Abstract
The innermost layer of the Drosophila eggshell, the vitelline membrane, provides structural support and positional information to the embryo. It is assembled in an incompletely understood manner from four major proteins to form a homogeneous, transparent extracellular matrix. Here we show that RNAi knockdown or genetic deletion of a minor constituent of this matrix, Palisade, results in structural disruptions during the initial synthesis of the vitelline membrane by somatic follicle cells surrounding the oocyte, including wide size variation among the precursor vitelline bodies and disorganization of follicle cell microvilli. Loss of Palisade or the microvillar protein Cad99C results in abnormal uptake into the oocyte of sV17, a major vitelline membrane protein, and defects in non-disulfide cross-linking of sV17 and sV23, while loss of Palisade has additional effects on processing and disulfide cross-linking of these proteins. Embryos surrounded by the abnormal vitelline membranes synthesized when Palisade is reduced are fertilized but undergo developmental arrest, usually during the first 13 nuclear divisions, with a nuclear phenotype of chromatin margination similar to that described for wild-type embryos subjected to anoxia. Our results demonstrate that Palisade is involved in coordinating assembly of the vitelline membrane and is required for functional properties of the eggshell.
Keywords: Drosophila, vitelline membrane, sV17, sV23, CG9050, Palisade, Cad99C, microvilli, extracellular matrix assembly, ends-out targeting
INTRODUCTION
Assembly of extracellular matrices as diverse as tendons, basement membranes, and the pearl oyster shell requires specific cell-matrix interactions, self-assembly processes, accessory proteins modulating assembly and morphology of networks, and post-secretory proteolysis and cross-linking to generate structures with physical and functional properties appropriate to a given tissue (Kalluri, 2003; Zhang et al., 2006; Zhang et al., 2005). The Drosophila eggshell provides a valuable genetic model for studying regulated matrix assembly during development (Waring, 2000). Synthesized by somatic follicle cells surrounding the oocyte, the eggshell plays roles in sperm entry, gas exchange, physical protection of the egg, and embryonic patterning. It has a complex architecture, with a homogeneous vitelline membrane layer immediately adjacent to the oocyte, surrounded sequentially by a wax layer, inner chorionic layer, endochorion, and exochorion (Margaritis et al., 1980; Papassideri et al., 1991). The vitelline membrane is of particular interest as, in addition to its structural roles, it serves as a stable repository for positional information used in locally activating a receptor tyrosine kinase that patterns the embryo termini (Stevens et al., 2003), and may perform a similar role in establishment of the embryonic dorsoventral axis. Such roles may be analogous to the storage of growth factors and localized glycoproteins within the extracellular matrix of vertebrate tissues (Taipale and Keski-Oja, 1997; Tzarfaty-Majar et al., 2001), and argue for the importance of understanding the molecular interactions involved in the assembly and function of this eggshell layer.
The vitelline membrane is the first eggshell layer to be synthesized, in Stages 8-10 of oogenesis. It is first visible as a discontinuous layer of precursor vitelline bodies within the perivitelline space, comprising aggregates of secreted eggshell proteins (Mahowald and Kambysellis, 1980; Trougakos et al., 2001). The vitelline bodies are physically separated from one another by microvilli that extend from the oocyte and follicle cells, spanning the perivitelline space. Disorganization of the follicle cell microvilli caused by loss of the fly PCDH15 protocadherin orthologue, Cad99C, results in disordered vitelline membrane formation (D’Alterio et al., 2005; Schlichting et al., 2006), suggesting that the microvilli may act as a scaffold on which the vitelline bodies are assembled. These microvilli shorten in Stage 10B as the vitelline bodies coalesce to form a continuous vitelline membrane, which then thins over the remainder of oogenesis.
At a molecular level, four major structural proteins of the vitelline membrane have been identified (sV17, sV23, VM32E, VM34C) (Waring, 2000). These proteins are small (116-168 amino acids), rich in proline and alanine residues, and share a highly conserved 38 amino acid VM domain. Their assembly into the vitelline membrane involves stage-specific proteolytic processing (Pascucci et al., 1996) and disulfide cross-linking (Andrenacci et al., 2001; LeMosy and Hashimoto, 2000; Manogaran and Waring, 2004; Petri et al., 1979) during oogenesis, and non-disulfide cross-linking occurring rapidly following ovulation and likely involving cross-linking of tyrosines (Heifetz et al., 2001; Petri et al., 1976). The VM domain is essential for disulfide cross-linking, containing the 3 cysteines found in each of these proteins. Partial deletions of this domain in sV23 and in VM32E result in proteins that are poorly secreted and unstable (sV23 only) and that are unable to stably incorporate into the matrix (Andrenacci et al., 2001; Manogaran and Waring, 2004). The later tyrosine-based cross-linking depends upon the earlier alignment of cross-linking sites during assembly of the vitelline membrane proteins. In particular, the N-terminal domain of sV23, although later cleaved away from the mature protein, appears to be critical for the initial alignment of vitelline membrane proteins into an array or polymer (Manogaran and Waring, 2004).
