Abstract
The adenylate-forming enzymes, including acyl-CoA synthetases, the adenylation domains of non-ribosomal peptide synthetases (NRPS), and firefly luciferase, perform two half-reactions in a ping-pong mechanism. We have proposed a Domain Alternation mechanism for these enzymes whereby, upon completion of the initial adenylation reaction, the C-terminal domain of these enzymes undergoes a 140° rotation to perform the second thioester-forming half reaction. Structural and kinetic data of mutant enzymes support this hypothesis. We present here mutations to Salmonella enterica Acetyl-CoA Synthetase (Acs) and test the ability of the enzymes to catalyze the complete reaction and the adenylation half-reaction. Substitution of Lys609 with alanine results in an enzyme that is unable to catalyze the adenylate reaction, while the Gly524 to leucine substitution is unable to catalyze the complete reaction yet catalyzes the adenylation half-reaction with activity comparable to the wild-type enzyme. The positions of these two residues, which are located on the mobile C-terminal domain, strongly support the domain alternation hypothesis. We also present steady-state kinetic data of putative substrate-binding residues and demonstrate that no single residue plays a dominant role in dictating CoA binding. We have also created two mutations in the active site to alter the acyl substrate specificity. Finally, the crystallographic structures of wild-type Acs and mutants R194A, R584A, R584E, K609A, and V386A are presented to support the biochemical analysis.
Keywords: Adenylate-forming Enzymes, Non-Ribosomal Peptide Synthetase, Domain Alteration, X-ray Crystallography, CoA-ligase
The adenylate-forming superfamily is composed of a diverse group of enzymes that utilize ATP and an acyl- or aryl-carboxylate to form an adenylate intermediate. The superfamily consists of three subfamilies: the acyl- or aryl-CoA synthetases (also called ligases), the adenylation domains of the non-ribosomal peptide synthetases (NRPSs) (1–3), and the firefly luciferases. Between the subfamilies there is 20 to 40% sequence homology (4, 5). The acyl-adenylate intermediate usually reacts with the pantetheine thiol of an acyl carrier protein or CoA to form a thioester. In the case of the firefly luciferase subfamily, however, the adenylate reacts in an oxidative decarboxylation to yield an activated carbonyl that decomposes to emit light. The average size of these enzymes is 500–700 amino acids in length. The structures of several family members demonstrate that the enzymes contain a large 400–550 residue N-terminal domain and a smaller ~130 residue C-terminal domain.
The 72-kDa Acetyl-CoA synthetase (Acs, EC 6.2.1.1) from Salmonella enterica is part of the adenylate-forming superfamily (6). This enzyme allows bacteria to use acetate as a sole carbon and energy source. In low acetate concentrations, Acs is responsible for acetyl-CoA synthesis while at higher concentrations, acetyl-CoA production is carried about by acetate kinase and phosphotransacetylase (7). Synthesis of acetyl-CoA is carried out in two steps. In the first half-reaction Acs combines acetate with ATP to form an acyl-adenylate. In the second half-reaction, the thiol of CoA attacks the acyl carbon displacing the AMP leaving group to form the product acetyl-CoA (Scheme 1).
Scheme 1.
Members of the adenylate-forming superfamily contain ten regions of highly conserved residues that have been named A1–A10 (2). A conserved residue, part of the A8 region, forms a hinge that bridges the N-terminus to the C-terminus of these enzymes (6). This conserved hinge residue is usually an aspartic acid (Asp517 for Acs) however a lysine residue has also been observed (8). The A10 conserved region contains a lysine at the C-terminus of the adenylate-forming enzymes with a consensus sequence of PXXXXGK (Lys609 in Acs). This residue is known to be important for the adenylate-forming half-reaction based upon kinetic studies of propionyl-CoA synthetase and firefly luciferase (9, 10). The A5 conserved sequence of YGXTE is found at the ATP binding pocket, which can be seen in the 4-chlorobenzoate:CoA ligase/synthetase (CBAL) (11), PheA (12), and firefly luciferase (8) structures. In Acs, this sequence is modified to W413WQTE; Trp413 is orientated away from the active site, while Trp414 truncates the acyl binding pocket, accommodating the small acyl substrate acetate. Finally, there is a highly conserved region with the consensus sequence of (S/T)(S/T/G)G(S/T)(S/T)GXPKG (Thr264-Gly273 of Acs). This glycine- and Ser/Thr-rich sequence of the A3 region is thought to play a role in orientating the three phosphates of ATP prior to the first half-reaction (13) in a manner that is somewhat analogous to the Walker P-loop of certain nucleotide hydrolases (14).
The conserved hinge residue is proposed to be the pivot point for a C-terminal rotation of ~140° allowing for both half-reaction to occur (6). This Domain Alternation hypothesis arises from biochemical and crystallographic studies of a number of adenylate-forming enzyme. The three dimensional structures of PheA (12), DhbE (15), CBAL (11), and Eukaryotic Acs (16) have been solved in the proposed adenylate-forming conformation. Firefly luciferase has been solved in the adenylate forming conformation (17), as well as in an intermediate conformation of unknown significance (8). Prokaryotic Acs (6) was solved with the C-terminus rotated ~140° and in the presence of CoA relative to the orientation seen in the original PheA structure.
We present here a series of structural and functional studies of site-directed mutants of Acs. Residues from both the N- and C-terminal domains were chosen to explore the impact of side chain substitution on the steady state kinetics of ATP and CoA. In addition to residues that interact with the CoA, we have also mutated the hinge aspartic acid residue of the A8 region to glycine and proline in an attempt to increase and decrease, respectively, the conformational flexibility of this residue. Finally, in light of the relative promiscuity of the members of this family to recognize multiple acyl containing substrates (9, 18, 19), several residues were mutated to alter the acyl substrate specificity. Crystallographic structures of the ligand bound wild-type enzyme, as well as the R194A, R584A, R584E, K609A and V386A mutant proteins are also presented.
Methods
Materials
Adenosine-5'-propylphosphate was generously provided by Dr. Alex Horswill and Dr. Jorge Escalante-Semerena (9). NADH, sodium acetate, pyruvate, CoA, AMP, HEPPS, BTP,and HEPPS, as well as myokinase, pyruvate kinase, and lactate dehydrogenase, used in the coupled enzyme assay, were from Sigma-Aldrich (Milwaukee, WI). The 32P-PPi was obtained from Perkin-Elmer Life Sciences (Shelton, CT). TCEP was from Hampton Research (Aliso Viejo, CA). Polyethylene glycol (8000 mw) was from Fluka (Milwaukee, WI).
Acs Mutagenesis
Mutations were made to wild-type Acs/pTYB1 construct (6), using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutagenic primers were designed to introduce silent restriction sites for preliminary identification of mutants. Plasmid sequences were confirmed to contain the mutation by DNA sequencing of the Acs gene at Roswell Park Cancer Institute, Buffalo, N.Y.
