure 1.
Structures of polymerizable peptide derivatives and HPMA copolymer–peptide conjugates. PB, HPMA copolymer backbone labeled with a PEG3400-biotin graft; HPMA copolymer–peptide conjugates consist of three components: an HPMA copolymer backbone, a PEG-biotin graft used as a label for immunoassay, and peptide ligands with spacers of different lengths, A1, A2, A3, and A4. PB-A-B1, PB-A2-B1, PB-A3-B1, and PB-A4-B1, peptide B1 attached to the HPMA copolymer backbone via spacer A, A2, A3, and A4. PB-A2-P1, peptide P1 attached to the HPMA copolymer backbone via spacer A2.
EXPERIMENTAL PROCEDURES
Materials
SCD2119 and rsCR2.1-421 were prepared as previously described. TentaGel S NH2 was purchased from Peptides International, Inc.. 2-chlorotrityl chloride resin, MBHA (4-methylbenzhydrylamine) resin and all N-α-Fmoc protected amino acids were purchased from Novabiochem.
Synthesis of One-bead One-compound Peptide Library
TentaGel S NH2 (2 g; 90 μm, 0.24 mmol/g substitution) was used as the solid support for the synthesis of a heptapeptide library. The library was synthesized with standard solid phase Fmoc chemistry using the “split and mix” method. TentaGel S NH2 was first swollen to equilibrium in DMF overnight and then evenly distributed into 19 vials. Different N-α-Fmoc protected amino acids (19 natural amino acids excluding cysteine) were loaded onto the resin in the presence of DIC/HOBt. The completeness of reaction was monitored with bromophenol blue. After the first amino acid was loaded, all the resin was mixed together and washed with DMF. The Fmoc protection was removed by incubation twice with 25% piperidine/DMF and the resin was washed with DMF. The resin was again distributed evenly into 19 vials and coupling of the remaining amino acids was repeated as described above. The side-chain protection was removed by incubation with phenol/thioanisole/H2O/EDT/TFA (0.75:0.5:0.5:0.25:10, w/v/v/v/v) at room temperature for 2.5 h. Finally, the resin was washed serially with DMF, methanol, DCM, DMF, 30% H2O/DMF, 60% H2O/DMF, 100% H2O, and PBS. The resin was stored in 0.05% sodium azide/PBS at 4°C.
Fluorescent Labeling of rsCR2.1-4
rsCR2.1-4 was labeled with two different F/P (number of fluorophores per protein) ratios, 1 and 5. To a solution of rsCR2.1-4 in 200 μl PBS (0.5 mg/ml), 20 μl of 1 M NaHCO3 (pH 9.0) and 23.3 μl of FITC in DMSO (10 mg/ml) was added and the reaction was stirred for 1 h (F/P ≈ 1) or 5 h (F/P ≈ 5) at room temperature in the dark. To remove free FITC, the reaction mixture was diluted to 0.5 ml with PBS and immediately applied to a PD-10 column (Amersham Biosciences) pre-equilibrated with PBS buffer. The concentration of the fluorescently-labeled protein and labeling efficiency were determined using UV spectrometry.
Two-step Fluorescence Screening of Peptide Ligands of CD21 Receptor with FITC-labeled rsCR2.1-4
First step: screening of fluorescence positive beads with bound receptor
TentaGel beads containing the peptide library (2 ml; approximately 1× 106 beads) were transferred to a polypropylene column. After washing with water, the beads were blocked with 0.1 % gelatin in water, followed by washing with 0.1 % TPBS. Then, the beads were incubated with FITC-labeled rsCR2.1-4 (F/P ≈ 1, 0.02 μM) for 1 h with gentle shaking. The unbound receptor was washed off with TPBS. The beads were then transferred to several Petri dishes and fluorescently bright beads were picked under fluorescence microscope (Nikon ECLIPSE E800); totally 48 beads were selected.
Second step: screening of fluorescence negative beads after treating with guanidium hydrochloride (Gu.HCl)
Selected beads were divided into two groups on the basis of their physical appearance: opaque fluorescence (27 beads) and transparent fluorescence (21 beads). Both groups of beads were treated with Gu.HCl (6 M, pH 1.0) for 3 h. Because Gu.HCl can disrupt the binding of receptor to corresponding peptide ligands, it was used to differentiate autofluorescence of TentaGel beads. It was found that beads with initial opaque fluorescence lost the fluorescence after treatment with Gu.HCl; however, the fluorescence of beads with initial transparent fluorescence was retained, an indication of autofluorescence. The fluorescence of opaque fluorescence beads was recovered after washing the beads with PBS and incubation with FITC labeled rsCR2.1-4, but with lower intensity.
Synthesis of Peptides
Peptides, YILIHRN (B1), PTLDPLP (B2), and LVLLTRE (B3) were synthesized manually on a solid support of 2-chlorotrityl chloride resin using standard Fmoc chemistry. The synthesized peptides were cleaved from resin with a mixture of TFA:H2O:TIS (95:2.5:2.5, v/v/v) for 4 h at room temperature. All synthesized peptides were purified with RP-HPLC and verified with MALDI-TOF MS. MALDI-TOF MS calculated for B1 (MH+) 928.53, found 928.61; MALDI-TOF MS calculated for B2 (MH+) 1702.73, found 1702.78; MALDI-TOF MS calculated for B3 (MH+) 843.52, found 843.72.
Determination of Peptide Binding Constants with Fluorescence Quenching
A series of mixtures of the labeled receptor rsCR2-FITC at 0.02 μM, and peptides, B1, B2, and B3, at different concentrations were prepared in 150 μl PBS. The mixtures were incubated at room temperature for 5 h. The fluorescence intensity of each sample was measured in duplicates with a LS-55 luminescence spectrometer (Perkin-Elmer) with excitation and emission wavelengths of 495 and 515 nm, respectively. The fluorescence background caused by the peptides was negligible within the concentration range used. The association constant was estimated by fitting the quenching data to Eq. 1 with software KaleidaGraph (version 3.0).36
| (Eq. 1) |
| (Eq. 2) |
where, Q is the quenching value defined in Eq. 2, and Qm represents maximal fluorescence quenching, Ka is the association constant of peptide binding, and [P] is the concentration of free peptide. F0 and F are fluorescence intensities of fluorescently-labeled rsCR2.1-4 in the absence and presence of peptide ligands, respectively.
Synthesis of Polymerizable Peptide Derivatives
Polymerizable N-methacryloylated peptides containing spacers of repeating units of 8-amino-3,6-dioxaoctanoic acid (A), MA-A-B1, MA-A2-B1, MA-A3-B1, MA-A4-B1, MA-A2-P1 and MA-A3-P1 (P1, RMWPSSTVNLSAGRR, was selected by phage display15) were synthesized (see structures in Figure 1) on MBHA resin using standard Fmoc chemistry. After the peptides were synthesized on resin, they were extended with additional spacers: A, A2, A3, and A4 spacers were added by coupling with one, two, three, or four repeats of Fmoc-A (9-fluorenylmethoxycarbonyl-8-amino-3,6-dioxaoctanoic acid, Peptides International) and capped with methacryloyl chloride. All peptide monomers were purified with RP-HPLC and were verified with MALDI-TOF MS. MALDI-TOF MS calculated for MA-A-B1 (MH+) 1140.65, found 1140.70; MALDI-TOF MS calculated for MA-A2-B1 (MH+) 1285.72, found 1285.79; MALDI-TOF MS calculated for MA-A3-B1 (MH+) 1430.79, found 1430.82; MALDI-TOF MS calculated for MA-A4-B1 (MH+) 1575.87, found 1575.87; MALDI-TOF MS calculated for MA-A2-P1 (MH+) 2076.06, found 2076.11; MALDI-TOF MS calculated for MA-A3-P1 (MH+) 2220.13, found 2220.25.