Of the major structural proteins, only sV23 has been studied as a null mutant resulting in 100% collapsed eggs that remain unfertilized (Savant and Waring, 1989). Several other genes are required for vitelline membrane assembly, although their roles remain unclear. Eggshell collapse and defects in vitelline membrane cross-linking are seen in a subset of mutations in three oocyte surface proteoglycans, Nasrat, Polehole, and Nudel, which have additional roles in embryonic patterning (Cernilogar et al., 2001; Degelmann et al., 1990; Hong and Hashimoto, 1996; LeMosy and Hashimoto, 2000; Turcotte and Hashimoto, 2002). Two additional genes, yellow-g and alpha-methyldopa hypersensitive, are required for vitelline membrane integrity and have been postulated to have roles in tyrosine-based cross-linking (Claycomb et al., 2004; Konrad et al., 1993). Isolated loss of Nudel’s protease activity, also required for dorsoventral patterning, leads to a specific and complete defect in tyrosine cross-linking, and a milder phenotype of vitelline membrane permeability and fragility without spontaneous eggshell collapse (LeMosy and Hashimoto, 2000). With the exception of Nudel protease, known to become active only at the time of tyrosine cross-linking, it is not clear what processes of eggshell assembly are first affected by mutation of these proteins.
As a complement to ongoing studies in other labs of the major structural components of the vitelline membrane, we have become interested in identifying minor proteins that may perform important structural or regulatory functions during eggshell formation (Fakhouri et al., 2006), e.g., in the alignment of vitelline membrane proteins into an organized array. Here we describe one such protein that we have named Palisade because its loss results in disruption of the palisade-like arrangement of vitelline bodies and follicle cell microvilli within the perivitelline space during vitelline membrane secretion. We show that Palisade is an integral component of the vitelline membrane whose knockdown or deletion affects the cross-linking and localization of major vitelline membrane proteins, consistent with a role in coordinating their assembly. Additionally, knockdown of Palisade results in a majority of embryos that initiate development but arrest prior to cellularization, supporting a developmental role for the vitelline membrane, possibly in gas exchange, that could not be visualized in previous mutations that result in early collapse of all eggs.
MATERIALS AND METHODS
Fly strains
The wild-type strains were Oregon R and w1118, the latter used for production of transgenic flies. The Gal4 driver strains PG45/FM7 (Bourbon et al., 2002), Act5C-Gal4/CyO (Ito et al., 1997), and tubP-Gal4/TM3 (Lee and Luo, 1999) were provided by D. Stein; the sV23-null mutant strain fs(2)QJ42/CyO (Savant and Waring, 1989) was provided by G. Waring; and the Cad99C-null mutant strains Cad99C21-5/TM6B and Cad99C21-8/TM6B (D’Alterio et al., 2005) were provided by D. Godt.
Generation and analysis of the UAS-psd-RNAi transgene
The transgenic RNAi construct was based on the “splice-activated hairpin” method (Lee and Carthew, 2003; Reichhart et al., 2002). We first amplified the 816 bp CS-2 intron from a plasmid provided by D. Gubb, using a 5′ primer containing a Not I site and 3′ primer containing Nhe I, Kpn I, Xba I and Hind III sites, and amplified a 460 bp psd sequence corresponding to nucleotides 150-610 in the GM32356 psd cDNA sequence, using a 5′ primer containing a Not I site and a 3′ primer containing Bgl II and Sal I sites. A 3-way ligation between Not I- Hind III cut CS-2 intron, Sal I- Not I cut psd fragment, and Sal I- Hind III cut pGEM-11Zf(+), generated an intermediate construct in which a 460 bp antisense-oriented psd fragment is upstream of the CS-2 intron. The same 460 bp sequence was re-amplified from the GM32356 template using a 5′ primer containing an Nhe I site and a 3′ primer containing an Xba I site, and inserted into the Nhe I and Xba I sites of the previous construct. A cassette containing antisense-intron-sense sequences was cut out using Bgl II and Xba I and inserted into the pUAST vector, provided by N. Perrimon and chosen to enable Gal4-driven expression in somatic follicle cells without germline expression (Brand and Perrimon, 1993; Rorth, 1998). Nineteen independent transgenic lines were generated by standard methods (Spradling, 1986); four of these were functionally tested, and the strongest two were used for subsequent studies. For functional studies of psd, females carrying one copy each of UAS-psd-RNAi and a Gal4 driver were placed at 23-25° C with wild-type OR males in either heavily yeasted vials, to stimulate oogenesis, or in egg collection cups. Constitutive high-level expression of UAS-psd-RNAi had no observable impact on viability or male fertility, consistent with psd being required only in the ovary.