Expression and Purification of wild-type Acs and mutants
Wild-type and Mutant Acs proteins were purified as reported previously (6). LB containing 50 mg/ml ampicillin was inoculated with BL21(DE3) cells harboring the expression plasmid and grown overnight at 37°C in a shaker. The 25 ml culture was used to inoculate a 1 L flask. Cells were grown at 16°C. At mid log phase (OD600=0.6) cells were induced with 1 mM IPTG and allowed to grow for 12–15 hrs. Cells were harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C until needed. Purification was achieved utilizing the IMPACT protein purification system (New England Biolabs, Ipswich MA). Acs is produced as a C-terminal fusion protein with a chitin-binding domain, joined by a modified intein that is succeptible to thiol-dependent cleavage (20). The protein is adsorbed to a chitin affinity column and incubated for 40 h at 4°C on column with 30 mM DTT. Following thiol-dependent cleavage of the chitin binding domain-intein fusion protein, the 652-residue protein with no extra residues derived from the expression vector is eluted from the column. After purification, protein was dialyzed to 10 mM EPPS (pH 8.0) and 0.2 mM TCEP. Wild-type and mutant proteins were then concentrated to > 10 mg/ml and frozen as small aliquots in liquid N2 until needed (21).
Kinetic Determination of Apparent KM, kcat and kcat/KM
A NADH consumption assay (9) was used which contained 50 mM HEPES buffer, pH 7.5, 5 mM MgCl2, 1 mM TCEP, 3 mM phosphoenolpyruvate, 5 units myokinase, 1 unit pyruvate kinase, 1.5 units lactate dehydrogenase, and 0.1 mM NADH. In this assay, the production of AMP is detected through coupled assays with Myokinase, which catalyzes the production of 2 ADP molecules from AMP and ATP, Pyruvate Kinase, which converts phosphoenolpyruvate and ADP to pyruvate and ATP, and lactate dehydrogenase, which converts pyruvate to lactate with the oxidation of NADH to NAD+. The loss of NADH is monitored with ε340 of 6220 M−1 min−1. Apparent kinetic constants were determined by varying one substrate while holding the remaining two constant. To determine kinetic constants for ATP, CoA and Acetate were held constant at 1.0 mM and 20 mM, respectively, and ATP was varied from 5 to 1000 µM. To determine kinetics for CoA, ATP and Acetate were held constant at 2.5 mM and 20 mM, and CoA was varied from 50 to 800 µM. Assays were preincubated at 37°C in disposable cuvettes in an Agilent 8453 spectrophotometer with a circulating water-jacket. The reaction was initialized with 0.052 µM of wild-type Acs or 0.104 µM mutant Acs. Acs proteins were diluted in 250 mM HEPES buffer, pH 7.5 before assay measurement. The reaction was monitored at 340 nm for 1 min. Initial velocity data were analyzed using the program Dynafit (22).
KM, kcat, and kcat/KM determination for Acetate, Propionate, and Glycine
The same coupled assay was used to determine the kinetic constants for acetate, propionate, and glycine. ATP and CoA were held constant at 2.5 mM and 1.5 mM, respectively, while acetate varied from 0.5 mM to 5 mM, propionate varied from 0.5 mM to 5 mM, and glycine varied from 1 mM to 15 mM. The reaction was initialized with 0.005 µM of wild-type Acs, V386A, or V310D. Proteins were diluted in 250 mM HEPES buffer, pH 7.5 before assay measurements.
Pyrophosphate exchange assay
The pyrophosphate-exchange assay was used to measure the activity of the adenylate-forming half-reaction by monitoring the incorporation of 32P-labeled pyrophosphate into ATP through cycles of the reversible adenylation half-reaction (23). The reaction contained 2 mM ATP, 0.2 mM NaPPi, 5 mM Acetate, 50 mM HEPES (pH 8.0), 100 mM NaCl, 10 mM MgCl2, and 1.5 µCi/ml of 32PPi. The reaction was initialized with 10 µM of enzyme and allowed to incubate at 37°C for 10 min. The reaction was quenched with a solution containing 1.2% active charcoal, 0.1 M unlabeled PPi, and 0.35 M perchloric acid. The charcoal pellet was washed twice with 1 ml H2O and then resuspended in 0.5 ml H2O for scintillation counting. Counts were converted to µmol/min/mg using a standard curve of known activity. Each assay was performed with five replicates along with a control for background.
Crystallization of wild-type Acs and mutant Acs
Crystals were grown via the vapor diffusion method using 6–15% PEG 8000, 9–13% ethylene glycol, and 50 mM BTP, pH 6.5. Wild-type Acs was incubated with 1 mM of AMP, CoA, and acetate prior to set up. 1 mM each of propyl-AMP, CoA, and TCEP was added to the mutant protein sample before set up. Crystals were cryoprotected by transferring through three solutions containing increasing amounts of ethylene glycol (14,18,24%) for approximately 1 minute each. The final cryoprotectant solution contained 25% PEG 8000, 24% ethylene glycol, and 50 mM BTP (pH 6.5). Each cryoprotectant solution contained 1.25 mM of ligand.
Crystallographic data collection and data refinement
Data collection for wild-type Acs and the mutants R194A and V386A were collected at the National Synchrotron Light Source at Brookhaven National Laboratory at beamlines X4A (R194A) and X12C (wild-type and V386A). Data for mutants R584A, R584E, and K609A were collected at the Cornell High Energy Synchrotron Source beamline F2. Two crystal forms were identified from identical crystallization conditions, the monoclinic form observed in our original structure (6) and a new orthorhombic form. Both crystal forms contained two molecules in the asymmetric unit. All data were processed with HKL2000 (24), except for the R584A mutant crystals, which were processed with MOSFLM and SCALA (25, 26), and converted from intensities to structure factors with TRUNCATE of the CCP4 package (25). All structures were solved by molecular replacement using MOLREP (27) of the CCP4 package using the wild-type Acs (1PG4) structure as a search model with all non-protein atoms removed. Refinement of all structures was performed with REFMAC5 (28) using weighted individual atomic B-factors. Manual model building was performed with TURBO (29) and COOT (30). Water molecules were included into spherical, or nearly spherical difference density that made chemically plausible interactions with protein atoms or neighboring waters. Near the completion of refinement, TLS anisotropic refinement (31) was included for the N- and C-terminal domains, which was monitored by a drop in the Rfree value. The final models all are missing the N-terminal four or five residues, the C-terminal five or six residues, and contain one disordered loop at positions 623 through 633; the density of this disordered loop was of variable quality and the exact number of omitted residues varied from two to ten. This loop and the disorder at the N- and C-termini were also observed to be disordered in the original structure (6).
Results
To explore the effects of specific residues on the activity of Acs, a series of point mutations were constructed. Mutant enzymes were purified and subjected to steady state kinetics to determine apparent kinetic constants for ATP and CoA. Additionally, the effect of several mutations on the adenylation half-reaction was examined with the PPi-exchange assay. The apparent kcat and KM values for both ATP and CoA are similar to constants that have been observed previously for both PrpE and CBAL (9, 13).