Synthesis of MA-PEG3400-biotin
MA-PEG3400-biotin was synthesized as described previously.15 Briefly, N-(3-aminopropyl)methacrylamide hydrochloride (MA-NH2) (2.7 mg) was dissolved in 200 μl DMF and 6.7 μl N,N-diisopropylethylamine (DIPEA) was added. The temperature of the solution was lowered to 0 °C with an ice bath. Biotin-PEG3400-NHS (PEG, poly(ethylene glycol), mol. wt. 3,400; NHS, N-hydroxysuccinimide ester; Nektar Therapeutics) dissolved in 200 μl DMF was added dropwise. The reaction was kept at 0°C for 30 min and then left overnight at room temperature. The crude product was diluted 1:1 with water and MA-PEG3400-biotin purified on a PD-10 column. Molecular weight as determined by MALDI-TOF MS was Mw = 3.89 kDa; Mn = 3.86 kDa; Mw/Mn = 1.01.
Synthesis of Biotin-labeled HPMA Copolymer–peptide Conjugates (see structures in Figure 1)
The synthesis of HPMA copolymer–peptide conjugate PB-GG-B1 (PB is HPMA copolymer backbone labeled with a PEG3400-biotin graft) is described as an example: HPMA (14.1 mg), MA-A-B1 (6 mg), MA-PEG3400-biotin (3.6 mg) (molar ratio, 94:5:1) and initiator AIBN (1.5 mg) were dissolved in 149 μl of DMSO in a glass ampoule. The solution was bubbled with nitrogen for 10 min and polymerized in the sealed ampoule at 50 °C for 24 h. After the reaction was finished, the reaction solution was diluted to 3 ml with H2O and dialyzed against water in a dialysis tubing with a molecular cutoff of 12–14 kDa for 3 days. After dialysis, the polymer water solution was lyophilized. PB, PB-A2-B2, PB-A3-B3, PB-A4-B1, PB-A2-P1, and PB-A3-P1 were synthesized similarly, except that PB contained no peptide. The polymers were analyzed with size-exclusion chromatography on an ÄKTA FPLC system (Pharmacia). The content of peptide was determined by amino acid analysis and the content of biotin was determined with EZ Biotin Quantitation Kit (PIERCE Biotech. Inc.).
ELISA of HPMA Copolymer–peptide Conjugates
The receptor rsCR2.1-4 (150 ng) was coated onto a MaxiSorp plate by overnight incubation at 4 °C. The plate was blocked with 4% milk at room temperature for 1.5 h, washed with TPBS, and incubated with HPMA copolymer–peptide conjugates (0.02 μM) for 30 min. Then, the plate was washed five times with TPBS, followed by incubation with streptavidin/HRP (Zymed Laboratories) for 30 min. The plate was washed 5 times with TPBS; then TMB substrate was added to the plate and incubated for 30 min. The absorbance was read at 450/630 nm.
RESULTS
Synthesis of One-bead One-compound Peptide Library
A heptapeptide library containing more than one million different peptides was synthesized with one-bead one-compound method. TentaGel S NH2 resin was used as solid support and peptides were synthesized with manual solid phase Fmoc peptide synthesis. It was noted that the coupling reactions of hydrophobic and bulky residues proceeded slower than those of small and hydrophilic residues. However, almost all reactions were able to finish within one hour as indicated by analysis with bromophenol blue. To ensure the quality of synthesized library, a control heptapeptide with a known sequence, HPLSSSV, was synthesized along with the heptapeptide library. The composition of control peptide was confirmed by automated Edman sequencing. Therefore, the purity of the synthesized heptapeptide library should be usable for the following biological screening.
Two-step Fluorescence Screening of Peptide Ligands for CD21 Receptor
To minimize the influence of FITC labeling on the receptor binding, rsCR2.1-4 with a low fluorescence label content (F/P ≈ 1) was used for screening peptide ligands of CD21 receptor. By fluorescently labeling the recombinant short CD21 receptor, rsCR2.1-4, the binding of receptor to its peptide ligands can be conveniently identified with fluorescence visualization. However, we found, in agreement with previous reports, that a very small number of TentaGel beads are autofluorescent,34, 35 which largely limited the fluorescence screening process. In this work, an innovative two-step fluorescence screening process was developed: in the first step, 48 most fluorescently bright beads were picked; in the second step, 27 fluorescence negative beads were picked after all the beads from first step were treated with Gu.HCl (6 M, pH 1). Gu.HCl is a strong protein chaotropic agent and it disrupts the binding between rsCR2.1-4 and bead-bound peptides, resulting in the disappearance of fluorescence. However, the fluorescence of beads, which possessed autofluorescence was not susceptible to Gu.HCl, and the fluorescence did not change after treatment with Gu.HCl. Therefore, fluorescence negative beads in the second step actually contained real peptide ligands of the receptor.
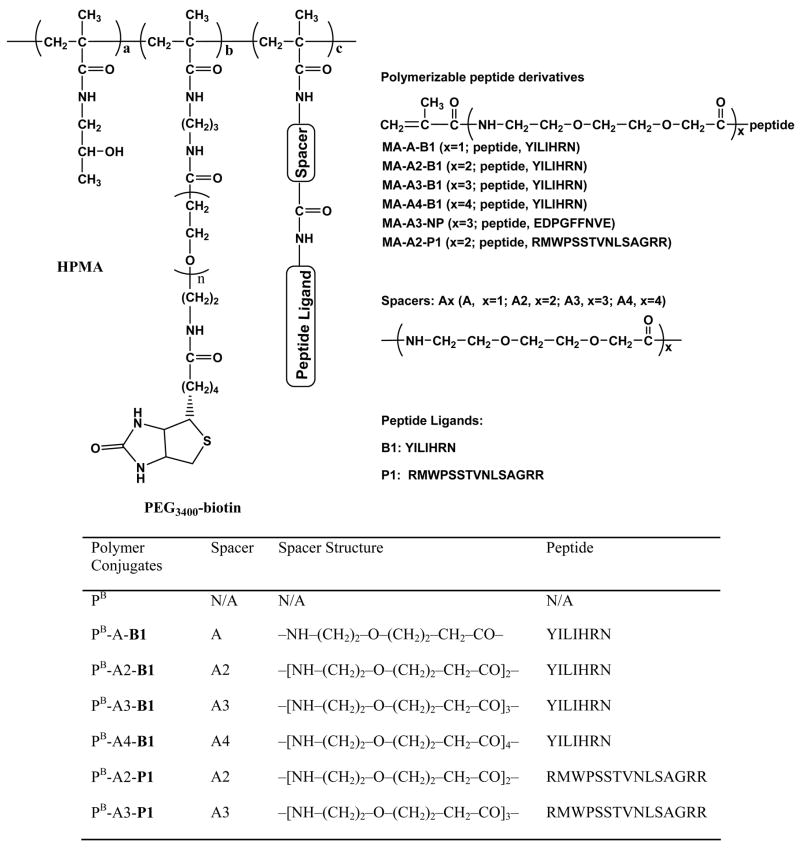
In fact, fluorescence of the selected beads in the first screening step showed two different appearances: opaque and transparent (Figure 2). Beads with opaque fluorescence seemed to have homogeneous fluorescence as well; on the contrary, beads with transparent fluorescence seemed to have heterogeneous fluorescence. All beads selected in the first step were divided into two groups on the basis of their fluorescence appearance: one group with opaque fluorescence and the other with transparent appearance. After both groups were treated with Gu.HCl, the fluorescence of all beads in the opaque group disappeared, while the fluorescence of the transparent group remained. Furthermore, the spatial fluorescence distribution of selected beads was characterized with surface plotting using the software of Image Pro Plus version 4.0 (C and F, Figure 2). It was found that spatial fluorescence distribution of fluorescence opaque beads and transparent beads was clearly different. The fluorescence distribution of transparent beads was heterogeneous with rugged surfaces, while that of opaque fluorescence beads was homogeneous with smooth surfaces. Therefore, it was deducted that fluorescence of transparent beads with heterogeneous distribution is probably the result of autofluorescence. Although there is not enough evidence to prove it, this fact may be used as cross-reference in fluorescence screening process.