Generation and analysis of the psd null allele
The psd gene (1147 bp) plus 115 bp 5′ and 58 bp 3′ flanking sequence were deleted by ends-out targeting (Gong and Golic, 2003, 2004). Flanking arms to be used in the targeting construct included a 5′ 3162 bp Arm A that included the CG13992 gene and a 3′ 3316 bp Arm B that included the sV17 gene (Fig. 1A), because of the tight packing of these genes. The arms were amplified using Platinum Taq DNA polymerase High Fidelity Mix (Invitrogen), cloned into TOPO vectors, and sequenced. Accurate clones were then cut out and sequentially ligated into the pw25 vector to flank the mini-white gene that will replace the psd gene after recombination. Following recovery of an X-chromosome insertion that was homozygous viable and fertile, we induced homologous recombination with FLP and I-Sce-I enzymes. Subsequent screening recovered 5 new insertions that exhibited female sterility either as homozygotes or when hemizygous over Df(2L)Exel7024, a deficiency that uncovers the psd gene. Two of these lines (psdMF56, psdMF104) were further verified by Southern blotting of genomic DNA from adult homozygotes with a probe representing the entire psd gene to lack any detectable psd-hybridizing sequences, and by generation of correctly-sized 5′ and 3′ PCR fragments using primers from outside the flanking arms in combination with mini-white gene sequences. Further phenotypic characterization and verification of absence of the Palisade protein in psdMF56 homozygous null ovaries by Western blotting and immunofluorescence staining is described in Results.
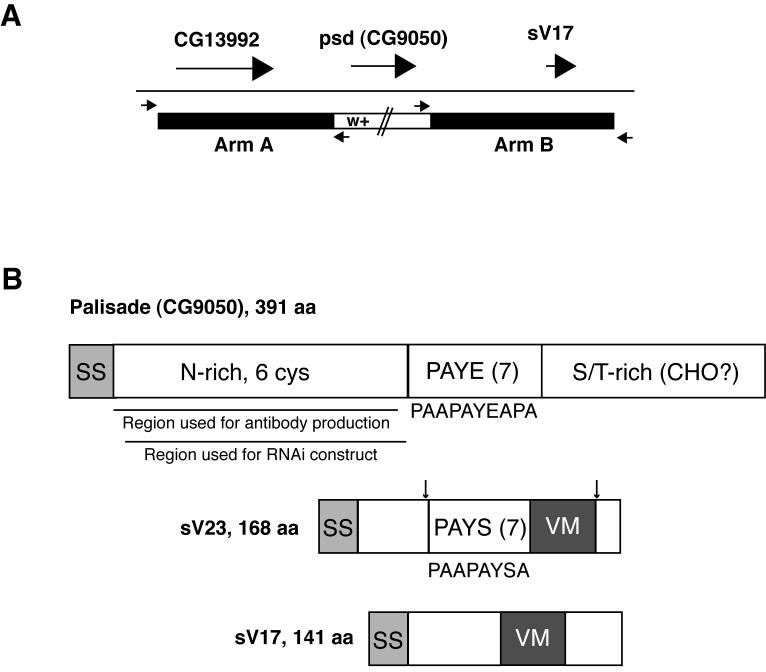
Fig. 1. Palisade gene and protein diagrams.