Mutation in Lys609 from the C-terminal domain
Previous studies suggested that the lysine of the A10 region is catalytically responsible for the first half-reaction (9, 10). This lysine has been observed in structures of the adenylate-forming conformation of PheA (12), DhbE (15), and Luciferase (17) to interact with the α-phosphate of the adenylate. S. enterica Acs shares this conserved lysine at position 609. Mutation of this lysine in PrpE has been shown to block specifically the adenylation half-reaction while leaving the thioester-forming half-reaction unaffected (9). The Acs enzyme from S. enterica (32) and mouse (33) have been shown to be regulated by acetylation of this C-terminal lysine; the enzyme is activated by the activity of a deacetylase of the sirtuin family. A mutation from a lysine to an alanine at position 609 was created in Acs. No detectable activity was observed for this mutant of Acs in steady-state kinetic experiments. The ability of this enzyme to catalyze the adenylation half-reaction was determined with PPi-exchange assay (see below).
Mutations in residues that interact with CoA or play a role in stabilization of the thioester-forming conformation
The previous structural characterization of Acs in the thioester-forming conformation in the presence of propyl-AMP and CoA (6) was used to identify residues that interact with CoA (Figure 1) and perhaps stabilize the second half-reaction conformation. These residues were targeted for analysis by site-directed mutagenesis. The results for the kinetics are shown in Table 1.
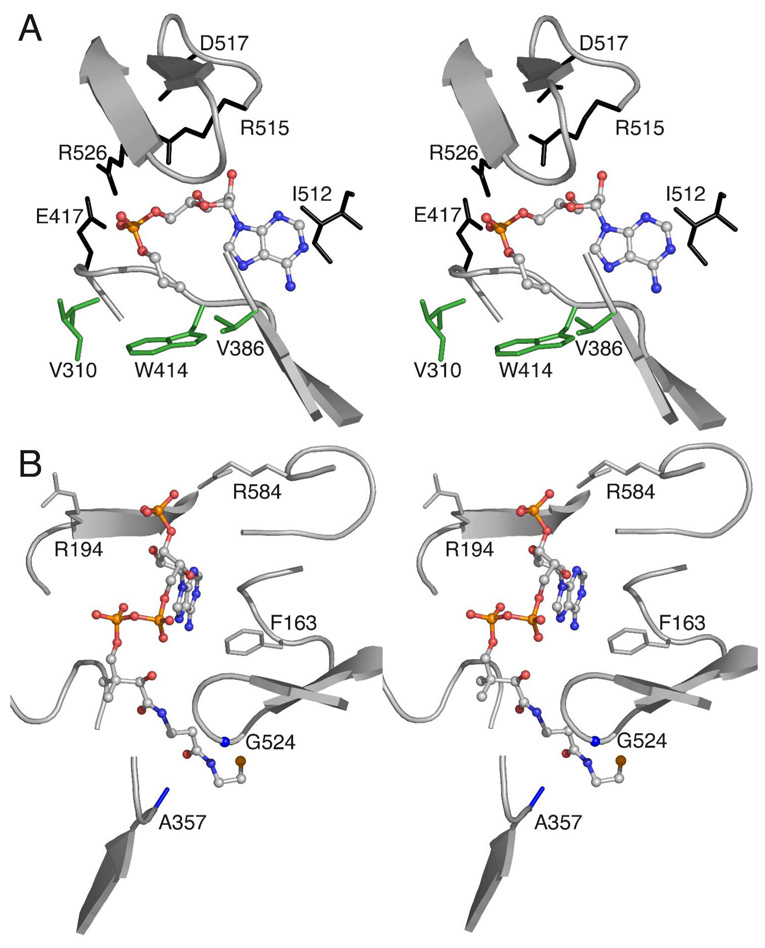
Figure 1. Active site interactions of Acs.
The structure of the wild-type Acs enzyme determined in the presence of propyl-AMP and CoA (1PG4) and amino acids that form the active site pocket are shown. A. The orientation of propyl-AMP bound to Acs in the thioester-forming conformation illustrates residues that interact that interact with the nucleotide. Interactions made to the nucleotide include Arg526 and Arg515, which interact with the phosphate group. The acetate binding pocket is formed by the three side chains of Val310, Val386, and Trp414 (shown in green). B. The CoA binding site. The nucleotide of CoA binds at the surface of the protein and the pantetheine group leads in to the adenylate site through a tunnel that is bordered by Ala357 and Gly524, two residues that were mutated to contain bulkier side chains. Arg584 was positioned to interact with the CoA 3' phosphate. Arg194 was oriented away from the phosphate but could interact through a torsional rotation.
Table 1.
Steady State Kinetics with Acs Mutations
| Enzyme | KM ATP (µM) |
kcat ATP (s−1) |
kcat/KM ATP (µM−1 s−1) |
KM CoA (µM) |
kcat CoA (s−1) |
kcat/KM CoA (µM−1 s−1) |
|---|---|---|---|---|---|---|
| Wild-type | 77.1 ± 13 | 100.6 ± 2.6 | 1.3 ± .2 | 50 ± 4.5 | 95.1 ± 1.9 | 1.9 ± .2 |
| Mutations that affect the adenylation half-reaction | ||||||
| K609Aa | -- | -- | -- | -- | -- | -- |
| Mutations of residues that interact with CoA or stabilize the thioester-forming conformation | ||||||
| R194A | 28.7 ± 2.2 | 39.9 ± 0.5 | 1.4 ± .1 | 141.7 ± 17.4 | 41.1 ± 0.7 | 0.3 ± .04 |
| R194E | 37.3 ± 5.7 | 41.7 ± 1.3 | 1.1 ± .2 | 107.2 ± 11.9 | 40.0 ± 1.5 | 0.4 ± .05 |
| R584A | 38.1 ± 5.5 | 46.7 ± 1.2 | 1.2 ± .2 | 358.3 ± 60.6 | 41.5 ± 1.1 | 0.1 ± .02 |
| R584E | 26.5 ± 2.3 | 36.4 ± 0.6 | 1.4 ± .1 | 426.0 ± 39.0 | 40.8 ± 0.8 | 0.09 ± .009 |
| R526A | 22.4 ± 3.5 | 37.4 ± 1.3 | 1.7 ± .3 | 205.0 ± 31.2 | 40.5 ± 0.6 | 0.2 ± .03 |
| Mutations that border the pantetheine binding tunnel | ||||||
| A357V | 23.8 ± 2.7 | 38.9 ± 0.5 | 1.6 ± .2 | 133.0 ± 18.0 | 35.0 ± 0.8 | 0.3 ± .04 |
| G524S | 63.8 ± 7.3 | 49.8 ± 1.6 | 0.8 ± .09 | 448.0 ± 90 | 43.1 ± 1.1 | 0.1 ± .02 |
| G524La | -- | -- | -- | -- | -- | -- |
| Mutations in the hinge residue, Asp517 | ||||||
| D517P | 44.0 ± 3.2 | 40.9 ± .7 | 0.9 ± .07 | 527.8 ± 60 | 43.8 ± 1.5 | 0.08 ± .009 |
| D517G | 243.0 ± 13.8 | 42.8 ± 0.6 | 0.2 ± .01 | 228.0 ± 31.7 | 42.4 ± 0.7 | 0.2 ± .03 |
No detectable activity was observed for K609A and G524L.