Figure 2.
Two-step fluorescence screening of OBOC peptide library: A, DIC image of TentaGel beads; B, fluorescence positive beads. Bead 1, 3 are fluorescence transparent; bead 2 is fluorescence opaque; C, surface plot of fluorescence of beads in C; D, the fluorescence of fluorescence opaque beads disappeared after treatment with Gu.HCl; E, the fluorescence of fluorescence transparent beads remained after treatment with Gu.HCl; F, surface plot of fluorescence of beads in E.
With the two-step screening process, we were able to select 27 beads that matched our criteria. Those beads were washed with TPBS and allowed to incubate with FITC labeled rsCR2.1-4 again. The regaining of fluorescence confirmed their binding to CD21 receptor, however with diminished intensity. Probably, the binding has been significantly weakened by the prior treatment with Gu.HCl. Finally, we selected four most fluorescently bright beads and analyzed the peptide sequences by Edman sequencing (Table 1). It was found that peptides B1 (YILIHRN), B3 (LVLLTRE), and B4 (IVFLLVQ) have similar structures: first four amino acids at N-terminus were composed of hydrophobic residues, mostly of leucine, isoleucine, and valine. One may speculate that these hydrophobic residues mediate hydrophobic interactions with the CD21 receptor.
Table 1.
Peptide ligands of CD21 receptor identified with OBOC
| Peptide | Sequence | Binding Constant, Ka (M−1) |
|---|---|---|
| B1 | YILIHRN | 4.6 × 106 |
| B2 | PTLDPLP | 1.0 × 106 |
| B3 | LVLLTRE | 2.1 × 106 |
| B4 | IVFLLVQ | NDa |
ND: not determined
Binding Constant Determination with Fluorescence Quenching
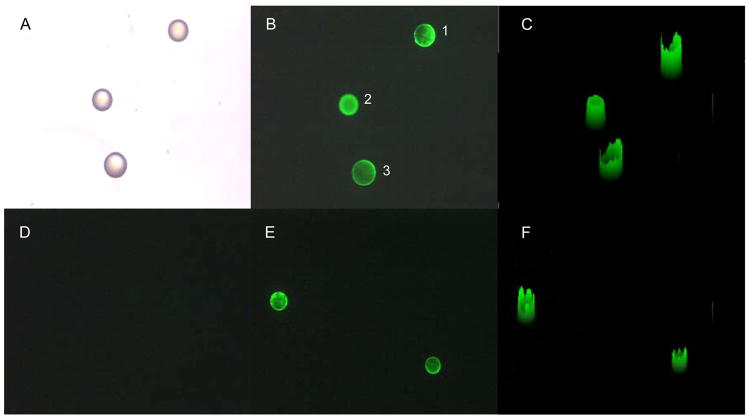
It was found that fluorescence quenching due to the binding of peptides was not apparent when the labeling of rsCR2.1-4 was low (F/P ≈ 1) (data not shown). When the labeling of rsCR2.1-4 was high (F/P ≈ 5), the quenching effect was sufficient for the determination of binding affinity. On the basis of X-ray crystallography of free CD21 (SCR1– 2) and the complex of CD21 (SCR1–2) and C3d, 37,38 it was found that two domains of SCR1 and SCR2 formed a V shaped structure and these two domains were glued together through extensive side-by-side hydrophobic interactions. The binding of C3d to CD21 receptor induced not only local but also global conformational changes, which suggested that the CD21 receptor (or at least SCR1–2) is a relatively flexible molecule and the binding of CD21 receptor to its ligands was an induced-fitting process.37 This induced-fitting process may change the microenvironment of fluorophore of FITC labeled rsCR2.1-4, resulting in change of fluorescence intensity, therefore providing a convenient way for measuring binding affinity of peptide-receptor interaction. Meanwhile, this induced-fitting process may also influence the time required to reach binding equilibrium. As indicated in our binding kinetics study of peptide YILIHRN, it took about three hours to reach quenching equilibrium, although more than half of maximal quenching was achieved within first half an hour (Figure 3). Consequently, in binding constant determination studies, fluorescence intensity was measured after the peptides and receptor were incubated for 5 h.
Figure 3.
Binding kinetics of peptide YILIHRN to rsCR2.1-4. YILIHRN (3 μM) was incubated with rsCR2.1-4 (0.02 μM) and the fluorescence quenching ratio was determined as a function of time.
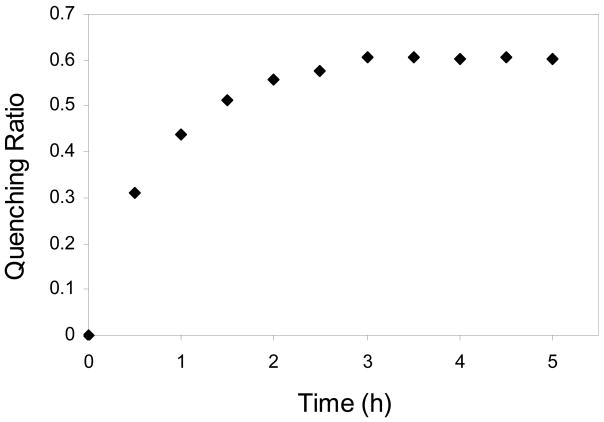
The binding affinity of B4 (IVFLLVQ) was not determined, because of its excessive hydrophobicity as implied by its sequence composition. The association constants of peptides B1, B2, and B3 were determined to be 4.6×106 M−1 (fitting error = 1.4; R = 0.97), 1.0×106 M−1 (fitting error = 0.3; R = 0.98), and 2.1×106 M−1 (fitting error = 0.3; R = 0.99), respectively (Figure 4).
Figure 4.
Fluorescence quenching of fluorescein labeled rsCR2.1-4 as a function of peptide concentration. Data are depicted as mean ± standard deviation from duplicate measurements. (○) peptide B1: YILIHRN; (□) peptide B2: PTLDPLP; (◇) peptide B3: LVLLTRE.
Synthesis and Characterization of HPMA Copolymer–peptide Conjugates
All HPMA copolymer conjugates were synthesized by radical copolymerization of HPMA with polymerizable derivatives of peptide and biotin using 2,2′-azobisisobutyronitrile (AIBN) as the initiator. The composition of the synthesized polymers is summarized in Table 3. The weight-average molecular weight was determined with size-exclusion chromatography on a Sepharose 6B column calibrated with PHPMA standards. Copolymerization with a small amount of MA-PEG3400-biotin provided a convenient way to monitor the interaction between HPMA copolymer–peptide conjugates and CD21 receptor using biotin/streptavidin-HRP detection system in ELISA experiments. Only 1 mol% of MA-PEG3400-biotin was used in the monomer feed and the polymer conjugates contained approximately one biotin per macromolecule (Table 3).
Table 3.