Arg194 and Arg584 interact with the CoA 3'-phosphate (6); these residues were mutated to alanine and glutamate. The kcat values for the enzymes containing each of the four mutations are reduced by roughly a factor of two. The KM values for ATP are reduced in each protein by an equivalent amount resulting in no change in kcat/KM with respect to ATP. The apparent kinetic constants for CoA, however, show an increase in KM for CoA ranging from 2- to 3-fold for the Arg194 mutations and 7- to 8-fold for the Arg584 mutations. At limiting CoA concentrations, the rate of kcat/KM are reduced as much as 20-fold.
Acs contains an Arg526 residue that is located on the C-terminal domain on the loop joining a two-stranded antiparallel β-sheet that follows the Asp517 hinge residue; this Arg is part of the A8 region and is highly conserved although not to the same degree as the hinge reisdue (Asp517) and Gly524. In the proposed thioester-forming conformation, the Arg526 forms an ionic interaction with the highly conserved Glu417 of the N-terminal domain (Figure 1A). In the crystal structure (6), the side chain also interacts with the phosphate of the adenylate intermediate. Although Arg526 does not interact directly with the CoA, we thought that it may stabilize the thioester-forming conformation through the interaction with Glu417. The R526A mutation results in a two-fold reduction in the apparent kcat values for both ATP and CoA, and a 9.5-fold reduction in kcat/KM for CoA.
Two residues were identified that abut the channel that the pantetheine group of CoA occupies when bound. The mutations made to these residues were designed to fill this channel to prevent CoA binding. The mutation of Ala357 to a valine exhibited only limited effects on CoA binding with a reduced kcat/KM for CoA by 6.3-fold. Thus, the pantetheine group of CoA can still pass through the tunnel in the presence of the larger side chain at position Ala357.
A more dramatic result was observed with the second mutation at the pantetheine binding tunnel. Gly524 is universally conserved in the A8 region of adenylate-forming enzymes, occurring on a loop that follows the hinge between the N- and C- terminal domains. The β-alanine portion of CoA passes below the Gly524 Cα position (Figure 1A). Gly524 was mutated to a serine and to a leucine to add increasingly larger side chains that might affect CoA binding. The G524S mutation had little effect on the apparent kinetic constants for ATP. In contrast, the kcat for CoA was reduced by a factor of two and, more significantly, the kcat/KM was reduced by nearly 20-fold, resulting in an increase in KM of nearly 9-fold. Increasing the size of the side chain at this position with the G524L mutation reduced activity below detectable levels. As with the K609A mutation, this enzyme was analyzed further to determine its ability to catalyze the adenylation half-reaction (see below).
Mutations that Targeted the Hinge Residue
Comparing the structures of adenylate forming enzymes in both conformations, a change in the backbone torsional angles is observed at a conserved residue located between the N- and C-terminal domains (6, 11). Two mutations were made to the hinge residue of the A8 region. A D517P mutation was made that would increase the rigidity of this hinge and a D517G mutation was made to increase the conformational flexibility of the hinge area. Both mutant proteins exhibited activity (Table 1). Again, the effetc on the ATP kinetic constants were more limited than the effects on CoA. The kcat values changed by only two-fold. Interestingly, the effects on the KM for ATP of the D517G mutant suggest that the C-terminal domain may have difficulty adopting a stable conformation for the adenylation half-reaction. The kinetics with CoA demonstrate that the proline and glycine mutant enzymes are able to catalyze the second half-reaction, however the affinity for CoA is reduced in both mutant enzymes. The D517P mutant protein has the lowest kcat/KM value observed for CoA.
Pyrophosphate exchange assay on hinge and inactive mutants
To examine which half-reaction was more dramatically effected by the mutations, we used the PPi-exchange assay to further examine several mutant enzymes (Table 2). The K609A mutation showed no activity with the PPi-exchange assay, while G524L, D517P, and D517G all showed activity similar to wild-type Acs. Of note, the G524L mutation, which showed no activity using the NADH consumption assay, can quite readily catalyze the adenylation half-reaction, demonstrating that the G524L mutation is disrupting only the second half-reaction, most likely by occluding the pantetheine tunnel.
Table 2.
PPi-exchange Assay results
| Enzyme | Activity (µmol ATP produced min−1 mg−1) |
|---|---|
| No enzyme | 3 ± 2 |
| Wild-type | 128 ± 5 |
| K609A | 4 ± 1 |
| G524L | 159 ± 39 |
| D517P | 194 ± 23 |
| D517G | 206 ± 18 |
Substrate specificity studies
The acetate binding pocket of Acs (Figure 1A) is formed by three residues, Val310, Val386, and Trp414 (6). We examined whether we could alter the substrate specificity of Acs by targeted mutation of these residues. Bacterial propionyl-CoA synthetases (9) contain an alanine in place of Val386 however retain the other two acyl pocket residues. We thus constructed a V386A mutant to attempt to alter the specific activity of Acs towards propionate. The V386A mutation decreased the catalytic efficiency for acetate and more than doubled its catalytic efficiency for propionate (Table 3). Thus, the single mutation of V386A switches the substrate preference from acetate to propionate.
Table 3.
Substrate Kinetics
| Enzyme | Substrate | KM (µM) | kcat (s−1) | kcat/KM (µM−1 s−1) | Relative Catalytic Eff.a |
|---|---|---|---|---|---|
| Wild-type | Acetate | 6047 ± 1024 | 276.8 ± 21.4 | 0.045 ± 0.008 | |
| Propionate | 9413 ± 1709 | 261.0 ± 20.8 | 0.027 ± 0.005 | ||
| Glycine | 9450 ± 1658 | 259.4 ± 13.3 | 0.027 ± 0.005 | ||
| V386A | Acetate | 6403 ± 701 | 183.5 ± 10.3 | 0.028 ± 0.003 | 0.6 |
| Propionate | 1539 ± 119 | 93.4 ± 2.4 | 0.060 ± 0.005 | 2.2 | |
| V310D | Acetate | 2464 ± 321 | 232.0 ± 12.3 | 0.090 ± 0.010 | 2.0 |
| Glycine | 8367 ± 753 | 165.5 ± 6.4 | 0.020 ± 0.002 | 0.7 |
Relative catalytic efficiency is defined as the ratio of kcat/KM for a particular substrate for the mutant enzyme compared to the wild-type.