Composition of HPMA copolymer–peptide conjugates
| Conjugates | Structure | Mwa (kDa) | Pdb | Peptide (mol%)c | Peptide #/chain | Biotin (mol%)d | Biotin #/chain |
|---|---|---|---|---|---|---|---|
| PB | PHPMA-(PEG-biotin) | 63.2 | 1.7 | 0 | 0 | 0.4 | 1.8 |
| PB-A-B1 | PHPMA-(A-B1)-(PEG-biotin) | 42.1 | 1.4 | 2.1 | 5.2 | 0.34 | 1.0 |
| PB-A2-B1 | PHPMA-(A2-B1)-(PEG-biotin) | 39.0 | 1.3 | 2.0 | 4.7 | 0.23 | 0.6 |
| PB-A3-B1 | PHPMA-(A3-B1)-(PEG-biotin) | 38.7 | 1.3 | 2.2 | 4.9 | 0.45 | 1.2 |
| PB-A4-B1 | PHPMA-(A4-B1)-(PEG-biotin) | 41.1 | 1.3 | 1.7 | 4.2 | 0.34 | 1.0 |
| PB-A2-P1 | PHPMA-(A2-P1)-(PEG-biotin) | 55.6 | 1.5 | 1.1 | 4.3 | 0.36 | 1.4 |
| PB-A3-P1 | PHPMA-(A3-P1)-(PEG-biotin) | 37.3 | 1.5 | 1.83 | 3.7 | 0.31 | 0.8 |
PB HPMA copolymer backbone labeled with a PEG3400-biotin graft
Mw: weight-average molecular weight as determined with size exclusion chromatography
Pd: Polydispersity, ratio of weight-average molecular weight over number-average molecular weight
determined with amino acid analysis
determined with EZ Biotin Quantitation Kit
The Effect of Spacer Length on Binding of HPMA Copolymer–peptide Conjugates to CD21 Receptor
The binding of HPMA copolymer–peptide conjugates with surface bound receptor was detected with ELISA using biotin/streptavidin/HRP as the reporting system. Nonspecific binding was blocked with 4% nonfat milk as it was found to produce the lowest background. Although milk contains various amount of endogenous biotin which may interfere with the biotin/streptavidin/HRP detection system, it didn’t seem to cause any problems in the binding studies in this work. If the endogenous biotin were to interact with the detection system, it can be masked with egg white and milk.39
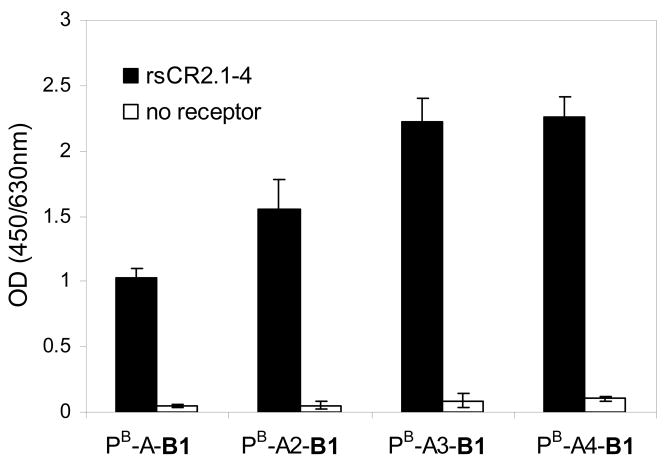
Peptide B1 was used as an example to study the binding properties of HPMA copolymer–peptide conjugates as B1 demonstrated the best binding affinity with good solubility. To evaluate whether spacer length has a significant effect on the binding of HPMA copolymer–B1 conjugates to CD21 receptor, a series of conjugates, PB-A-B1, PB-A2-B1, PB-A3-B1, and PB-A4-B1 (Figure 1), were prepared which differed in spacer length. Because of its flexibility and moderate hydrophilicity, 8-amino-3,6-dioxaoctanoic acid (A) was used as repeating units of spacers, A, A2 (dimer of A), A3 (trimer of A), and A4 (tetramer of A), between peptide and polymer backbone. The binding results of these four peptide conjugates with different spacers (B1 concentration 0.02 μM) are shown in Figure 5. All conjugates showed better binding to the surface with receptor than to the surface without receptor. Most importantly, a clear spacer effect on the surface binding was observed. The conjugate with the shortest spacer, PB-A-B1, showed the weakest binding. With the increase of spacer length (from A to A3), the binding increased significantly. This spacer effect on the binding may be ascribed to the decreased accessibility of a peptide after its conjugation with a polymer,40 but the accessibility of the peptide can be improved using proper spacer. A further increase in spacer length, from A3 to A4, did not result in an improvement of binding. It appears that spacer A3 was sufficient for presenting the targeting peptide B1; the peptide is fully accessible with either spacer A3 or A4.
Figure 5.
Surface binding results of HPMA copolymer–B1 conjugates with different spacer length. Data are depicted as mean ± standard deviation from duplicate measurements.
ELISA of HPMA Copolymer-B1 Conjugate Binding to Surfaces with Different Amounts of rsCR2.1-4
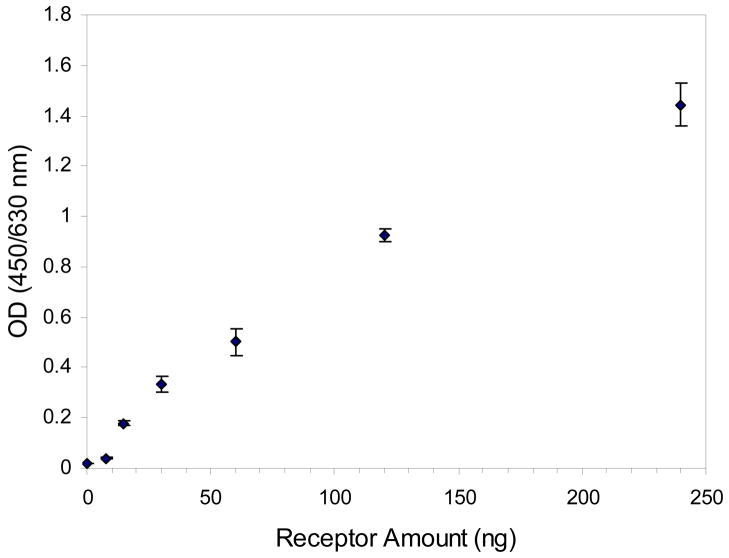
PB-A3-B1 was used to study its specific interaction with surface bound receptor, rsCR2.1-4. MaxiSorp plate was coated with different amounts of rsCR2.1-4 (0 – 240 ng), which resulted in varied density of surface bound receptor. The binding of fixed amount of PB-A3-B1 (0.02 μM) was detected with ELISA using biotin/streptavidin-HRP as the reporting system. The binding of PB-A3-B1 to surface bound receptor was directly related to the receptor density: the higher the receptor density, the higher the amount of surface bound PB-A3-B1 (Figure 6). When the MaxiSorp plate surface was coated with 7.5 ng of rsCR2.1-4, the binding of PB-A3-B1 was comparable to control surface containing no receptor; when surface was coated with a double amount of rsCR2.1-4 (15 ng), the binding signal increased almost ten times. However, subsequent doubling of the coated amount of rsCR2.1-4 only increased the binding signal less than twice. This direct response of PB-A3-B1 binding to surface bound receptor density corroborated the specific interaction between HPMA copolymer–peptide conjugate and receptor. High receptor density was favorable for the multivalent interaction between PB-A3-B1 and surface bound receptor.
Figure 6.
The relationship between the binding of PB-A3-B1 and amount of surface coating receptor. Data are depicted as mean ± standard deviation from duplicate measurements.
Comparison of Surface Binding of HPMA Copolymers Containing Peptides B1 and P1
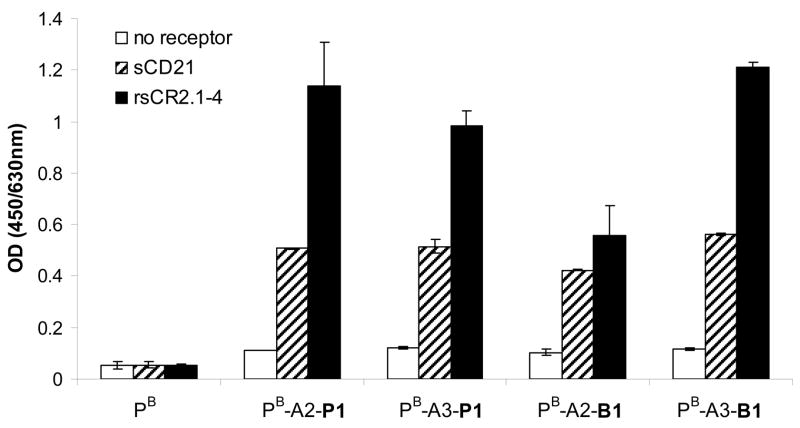
The binding properties of HPMA copolymer–P1 conjugates were studied previously.15 Here, their surface binding was compared with that of HPMA copolymer–B1 conjugates. Four HPMA copolymer–peptide conjugates, PB-A2-B1, PB-A3-B1, PB-A2-P1 and PB-A3-P1 were incubated with three different surfaces: control surface without any receptor, surface coated with rsCR2.1-4, and surface coated with sCD21,41 a soluble form of CD21 receptor. Control polymer without targeting peptides, PB, didn’t show any specific binding to either receptor coated surface; while conjugates, PB-A2-B1, PB-A3-B1, PB-A2-P1 and PB-A3-P1, all showed specific binding towards both rsCR2.1-4 and sCD21 coated surfaces (Figure 7). PB-A3-B1 showed higher binding than PB-A2-B1 because of spacer effect as discussed above, while PB-A2-P1 and PB-A3-P1 showed similar binding for the lacking of spacer effect.15 The binding of PB-A3-B1 was comparable to that of PB-A2-P1 and PB-A3-P1, as the binding constant of B1 (4.6 × 106 M−1) is only slightly higher than that of P1 (2.9 × 106 M−1).15 For unknown reasons, the binding of all four conjugates, PB-A2-B1, PB-A3-B1, PB-A2-P1, and PB-A3-P1, to rsCR2.1-4 coated surface is higher than the binding to sCD21 coated surface. Since B1 was screened with rsCR2.1-4, the specific binding of HPMA copolymer–B1 conjugates to sCD21 confirmed the successful identification of peptide ligands of CD21 receptor with our two-step fluorescence screening.
Figure 7.
Comparison of surface binding results of PB-A3-B1 and PB-A2-P1. Data are depicted as mean ± standard deviation from duplicate measurements.
DISCUSSION
Peptide Ligand Identification with One-bead One-compound Combinatorial Method
OBOC peptide libraries have been used to screen ligands for various targets, such as antibodies,23 protease,42 protein kinase, etc.24 Successful identification of targeting peptides for lymphoma with OBOC has also been reported, with the target of surface idiotype of lymphoma cell lines,43 and α4β1 integrin receptor in non-Hodgkin’s lymphoma.44 In this work, heptapeptide ligands of CD21 receptor, an alternative lymphoma target, were identified. Instead of focusing on validating the successful screening, this work further investigated the binding properties of one of the selected peptides, B1, after its conjugation with HPMA copolymer.
One-bead one-compound combinatorial method has found its applications not only in peptide libraries, but also in other libraries with oligomers such as peptoids, oligocarbamates, oligoureas, etc., and with small molecules, as long as those libraries meet three requirements: first, well established synthetic chemistry for library preparation; second, suitable screening method to distinguish desired products; third, appropriate decoding system for identification of selected compound.24 The scope of OBOC was further broadened with the appearance of novel ideas in library preparation such as one-bead two-compound library45, dimeric OBOC library,46 and topologically segregated bilayer beads for OBOC,47 new instrumentation like COPAS (Union Biometrica) for fluorescence screening,48 and creative coding-decoding systems.49, 50
There are several screening methods for OBOC libraries such as enzyme-linked colorimetric assay,23 whole cell binding assay,44 and autoradiography.51 The enzyme-linked colorimetric assay usually requires a reporter protein for detection. Therefore, beads which specifically bind to the reporter protein have to been removed before screening. Otherwise, false positive beads might be selected. Fluorescence screening has no such problems, but its application has been largely limited by the autofluorescence of TentaGel. To overcome this limitation, instead of using traditional organic fluorophores, improved visualization has been achieved by labeling target molecules with quantum dots, because quantum dots provide higher quantum yield, and, more importantly, they exhibit narrower and symmetrical emission peaks.34, 35 Another approach is to remove autofluorescent beads before the binding assay with the aid of fluorescence activated COPAS sorter.48 In this work, using a readily available organic fluorescein dye, this two-step fluorescence screening method proved to be successful for screening receptor bound beads. In addition, the discovery of the different fluorescence appearance of autofluorescent beads (transparent) and beads bound with fluorescently labeled receptor (opaque) provided a convenient method to expedite the fluorescence screening process. The different fluorescence appearance of selected beads in the first step also demonstrated different spatial fluorescence distribution as visualized with surface plotting (C and F, Figure 2). Hypothetically, the different fluorescence appearance is the simple reflection of two different fluorescence mechanisms.
Screening Results with OBOC
Four different heptapeptide ligands of CD21 receptor were identified with the OBOC combinatorial method (Table 1). Previously, five different pentadecapeptide ligands were identified with phage display15 (Table 2). As determined with a fluorescence quenching assay, the binding constants of peptide ligands selected with both methods were within the same range: with phage display, P1, 2.9×106 M−1; P2, 4.7×106 M−1; P3, 2.2×106 M−1, and with OBOC, B1, 4.6×106 M−1; B2, 1.0×106 M−1; B3, 2.1×106 M−1. Pentadecapeptide ligands did not demonstrate a much higher binding affinity than heptapeptides, probably because larger peptides have to overcome an extra entropy loss compared with smaller peptides in their binding processes. All pentadecapeptide ligands showed similar but much larger quenching ratio, ~ 70%; while heptapeptide ligands only quenched 20 – 50% fluorescence. This probably suggests that larger peptide ligands, pentadecapeptides, induced a larger conformational change of the receptor upon binding with ligand, most probably because their binding sites covered a larger area than the heptapeptide binding area. It is interesting to note, that the peptide (B1) possessed both the highest binding constant (4.6×106 M−1) among the selected heptapeptides and the highest quenching ratio (~ 50%). In contrast, the peptide (B2) having the lowest binding constant (1.0×106 M−1) also showed the lowest quenching ratio (~ 20%), an indication of possible relationship between binding affinity of heptapeptides and quenching ratio.
Table 2.
Comparison of peptide sequences obtained from phage display and OBOC
| Phage Display | OBOC (this work) |
|---|---|
| RMWPSSTVNLSAGRR | |
| PNLDFSPTCSFRFGCa | PTLDPLPa |
| GRVPSMFGGHFFFSR | |
| RLAYWCFSGLFLLVCa | LVLLTRE, IVFLLVQ, YILIHRNa |
| PVAAVSFVPYLVKTY |
The similar residues of peptides selected from phage display and OBOC are indicated by boldface italic type.
The five pentadecapeptides, selected by phage display,15 did not demonstrate much homology. However, three out of four OBOC selected heptapeptides, YILIHRN, LVLLTRE, and IVFLLVQ, possessed some shared features: first four amino acids at the N-terminus were mainly hydrophobic residues, leucine, isoleucine, and valine. By comparing the peptide sequences from OBOC with those from phage display, it was found the hydrophobic section of peptides B1, B2, and B3 are similar to segment LFLLV of peptide RLAYWCFSGLFLLVC which was selected from phage display (similar residues are indicated in italic boldface type).15 Such resemblance is a significant indication of peptide binding site, because in peptide RLAYWCFSGLFLLVC, LFLLV resides within two cysteines which can form a cyclic structure and such structure is usually critical in binding.52 Additionally, it was also found that peptide B2 (PTLDPLP) has plausible similarity to peptide PNLDFSPTCSFRFGC selected from phage display.