Analysis of the amino acid binding pockets of NRPS adenylation domains (12, 34) identifies a conserved aspartic acid that interacts with the substrate α-amino group. The homolgous residue in Acs is Val310, which forms part of the acetate binding pocket. We replaced Val310 of Acs with aspartic acid to create an enzyme that might accommodate the amino acid glycine as an acyl substrate. The V310D mutation increased the catalytic efficiency for acetate while the catalytic efficiency of this mutant enzyme for glycine remained similar to that of wild-type. This mutation is thus unable to improve the Acs activity with an α-amino acid.
Structural determination of wild-type and mutant Acs
To supplement the kinetic analyses and demonstrate that no large scale structural rearrangements have occurred, the proteins were used in crysallographic studies. A new wild-type crystal structure was obtained along with the structures of the R194A, R584A, R584E, K609A, and V386A mutant enzymes. Suitable crystals of the remaining proteins were not obtained; because all of the other mutant proteins retain activity in either the complete or PPi-exchange assay, we believe that the failure to obtain crystals does not result from large-scale structural changes or improper protein folding. The structures, obtained in two crystals forms, all are in the proposed thioester-forming conformation. The crystals diffract to moderate resolution (Table 4) and the molecular models refined with good geometry and statistics (Table 5).
Table 4.
Crystallographic statistics for wild-type and mutant Acs.
| Wild-type AMP + | R194A | R584A | R584E | K609A CoA + | V386A CoA + | |
|---|---|---|---|---|---|---|
| Acetate + CoA | Propyl-AMP | Propyl-AMP | Propyl-AMP | PropylAMP | Propyl-AMP | |
| Resolution | 25 − 2.6 Å | 40 − 2.1 Å | 30 − 2.2 Å | 35 − 2.4 Å | 35 − 2.3 Å | 35 − 2.0 Å |
| Space Group | P212121 | P212121 | P212121 | P212121 | P212121 | P21 |
| Unit Cell | a= 90.9 Å | a= 91.7 Å | a= 91.8 Å | a= 92.0 Å | a= 91.6 Å | a= 59.9 Å |
| b= 94.9 Å | b= 95.5 Å | b= 95.4 Å | b= 95.5 Å | b= 94.5 Å | b= 143.0 Å | |
| c= 161.8 Å | c= 163.5 Å | c= 164.7 Å | c= 164.2 Å | c= 167.5 Å | c= 71.3 Å | |
| β= 90.9 | ||||||
| Rmerge a | 7.2% (22.4%) | 5.5% (27.9%) | 6.2% (31.4%) | 10.6% (36.4%) | 8.5% (39.9%) | 4.7% (21.3%) |
| Completenessa | 99.6% (100%) | 91.8% (58.4%) | 92.0% (61.6%) | 93.4% (81.0%) | 95.2% (81.0%) | 96.9% (85.1%) |
| I/σa | 17.5 (3.0) | 17.5 (2.4) | 20.1 (4.7) | 11.7 (2.0) | 11.8 (1.9) | 17.3 (4.2) |
| # Observations | 224405 | 259320 | 313104 | 173551 | 202699 | 200915 |
| # Reflections | 44408 | 77387 | 67992 | 55167 | 62317 | 59117 |
The number in parentheses represents the value for the highest resolution shell.
Table 5.
Refinement statistics for wt and mutant Acs
| Wild-type AMP + | R194A | R584A | R584E | K609A CoA + | V386A CoA + | |
|---|---|---|---|---|---|---|
| Acetate + CoA | Propyl-AMP | Propyl-AMP | Propyl-AMP | PropylAMP | Propyl-AMP | |
| Rcrysta | 18.6% (22.6%) | 18.0% (20.3%) | 17.8% (28.1%) | 18.8% (23.0%) | 18.8% (23.5%) | 16.8% (17.7%) |
| Rfreea | 24.7% (34.1%) | 22.0% (26.6%) | 23.3% (37.0%) | 24.5% (32.8%) | 23.7% (27.5%) | 21.1% (24.3%) |
| Wilson B-factor | 38.3 Å2 | 24.9 Å2 | 26.3 Å2 | 31.2 Å2 | 29.6 Å2 | 27.8 Å2 |
| Average B-factor (all atoms) | 23.9 Å2 | 24.0 Å2 | 23.2 Å2 | 24.2 Å2 | 24.6 Å2 | 25.3 Å2 |
| Average B-factor (N-term)b | (A) 20.2 Å2 | (A) 20.8 Å2 | (A) 20.2 Å2 | (A) 22.3 Å2 | (A) 20.8 Å2 | (A) 22.0 Å2 |
| (B) 22.5 Å2 | (B) 24.3 Å2 | (B) 25.0 Å2 | (B) 24.6 Å2 | (B) 26.2 Å2 | (B) 26.4 Å2 | |
| Average B-factor (C-term)b | (A) 36.4 Å2 | (A) 33.1 Å2 | (A) 28.3 Å2 | (A) 30.2 Å2 | (A) 31.8 Å2 | (A) 28.9 Å2 |
| (B) 2.1 Å2 | (B) 24.4 Å2 | (B) 22.4 Å2 | (B) 25.7 Å2 | (B) 25.6 Å2 | (B) 29.8 Å2 | |
| Average B-factor (Solvent) | 18.9 Å2 | 29.8 Å2 | 26.8 Å2 | 20.7 Å2 | 23.4 Å2 | 25.8 Å2 |
| Solvent Molecules | 162 | 625 | 600 | 239 | 384 | 316 |
| Average B-factor (Propyl-AMP)b,c | (A) 15.3 Å2 | (A) 14.6 Å2 | (A) 12.3 Å2 | (A) 17.2 Å2 | (A) 15.6 Å2 | (A) 18.3 Å2 |
| (B) 16.5 Å2 | (B) 16.9 Å2 | (B) 14.0 Å2 | (B) 19.7 Å2 | (B) 19.2 Å2 | (B) 20.7 Å2 | |
| Average B-factor (CoA)b | (A) 46.3 Å2 | (A) 48.3 Å2 | (A) 45.9 Å2 | |||
| (B) 44.4 Å2 | (B) 53.9 Å2 | (B) 44.8 Å2 | ||||
| rms deviations (lengths, angles) | 0.014 Å, 1.53° | 0.011 Å, 1.14° | 0.011 Å, 1.23° | 0.013 Å, 1.40° | 0.012 Å, 1.33° | 0.012 Å, 1.35° |
The number in parentheses represents the value for the highest resolution shell.
A and B refer to the two protein chains within the asymmetric unit.
For the wild-type enzyme, the average B-value is shown for AMP. The average B-value for acetate atoms is 55.6 and 60.5 for the A and B subunits.
Whereas the prior Acs crystal structure was determined with CoA and propyl-AMP, the current wild-type protein was incubated with acetate, AMP, and CoA to attempt to trap an adenylate-like complex. Despite repeated efforts, we have never been able to crystallize the bacterial Acs enzyme in the adenylate-forming conformation; the yeast Acs enzyme, which exhibits 45% sequence identity with the bacterial enzyme, was determined in the adenylateforming conformation by Jogl and Tong (16).
The crystals of the mutant proteins were grown in the presence of propyl-AMP and CoA. Ligand density is shown in Figure 2. As was observed in the original structure (6), the density of the CoA demonstrates that the cofactor is very well-ordered at the nucleotide however the pantetheine moiety is not well ordered. In the omit maps generated for the wild-type (Figure 2A) and the K609A (Figure 2C) structures, density is present for the pantetheine although it is not continuous at a level of 2.5σ. In the V386A mutant (Figure 2B), there is no significant density for the pantetheine group at a contour level of 2.5σ although the CoA nucleotide is well ordered.
Figure 2. Ligand density for the wild-type and mutant structures.
Omit maps were generated by removing the ligands followed by a cycle of refinement. The maps of the coefficients Fo-Fc were contoured at 2.5σ and shown for the CoA ligand of wild-type (A), V386A (B), and K609A (C). Maps were generated in the same way for the propyl-AMP inhibitor of the R194A (D), R584A (E), and R584E (F) mutants as well. Neighboring protein atoms, including Arg191, which interacts with the 5' diphosphate of CoA, are labeled in panels (A) and (D).
The new wild-type structure superimposes with the original structure (6) with an rmsd for all Cα positions of 0.37 Å over all 637 residues. Acetate was present in the active site and was modeled with one oxygen interacting with the phosphate oxygen. At the 2.6Å resolution of the wild-type structure, the precise orientation of the acetate molecule and the positions of the methyl group and the non-interacting oxygen atom are unclear.
The mutant structures are all very similar to the original structure, exhibiting the proposed thioester-forming conformation (Figure 3). The rmsd values of the mutants vs. the new wild-type structures for all Cα positions (ranging from a total of 633–638 atoms for the different comparisons) range from 0.19–0.37Å. The density for all mutant residues is unequivocal. (It should be noted that the solvent exposed side chain of Lys609 of the A10 region is disordered in all Acs structures and thus cannot be used to confirm the K609A mutation.) The V386A mutant illustrates how the larger binding pocket is able to accommodate the propyl side chain (Figure 4). The propyl group of the inhibitor bound in the active site adopts a more extended conformation with the torsion angle of 172° around the C1–C2 bond. In contrast, in the wild-type structures, this torsion angle is 32°, demonstrating that the propyl group adopts a more compact orientation to fit into the active site.
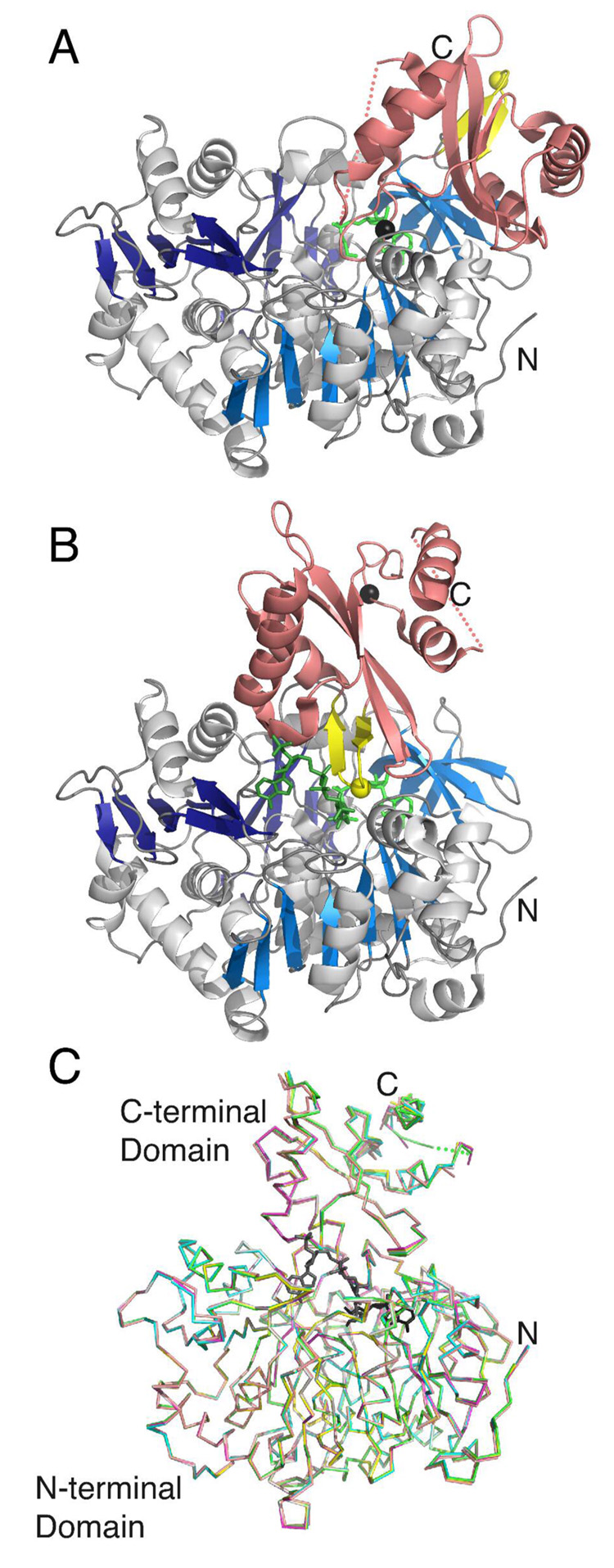
Figure 3. Three dimensional structure of Acs and the proposed conformational change.
Acs is shown in the A. adenylate-forming conformation bound to propyl-AMP and B. thioester-forming conformation bound to CoA and propyl-AMP. The N-terminal domains are shown with blue strands while the C-terminal domains is colored pink. The A8 region of the C-terminal domain is shown in yellow, with Gly524 depicted as a single yellow sphere. The Cα position of Lys609 from the A10 region is shown as a black sphere. While the thioester-forming conformation shown in B represents a crystallographically observed structure for Acs (1PG4), the adenylate-forming conformation represents a computational model that was produced by rotating the C-terminal domain of the Acs protein onto the C-terminal domain of the CBAL structure (1T5D), which was determined bound to 4CBA. The N- and C-terminal residues are labeled N and C, and the gap around residues 623–632 is shown with a dashed line. C. The Superposition of the Cα chain of the five mutant enzymes and wild-type Acs bound to acetate, AMP and CoA. The ligands from the wild-type enzyme are shown in black, while the wild-type protein is depicted with green. The mutant proteins are represented in the following colors: K609A (teal), R194A (magenta), R584A (yellow), R584E (grey), and V386A (peach).
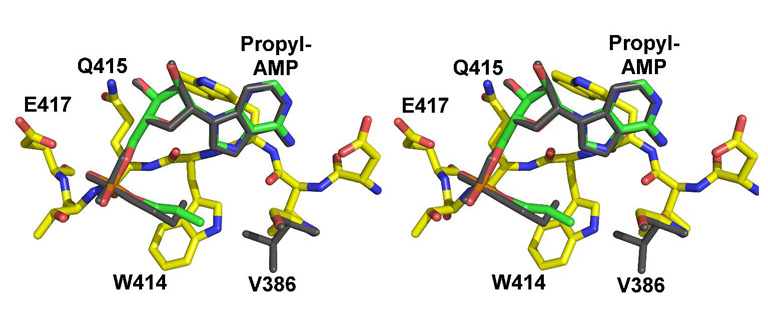
Figure 4. Superposition of the acyl binding pockets of wild-type and V386A mutants.