Spacer Effect on Binding of HPMA Copolymer–peptide Conjugates
There is evidence that the accessibility of a peptide can be influenced by conjugation with a polymer: the conjugation of a decapeptide (PYWKWQYKYD) with PEG 20,000 resulted in complete loss of reactivity; while its conjugation with PEG 5,000 maintained its activity.40 This was confirmed by the spacer effect in this work: the binding of HPMA copolymer–B1 conjugates increased significantly with the increase of spacer length from A (8-amino-3,6-dioxaoctanoic acid) to A3 (tris(8-amino-3,6-dioxaoctanoic acid)) (Figure 5). However, in our previous work, HPMA copolymer–P1 conjugates, PB-GG-P1 and PB-A2-P1, demonstrated similar binding to rsCR2.1-4 coated surface, although spacer A2 (8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoic acid) in PB-A2-P1 is four times as long as GG (glycylglycine) spacer in PB-GG-P1,15 suggesting that the length of the spacer has little effect on the binding of HPMA copolymer–P1 conjugates. Therefore, the spacer length showed different influences on the accessibility and the binding properties of peptides B1 in their HPMA copolymer conjugates. Hypothetically, this difference is ascribed to their different physical properties such as size and hydrophobicity/hydrophilicity of peptides. Pentadecapeptide P1 (RMWPSSTVNLSAGRR) is twice as long as heptapeptide B1 (YILIHRN), thus better chance to expose itself. In addition, HPMA copolymer–B1 conjugates may undergo conformational change due to the amphipathic composition of B1, which may also contribute to the accessibility of B1.
Structural Factors Influencing the Biorecognizability of HPMA Copolymer–peptide Conjugates
Undoubtedly, the biorecognizability of polymer–peptide conjugates is largely dictated by the intrinsic binding affinity between peptide ligands and its target molecules. Meanwhile, structural factors like multivalency for interaction,12, 15 peptide accessibility, and polymer conformation53 may also have significant influence on the biorecognizability. The importance of multivalent interaction was clearly demonstrated in the binding studies of HPMA copolymer–P1 conjugates containing various amount of P1 peptide.15 Being relatively small and chemically stable, peptides can be readily derivatized and incorporated into a polymers with multiple copies.54 Thus, multivalency become one major advantage for the designing of polymer–peptide conjugates for targeted drug delivery. The accessibility of a peptide could be significantly weakened by the conjugation with polymers, consequently resulting in decrease or total loss of binding activity.40 It can be greatly improved with the introduction of spacers with sufficient length. The binding of HPMA copolymer–B1 conjugates increased with the increase of spacer length from A to A3.
The conformation of HPMA copolymer might change drastically after it was conjugated with multiple copies of peptide depending on the physical properties of the peptide and the content of peptide. Peptide YILIHRN (B1) has an amphipathic structure with an N-terminal hydrophobic section (YILI) and a C-terminal hydrophilic section (HRN). It was found that the HPMA copolymer–B1 conjugate has the tendency to self-associate with the increase of peptide content (to be published). In this work, HPMA copolymer–B1 conjugates contain about 2% of peptide (approximately 5 peptide per polymer chain) and the degree of self-association was low. As PB-A3-B1 demonstrated comparable binding to PB-A3-P1, the effect of self-association on the binding seemed to be limited. To evaluate the effect of self-association on the binding, various amount of acrylic acid (10%, 20%, and 30%) were copolymerized with HPMA to disrupt the self-association process as a result of electrostatic expulsion. However, this effort was fruitless as the surface binding of HPMA copolymer–B1 conjugates containing acrylic acid was totally abolished, indicating that acrylic acid interfered with the binding process.
One-bead One-compound Peptide Library versus Phage Display Peptide Library
Peptide ligands of CD21 receptor had been identified with OBOC, and, previously, with phage display.15 Phage display is a biological combinatorial method: by engineering the DNA structure of phage, a phage peptide library is constructed, so that millions of different peptides are displayed on the phage capsid and are accessible for interaction with target proteins.29 OBOC is a chemical combinatorial method: with the procedure of “split and mix”, an oligopeptide library could be constructed on the solid support such that each bead contains only one single peptide followed by on-resin immunological assays which enable the identification of peptide ligands of receptor.24 Both OBOC method and phage display have been extensively used for selection and identification of oligopeptide ligands for tumor targeting.14
There are reports that peptide ligands with similar sequences were identified with both phage display and OBOC.55, 56 However, full understanding of the advantages and the disadvantages of both phage display library and OBOC library is helpful for choosing appropriate combinatorial techniques for a specific screening purpose. With phage display, molecules of different sizes from oligopeptides to proteins can be displayed on phage coat protein. Phage display can also be conveniently used for identification of ligands for tumor tissue in vivo57 and even with patient body.58 The advantage of using OBOC to prepare peptide library lies in its extraordinary control on the building blocks of the peptide library. The peptide sequence could be modified with D-amino acids or other unnatural amino acids to introduce desired properties. With the intercalation of structural determinants, the peptide structure on resin could be predefined: cyclic structure formation with disulfide bonds of two cysteines44 and with side chains of lysine and glutamine;25 turn structure formation with proline.44
CONLUSIONS
With a two-step fluorescence screening process, the limitation caused by autofluorescence of TentaGel was avoided, and four heptapeptide ligands of CD21 receptor were successfully identified with one-bead one-compound combinatorial method. The dissociation constants of all peptides except B4 were determined with a fluorescence quenching assay to be within the micromolar range. HPMA copolymer conjugated with selected peptide, B1, demonstrated specific binding towards CD21 receptor. The binding between HPMA copolymer–B1 conjugate and CD21 receptor was found to be dependent on the length of spacer between peptide and polymer backbone: spacers with sufficient length (A3 and A4) were able to significantly enhance the binding. With proper spacer, HPMA copolymer equipped with selected heptapeptides seems to be a promising targeted anticancer drug carrier for chemotherapy of lymphoma. Combined with binding studies of HPMA copolymer containing targeting peptide derived from phage display,15 this work should be informative for designing and optimizing other polymeric drug carriers containing targeting peptide in terms of structural factors such as peptide multivalency, peptide accessibility, and polymer conformation.
Acknowledgments
This work was supported in part by NIH Grant CA88047 from the National Cancer Institute.
ABBREVIATIONS
- A
8-amino-3,6-dioxaoctanoic acid
- A2
8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoic acid
- A3
8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoic acid
- A4
8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoyl-8-amino-3,6-dioxaoctanoic acid
- AIBN
2,2′-azobisisobutyronitrile
- B1
YILIHRN
- B2
PTLDPLP
- B3
LVLLTRE
- B4
IVFLLVQ
- DIPEA
N,N-diisopropylethylamine
- DMSO
dimethyl sulfoxide
- ELISA
enzyme linked immunosorbent assay
- Fmoc
9-fluorenylmethoxycarbonyl
- GG
glycylglycine
- HPMA
N-(2-hydroxypropyl)methacrylamide
- HRP
horse radish peroxidase
- NHS
N-hydroxysuccinimide ester
- MA
methacryloyl
- MA-GG-OH
N-methacryloylglycylglycine
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- P
HPMA copolymer backbone
- PB
HPMA copolymer backbone labeled with a PEG3400-biotin graft
- P1
RMWPSSTVNLSAGRR
- P2
PNLDFSPTCSFRFGC
- P3
GRVPSMFGGHFFFSR
- PBS
phosphate-buffered saline
- PEG
poly(ethylene glycol)
- PHPMA
polyHPMA
- rsCR2.1-4
recombinant short (truncated) CD21 receptor containing SRCs 1 to 4
- SCR
short consensus repeat
- sCD21
soluble CD21 receptor
- TFA
trifluoroacetic acid
- TIS
triisopropylsilane
- TMB
3,3′,5,5′-tetramethylbenzidine
- TPBS
PBS containing 0.1% Tween 20
References
- 1.Kopeèek J. Soluble biomedical polymers. Polim Med. 1977;7(3):191–221. [PubMed] [Google Scholar]
- 2.Kopeèek J, Kopeèková P, Minko T, Lu Z. HPMA copolymer-anticancer drug conjugates: design, activity, and mechanism of action. Eur J Pharm Biopharm. 2000;50(1):61–81. doi: 10.1016/s0939-6411(00)00075-8. [DOI] [PubMed] [Google Scholar]
- 3.Dreher MR, Liu W, Michelich CR, Dewhirst MW, Yuan F, Chilkoti A. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J Natl Cancer Inst. 2006;98(5):335–44. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 4.Maeda H, Seymour LW, Miyamoto Y. Conjugates of anticancer agents and polymers: advantages of macromolecular therapeutics in vivo. Bioconjug Chem. 1992;3(5):351–62. doi: 10.1021/bc00017a001. [DOI] [PubMed] [Google Scholar]
- 5.Shiah JG, Dvo ák M, Kopeèková P, Sun Y, Peterson CM, Kopeèek J. Biodistribution and antitumour efficacy of long-circulating N-(2-hydroxypropyl)methacrylamide copolymer-doxorubicin conjugates in nude mice. Eur J Cancer. 2001;37(1):131–9. doi: 10.1016/s0959-8049(00)00374-9. [DOI] [PubMed] [Google Scholar]
- 6.Rathi RC, Kopeèková P, Øíhová B, Kopeèek J. N-(2-Hydroxypropyl)methacrylamide copolymers containing pendent saccharide moieties. Synthesis and bioadhesive properties. J Polym Sci Part A: Polym Chem. 1991;29:1895–1991. [Google Scholar]
- 7.Wróblewski S, Berenson M, Kopeèková P, Kopeèek J. Potential of lectin-N-(2-hydroxypropyl)methacrylamide copolymer-drug conjugates for the treatment of pre-cancerous conditions. J Control Release. 2001;74(1–3):283–93. doi: 10.1016/s0168-3659(01)00338-8. [DOI] [PubMed] [Google Scholar]
- 8.Omelyanenko V, Kopeèková P, Gentry C, Shiah JG, Kopeèek J. HPMA copolymer-anticancer drug-OV-TL16 antibody conjugates. 1. Influence of the method of synthesis on the binding affinity to OVCAR-3 ovarian carcinoma cells in vitro. J Drug Target. 1996;3(5):357–73. doi: 10.3109/10611869608996827. [DOI] [PubMed] [Google Scholar]
- 9.Shiah JG, Sun Y, Kopeèková P, Peterson CM, Straight RC, Kopeèek J. Combination chemotherapy and photodynamic therapy of targetable N-(2-hydroxypropyl)methacrylamide copolymer-doxorubicin/mesochlorin e(6)-OV-TL 16 antibody immunoconjugates. J Control Release. 2001;74(1–3):249–53. doi: 10.1016/s0168-3659(01)00325-x. [DOI] [PubMed] [Google Scholar]
- 10.Lu ZR, Kopeèková P, Kopeèek J. Polymerizable Fab′ antibody fragments for targeting of anticancer drugs. Nat Biotechnol. 1999;17(11):1101–4. doi: 10.1038/15085. [DOI] [PubMed] [Google Scholar]
- 11.Lu ZR, Shiah JG, Kopeèková P, Kopeèek J. Polymerizable Fab′ antibody fragment targeted photodynamic cancer therapy in nude mice. STP Pharma Sci. 2003;13(1):69–75. [Google Scholar]
- 12.Tang A, Kopeèková P, Kopeèek J. Binding and Cytotoxicity of HPMA Copolymer Conjugates to Lymphocytes Mediated by Receptor-Binding Epitopes. Pharmaceutical Res. 2003;20(3):360–367. doi: 10.1023/a:1022639701388. [DOI] [PubMed] [Google Scholar]
- 13.Line BR, Mitra A, Nan A, Ghandehari H. Targeting tumor angiogenesis: comparison of peptide and polymer-peptide conjugates. J Nucl Med. 2005;46(9):1552–60. [PubMed] [Google Scholar]
- 14.Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66(3):184–99. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 15.Ding H, Prodinger WM, Kopeèek J. Identification of CD21-Binding Peptides with Phage Display and Investigation of Binding Properties of HPMA Copolymer-Peptide Conjugates. Bioconjug Chem. 2006;17(2):514–23. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 16.Press OW, Leonard JP, Coiffier B, Levy R, Timmerman J. Immunotherapy of Non-Hodgkin’s lymphomas. Hematology (Am Soc Hematol Educ Program) 2001:221–40. doi: 10.1182/asheducation-2001.1.221. [DOI] [PubMed] [Google Scholar]
- 17.Prodinger WM. Complement receptor type two (CR2, CR21): a target for influencing the humoral immune response and antigen-trapping. Immunol Res. 1999;20(3):187–94. doi: 10.1007/BF02790402. [DOI] [PubMed] [Google Scholar]
- 18.Rask R, Rasmussen JM, Hansen HV, Bysted P, Svehag SE. Complement C3d, g/Epstein-Barr virus receptor density on human B-lymphocytes estimated by immunoenzymatic assay and immunocytochemistry. J Clin Lab Immunol. 1988;25(3):153–6. [PubMed] [Google Scholar]
- 19.Tang A, Kopeèek J. Presentation of epitopes on genetically engineered peptides and selection of lymphoma-targeting moieties based on epitope biorecognition. Biomacromolecules. 2002;3(3):421–31. doi: 10.1021/bm015606+. [DOI] [PubMed] [Google Scholar]
- 20.Hess MW, Schwendinger MG, Eskelinen EL, Pfaller K, Pavelka M, Dierich MP, Prodinger WM. Tracing uptake of C3dg-conjugated antigen into B cells via complement receptor type 2 (CR2, CD21) Blood. 2000;95(8):2617–23. [PubMed] [Google Scholar]
- 21.Prodinger WM, Schoch J, Schwendinger MG, Hellwage J, Parson W, Zipfel PF, Dierich MP. Expression in insect cells of the functional domain of CD21 (complement receptor type two) as a truncated soluble molecule using a baculovirus vector. Immunopharmacology. 1997;38(1–2):141–8. doi: 10.1016/s0162-3109(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, Enstrom AM, Lam KS. Combinatorial peptide library methods for immunobiology research. Exp Hematol. 2003;31(1):11–30. doi: 10.1016/s0301-472x(02)01008-1. [DOI] [PubMed] [Google Scholar]
- 23.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 24.Lam KS, Lebl M, Krchnak V. The “One-Bead-One-Compound” Combinatorial Library Method. Chem Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 25.Lam KS, Lehman AL, Song A, Doan N, Enstrom AM, Maxwell J, Liu R. Synthesis and screening of “one-bead one-compound” combinatorial peptide libraries. Methods Enzymol. 2003;369:298–322. doi: 10.1016/S0076-6879(03)69017-8. [DOI] [PubMed] [Google Scholar]