The models were superimposed using the Cα positions of all residues. Atoms from the V386A model are colored blue (nitrogens), red (oxygens) and yellow (protein carbons) or green (ligand carbons). The propyl-AMP ligand and Val386 from the wild-type structure are shown in dark grey and illustrate the occluded orientation torsional angles of the propyl group.
Structures were also determined for the R194A, R584A, and R584E mutant enzymes. Although CoA was included in the crystallization and cryo-protection solutions, no significant density was observed for the CoA ligands. The propyl-AMP, in contrast, was well ordered in all three mutants (Figure 2). Although all three mutant enzymes can bind CoA, as evidenced by the activity–albeit reduced–in the steady state kinetics, the affinity of these enzymes is reduced for CoA such that the CoA molecules are not bound with sufficient occupancy to be included in the refined model.
Discussion
We have proposed that the adenylate-forming enzymes adopt two different conformations to catalyze the two half-reactions. In the domain alternation hypothesis, the members of this family are believed to adopt the conformation seen in the crystal structures of PheA (12), CBAL (11), and yeast Acs (16), among others (15, 17) during the catalysis of the initial adenylation half-reaction. The C-terminal domain of these enzymes then rotates by ~140° around the hinge residue (at Asp517 in Acs) to adopt a second conformation for the second half-reaction. This thioester-forming conformation has been observed in the bacterial Acs structures as well as in a long-chain acyl-CoA synthetase structure (35). That this thioester-forming conformation has now been observed in a second crystal form of Acs strongly suggests that the conformation is not stabilized by crystal contacts but rather represents a stable protein conformation that is likely part of the reaction mechanism.
The biochemical analysis of several mutant enzymes strongly supports our hypothesis of the conformational change. In particular, the K609A and G524L mutants that are located on opposite faces of the mobile C-terminal domain specifically affect the different half-reactions. Both mutants are completely inactive when assayed for the full reaction. The K609A mutant, however, is defective in the adenylation half-reaction while the G524L mutant is able to catalyze the PPi-exchange assay quite effectively suggesting that the defect in the complete reaction occurs in the thioester-forming half-reaction. Gly524, part of the A8 region, is universally conserved in the adenylate-forming enzymes. In the crystal structures that have been determined in the adenylate-forming conformation, the Gly524 residue is 25–30Å from the active site. This clearly supports the need for the C-terminal domain to rotate into the position observed in the Acs structure during the thioester-forming half-reaction.
Additional studies by us and others have also identified mutations on the C-terminal domains of adenylate forming enzymes that specifically affect the individual half-reactions. Mutation of the Lys609 homolog in PrpE was shown biochemically to be inhibited for the adenylation half-reaction while the thioester-forming half-reaction occurred with near wild-type activity (9). Branchini and colleagues and have shown that mutations in Lys529 (10) and Lys434 (36) of firefly luciferase can specifically disrupt the adenylation and oxidative decarboxylation half-reactions, respectively. While the Lys529 residue is homologous to Lys609 of Acs, Lys434 of luciferase is homologous to Asn521 of Acs. This residue is located within the A8 region that follows the hinge residue at Asp517. While Asn521 of Acs does not make any direct contacts to ligands at the Acs active site, the side chain of this residue is positioned 5.4 Å from the ribose ring oxygen. The Lys434 residue in firefly luciferase therefore appears to play a role in the second half-reaction upon adopting the conformation observed in the Acs structures. For the NRPS adenylation domain sub-family, we have also recently shown that a poorly conserved residue in the A8 region of the C-terminal domain can be mutated to specifically affect one half-reaction (37). Arg437 of the self-standing adenylation domain EntE was mutated to an aspartic acid and assayed in both the PPi-exchange assay and the complete transfer of the 2,3-dihydroxybenzoate substrate to the pantetheine cofactor of the acyl-carrier protein of EntB. While activity of the complete reaction was severely compromised, the mutant was still able to catalyze the adenylation half-reaction. This Arg437 residue precedes the universally conserved glycine residue (Gly524 of Acs). Together with the results presented here for G524L of Acs, the A8 region that forms a two-stranded loop that follows the hinge residue has now been shown in all three subfamilies of the adenylate-forming enzymes to be involved specifically in the second half-reaction.
Analysis of the original structure of Acs (6) demonstrated that Arg194 and Arg584 of Acs interact with the 3'-phosphate of CoA, while Gly524 and Ala357 border the pantetheine tunnel. Finally, Arg526 was proposed to stablize the thioester-forming conformation by its interactions with Glu417 and the adenylate phosphate. We mutated these residues to investigate their roles in steady state kinetics with respect to ATP and CoA. With the exception of G524L, which had no detectable activity, all mutations caused a ~two-fold reduction in kcat with no effect on kcat/KM with respect to ATP. The kcat values for CoA were similarly reduced by ~two-fold and kcat/KM values were reduced by 5- to 20-fold, reflecting a slower rate at limiting CoA concentrations. The reduced affinity for CoA demonstrates a role for these residues in binding CoA or stabilizing the thioester-forming conformation.
While it is always difficult to make any quantitative conclusions from the presence or absence of ligands in a crystallographic study, it is worth noting that although CoA was maintained at constant concentration in all of the proteins studied, no density was observed for the CoA molecule in the three Arg mutant enzymes. Although the CoA molecule was not observed, it can bind at some level as the enzymes retain activity. The long-chain acyl-CoA synthetase also was crystallized in the thioester-forming conformation and, in that case as well, CoA was not modeled into the crystal structure (35). CoA was, however, included in the crystallizations at 10 mM raising the possibility that for this enzyme, as for the Arg mutants of Acs, CoA bound with sufficient occupancy to drive the conformational change, yet not with a high enough occupancy to be seen in the electron density. Interestingly, arginine residues at 191, 194, and 584, which all interact with the CoA phosphates, are not part of the A1–A10 conserved motifs (2) and are not conserved even within the acyl- and aryl-CoA subfamily of the adenylate-forming enzymes. This raises the possiblity that different residues may be involved in binding the phosphates of CoA or the peptidyl-carrier protein bound phosphopantetheine.
The D517G and D517P mutants were designed to examine the hinge residue by increasing flexibility and increasing rigidity respectively. The D517G mutation reduced the catalytic efficiency for both substrates and was the only mutation studied that increased the KM for ATP. This mutation may thus have reduced conformational stability and may be compromised in stably adopting either conformation. While the catalytic efficiency for D517P for both substrates was reduced, we note that the apparent KM values were reduced for ATP and increased for CoA. The ϕ, ψ angles of the hinge residue Asp402 are −72.4°, −15.7° in the CBAL crystal structure (11), which was solved in the adenylate-forming conformation. In contrast, the ϕ, ψ angles for Asp517 of Acs were −103.6° and −170.9° in the thioester-forming conformation. As the backbone ϕ angle of proline residues are constrained to −60° ± 20°, the D517P mutant likely has difficulty adopting the second half-reaction conformation resulting in a higher KM for CoA and a lower KM for ATP. A similar result was obtained with the homolog 4-chlorobenzoate-CoA ligase. (Wu, Reger, Gulick, and Dunaway-Mariano, in preparation).
Our attempts at changing the specificity of the acetate binding pocket of Acs were mixed. We were able to improve the preference of the enzyme for propionate over acetate with a single mutation of Val386 to alanine. The structure of this mutant enzyme bound to the propyl-AMP ligand demonstrates that, while the propyl group adopts a more constrained orientation in the wild-type enzyme, the increase in the size of the acyl substrate binding pocket allows the propyl moiety to bind in an extended conformation. The V386A mutation was one of several mutations made in an Acs homolog from Methanothermobacter thermoautotrophicus (18). The V388A mutation of the MT-ACS1 protein conferred similar improvements in the activity with propionate; the increased binding pocket, however, was not sufficiently large to accommodate butyrate as the catalytic efficiency dropped by a factor of 20 with the additional methylene group into the acyl substrate. This study further examined the substrate specificity by mutating additional residues in the acetate-binding pocket to accommodate such substrates as butyrate, valerate, and hexanoate. As we had noted in the comparison of the structures of different adenylate-forming enzymes (11), the Trp414 of Acs forms one wall of the acyl-substrate binding pocket. Replacement of this Trp with a Gly opened the binding pocket of the MT-ACS1 enzyme to function with acyl substrates as large as octanoate (18).
In contrast to the success with the V386A mutation, the V310D mutation did not confer a catalytic preference for the amino acid glycine. Indeed, the catalytic efficiency with acetate improved slightly with this mutation. The inability to utilize glycine as a substrate more efficiently most likely failed because the pocket was not large enough to accommodate both the larger Asp side chain and the larger glycine substrate. The failure of the V310D mutant to catalyze the reaction with glycine suggests that further studies are needed to understand the relationship between substrate and the substrate-binding pocket.
Acknowledgments
We would like to thank Kristen Homick and Kelly Dearing for technical assistance with production of mutant Acs enzymes and Dr. Debra Dunaway-Mariano for helpful discussions.
This research was supported in part by NIH grant GM-068440 (A. M. G.). This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS) which is supported by the National Science Foundation under award DMR 0225180 and the National Institutes of Health through its National Center for Research Resources under award 5 P41 RR001646-23. Use of NSLS, the National Synchrotron Light Source at Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Abbreviations
- Acs
acetyl-CoA synthetase
- CBAL
4-chlorobenbenzoyl-CoA ligase form Alcaligenes sp. AL3007
- PheA
phenylalanine activating adenylation domain of the gramicidin synthetase A protein
- DhbE
2,3-dihydroxybenzoate activating domain of the bacillibactin siderophore synthetase
- NRPS
non-ribosomal peptide synthetase
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- EPPS
4-(2-Hydroxyethyl)-1-piperazinepropanesulfonic acid
- PrpE
the propionyl-CoA synthetase from Salmonella enterica
- HEPES
4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid
- BTP
1,3-Bis[tris(hydroxymethyl)methylamino]propane
- propyl-AMP
adenosine-5’-propylphosphate
- rmsd
root mean square deviation
- MT-ACS1
acetyl-CoA synthetase from the thermophilic bacterium Methanothermobacter thermoautotrophicus
- LB
Luria-Bertani Media
- IPTG
Isopropyl β-D-1-thiogalactopyranoside.
Footnotes
The structure factors and coordinates have been deposited in the protein data bank. Accession codes are: wild-type + AMP + CoA, 2P2F; R194A + propyl-AMP, 2P2M; V386A + propyl-AMP + CoA, 2P2B; R584A + propyl-AMP, 2P20; R584E + propyl-AMP, 2P2Q; K609A + propyl-AMP + CoA, 2P2J.
References
- 1.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 2.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 3.Challis GL, Naismith JH. Structural aspects of non-ribosomal peptide biosynthesis. Curr Opin Struct Biol. 2004;14:748–756. doi: 10.1016/j.sbi.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 5.Dunaway-Mariano D, Babbitt PC. On the origins and functions of the enzymes of the 4-chlorobenzoate to 4-hydroxybenzoate converting pathway. Biodegradation. 1994;5:259–276. doi: 10.1007/BF00696464. [DOI] [PubMed] [Google Scholar]
- 6.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5'-propylphosphate and coenzyme A. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 7.Starai VJ, Escalante-Semerena JC. Acetyl-coenzyme A synthetase (AMP forming) Cell. Mol. Life Scie. 2004;61:2020–2030. doi: 10.1007/s00018-004-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 9.Horswill AR, Escalante-Semerena JC. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41:2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
- 10.Branchini BR, Murtiashaw MH, Magyar RA, Anderson SM. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. doi: 10.1021/bi9928804. [DOI] [PubMed] [Google Scholar]
- 11.Gulick AM, Lu X, Dunaway-Mariano D. Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states. Biochemistry. 2004;43:8670–8679. doi: 10.1021/bi049384m. [DOI] [PubMed] [Google Scholar]
- 12.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. Embo J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang KH, Xiang H, Dunaway-Mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36:15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- 14.Saraste M, Sibbald PR, Wittinghofer A. The P-loop--a common motif in ATP-and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 15.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jogl G, Tong L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry. 2004;43:1425–1431. doi: 10.1021/bi035911a. [DOI] [PubMed] [Google Scholar]
- 17.Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 18.Ingram-Smith C, Woods BI, Smith KS. Characterization of the acyl substrate binding pocket of acetyl-CoA synthetase. Biochemistry. 2006;45:11482–11490. doi: 10.1021/bi061023e. [DOI] [PubMed] [Google Scholar]
- 19.Linne U, Schafer A, Stubbs MT, Marahiel MA. Aminoacyl-coenzyme A synthesis catalyzed by adenylation domains. FEBS Lett. 2007;581:905–910. doi: 10.1016/j.febslet.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 20.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self- cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 21.Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WG, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 23.Linne U, Marahiel MA. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry. 2000;39:10439–10447. doi: 10.1021/bi000768w. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski ZMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallography. 1988;21:916–924. [Google Scholar]
- 27.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1991. TURBO-FRODO; p. 86. [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 32.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 33.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 35.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem. 2004;279:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 36.Branchini BR, Southworth TL, Murtiashaw MH, Wilkinson SR, Khattak NF, Rosenberg JC, Zimmer M. Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry. 2005;44:1385–1393. doi: 10.1021/bi047903f. [DOI] [PubMed] [Google Scholar]
- 37.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]