- 26.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports--principles and applications. J Immunol Methods. 2002;267(1):13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 27.Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:149–69. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- 28.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–7. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 29.Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97(2):391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 30.Stahl S, Uhlen M. Bacterial surface display: trends and progress. Trends Biotechnol. 1997;15(5):185–92. doi: 10.1016/S0167-7799(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 31.Westerlund-Wikstrom B. Peptide display on bacterial flagella: principles and applications. Int J Med Microbiol. 2000;290(3):223–30. doi: 10.1016/S1438-4221(00)80119-8. [DOI] [PubMed] [Google Scholar]
- 32.Furka ASF, Asgedom M, Dibo G. Cornucopia of peptides by synthesis. Highlights of Modern Biochemistry, Proceedings of the 14th International Congress of Biochemistry; Prague, Czechoslovakia. 1988. pp. 47–47. [Google Scholar]
- 33.Furka ASF, Asgedom M, Dibo G. More Peptide by less labour. Poster presented at Xth International Symposium on Medicinal Chemistry; Budapest. 1988. [Google Scholar]
- 34.Olivos HJ, Bachhawat-Sikder K, Kodadek T. Quantum dots as a visual aid for screening bead-bound combinatorial libraries. Chembiochem. 2003;4(11):1242–5. doi: 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- 35.Garske AL, Denu JM. SIRT1 top 40 hits: use of one-bead, one-compound acetyl-peptide libraries and quantum dots to probe deacetylase specificity. Biochemistry. 2006;45(1):94–101. doi: 10.1021/bi052015l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin M, Nielsen K. Binding of the Brucella abortus lipopolysaccharide O-chain fragment to a monoclonal antibody. Quantitative analysis by fluorescence quenching and polarization. J Biol Chem. 1997;272(5):2821–7. doi: 10.1074/jbc.272.5.2821. [DOI] [PubMed] [Google Scholar]
- 37.Prota AE, Sage DR, Stehle T, Fingeroth JD. The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding. Proc Natl Acad Sci U S A. 2002;99(16):10641–6. doi: 10.1073/pnas.162360499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szakonyi G, Guthridge JM, Li D, Young K, Holers VM, Chen XS. Structure of complement receptor 2 in complex with its C3d ligand. Science. 2001;292(5522):1725–8. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- 39.Miller RT, Kubier P. Blocking of endogenous avidin-binding activity in immunohistochemistry: The use of egg whites. Appl Immunohistochem. 1997;5:63–66. [Google Scholar]
- 40.Kopecky EM, Greinstetter S, Pabinger I, Buchacher A, Romisch J, Jungbauer A. Effect of oriented or random PEGylation on bioactivity of a factor VIII inhibitor blocking peptide. Biotechnol Bioeng. 2006;93(4):647–55. doi: 10.1002/bit.20706. [DOI] [PubMed] [Google Scholar]
- 41.Fremeaux-Bacchi V, Kolb JP, Rakotobe S, Kazatchkine MD, Fischer EM. Functional properties of soluble CD21. Immunopharmacology. 1999;42(1–3):31–7. doi: 10.1016/s0162-3109(99)00026-0. [DOI] [PubMed] [Google Scholar]
- 42.Meldal M, Svendsen I, Breddam K, Auzanneau FI. Portion-mixing peptide libraries of quenched fluorogenic substrates for complete subsite mapping of endoprotease specificity. Proc Natl Acad Sci U S A. 1994;91(8):3314–8. doi: 10.1073/pnas.91.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam KS, Lou Q, Zhao ZG, Smith J, Chen ML, Pleshko E, Salmon SE. Idiotype specific peptides bind to the surface immunoglobulins of two murine B-cell lymphoma lines, inducing signal transduction. Biomed Pept Proteins Nucleic Acids. 1995;1(3):205–10. [PubMed] [Google Scholar]
- 44.Park SI, Manat R, Vikstrom B, Amro N, Song L, Lam KS. The use of one-bead one-compound combinatorial library method to identify peptide ligands for α4β1 integrin receptor in non-Hodgkin’s lymphoma. Lett Pep Sci. 2002;8:171–178. [Google Scholar]
- 45.Meldal M. The one-bead two-compound assay for solid phase screening of combinatorial libraries. Biopolymers. 2002;66(2):93–100. doi: 10.1002/bip.10229. [DOI] [PubMed] [Google Scholar]
- 46.Aggarwal S, Harden JL, Denmeade SR. Synthesis and screening of a random dimeric peptide library using the one-bead-one-dimer combinatorial approach. Bioconjug Chem. 2006;17(2):335–40. doi: 10.1021/bc0502659. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Peng L, Liu R, Xu B, Lam KS. Applications of topologically segregated bilayer beads in ‘one-bead one-compound’ combinatorial libraries. J Pept Res. 2005;65(1):130–8. doi: 10.1111/j.1399-3011.2005.00192.x. [DOI] [PubMed] [Google Scholar]
- 48.Kodadek T, Reddy MM, Olivos HJ, Bachhawat-Sikder K, Alluri PG. Synthetic molecules as antibody replacements. Acc Chem Res. 2004;37:711–718. doi: 10.1021/ar030145l. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Peng L, Liu R, Gill SS, Lam KS. Partial alloc-deprotection approach for ladder synthesis of “one-bead one-compound” combinatorial libraries. J Comb Chem. 2005;7(2):197–209. doi: 10.1021/cc049887b. [DOI] [PubMed] [Google Scholar]
- 50.Song A, Zhang J, Lebrilla CB, Lam KS. A novel and rapid encoding method based on mass spectrometry for “one-bead-one-compound” small molecule combinatorial libraries. J Am Chem Soc. 2003;125(20):6180–8. doi: 10.1021/ja034539j. [DOI] [PubMed] [Google Scholar]
- 51.Kassarjian A, Schellenberger V, Turck CW. Screening of synthetic peptide libraries with radiolabeled acceptor molecules. Pept Res. 1993;6(3):129–33. [PubMed] [Google Scholar]
- 52.Aina OH, Marik J, Liu R, Lau DH, Lam KS. Identification of novel targeting peptides for human ovarian cancer cells using “one-bead one-compound” combinatorial libraries. Mol Cancer Ther. 2005;4(5):806–13. doi: 10.1158/1535-7163.MCT-05-0029. [DOI] [PubMed] [Google Scholar]
- 53.Itoda K, Tamiya E, Yokoyama K. Evaluation of the molecular recognition of peptide-conjugated polymer. Anal Sci. 2003;19(1):185–7. doi: 10.2116/analsci.19.185. [DOI] [PubMed] [Google Scholar]
- 54.Sadler K, Zeng W, Jackson DC. Synthetic peptide epitope-based polymers: controlling size and determining the efficiency of epitope incorporation. J Pept Res. 2002;60(3):150–8. doi: 10.1034/j.1399-3011.2002.21009.x. [DOI] [PubMed] [Google Scholar]
- 55.Salmon SE, Lam KS, Lebl M, Kandola A, Khattri PS, Wade S, Patek M, Kocis P, Krchnak V, Thorpe D, et al. Discovery of biologically active peptides in random libraries: solution-phase testing after staged orthogonal release from resin beads. Proc Natl Acad Sci U S A. 1993;90(24):11708–12. doi: 10.1073/pnas.90.24.11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Healy JM, Murayama O, Maeda T, Yoshino K, Sekiguchi K, Kikuchi M. Peptide ligands for integrin alpha v beta 3 selected from random phage display libraries. Biochemistry. 1995;34(12):3948–55. doi: 10.1021/bi00012a012. [DOI] [PubMed] [Google Scholar]
- 57.Kolonin M, Pasqualini R, Arap W. Molecular addresses in blood vessels as targets for therapy. Curr Opin Chem Biol. 2001;5(3):308–13. doi: 10.1016/s1367-5931(00)00207-6. [DOI] [PubMed] [Google Scholar]
- 58.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8(2):121–7. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]