(A) Illustration of the strategy used for ends-out targeted replacement of the psd (CG9050) gene with the mini-white marker (see Methods). Large arrows represent psd and flanking genes, while small arrows represent primers used to verify the correct insertion of mini-white into the psd locus. (B) Comparison of Palisade protein structure to the sV23 and sV17 major vitelline membrane proteins. (Top) Palisade contains signal sequence (SS), an asparagine-rich domain including 6 of 8 cysteines in the protein (N-rich, 6 cys), a domain containing 7 imperfect copies of a 10 aa repeat (consensus listed below diagram) enriched in proline, alanine, tyrosine, and glutamic acid (PAYE(7)), and a domain enriched in serine and threonine (S/T-rich) and containing 11 of 15 predicted O-glycosylation sites (CHO?). Also shown are regions used for antibody and UAS-psd-RNAi construct generation. (Middle and bottom) sV23 and sV17 share the VM domain, missing in Palisade, while sV23 also contains a repeat region containing 7 copies of an imperfect 8 aa repeat (consensus listed below diagram) enriched in proline, alanine, tyrosine and serine (PAYS(7)). In sV23, approximate positions of N- and C-terminal cleavages are indicated by arrows; sV17 also undergoes cleavage following secretion but sites have not been demonstrated (Manogaran and Waring, 2004; Pascucci et al., 1996).
Antibodies and immunostaining
A trpE fusion protein containing Palisade amino acids 26-184 was prepared as described (LeMosy et al., 1998). Polyclonal antibodies were raised in two mice by the UGA Monoclonal Antibody Facility, and were affinity-purified by selection on fusion protein linked to Affigel-10/15 matrix (Bio-Rad), followed by depletion of trpE-specific antibodies on trpE-Affigel, and used at 1:100 for immunostaining. G. Waring provided rabbit polyclonal sV17 and sV23 antibodies (Pascucci et al., 1996), and D. Godt provided guinea pig anti-Cad99C (D’Alterio et al., 2005). Fixed ovary cryosections were used for immunostaining of the eggshell proteins Palisade, sV17 and sV23 because of poor access to the vitelline membrane in whole-mount preparations, and were detected with FITC-conjugated secondary antibodies (Jackson ImmunoResearch; used at 1:200) or Alexa-488-conjugated secondary antibodies (Molecular Probes; 1:300) and counterstained with Texas Red-phalloidin as described (LeMosy et al., 1998). The diluted sV17 and sV23 antibodies (used at 1:800) were preabsorbed against fixed, devitellinized wild-type embryos prior to staining ovary cryosections. Whole-mount ovaries gave superior preservation of follicle cell microvilli for immunostaining of Cad99C (1:3000 dilution) with Alexa-488-conjugated secondary antibody detection and Texas Red-phalloidin counterstain. Reagents used for embryo staining included DROP1.1 sperm tail antibody (1:1500) provided by T. Karr (Karr, 1991), the in situ cell death detection kit, AP (Roche), propidium iodide (4 μg/ml), and cleaved caspase-3 (Asp175) 5A1 and cleaved lamin A (Asp230) rabbit antibodies (Cell Signaling Technology). Embryos were fixed and stained as described (LeMosy et al., 1998); for panels D-F in Figure 4, embryos were from 0-18 hour collections that were further aged one hour before fixation to ensure that no newly fertilized embryos were represented, while for panels G-H in Figure 4, embryos were from a 0-4 hour collection that was fixed immediately. Imaging was performed using a Zeiss LSM510 confocal immunofluorescence microscope in the MCG Cell Imaging Core Facility. For examination of Cad99C staining, only Stage 10A egg chambers were used for optimal length and organization of microvilli, cross-sections were used (showing single row of adjacent follicle cells as judged by F-actin staining of their borders), and the most clear single-plane views of microvillar organization for each egg chamber were captured.
Western blot analysis
Counted egg chambers or laid eggs (with chorions left on) were homogenized in Laemmli sample buffer either with or without 100 mM DTT, rapidly frozen, and stored at -70° C. Before loading on gels, samples were boiled and insoluble material pelleted; in the case of samples boiled without DTT, the supernatant was transferred to a new tube and 100 mM DTT added prior to loading. Typically, 10-20 egg chambers or laid eggs were loaded per lane on 13% SDS-PAGE gels, and proteins were subsequently transferred to nitrocellulose using a semi-dry blotter (NovaBlot; GE Healthcare). Western blotting was performed using 5% nonfat dry milk/0.1% TX-100/TBS as blocking solution and developed using SuperSignal West Pico chemiluminescent substrate (Pierce). The antibodies included affinity-purified Palisade mouse antibodies (1:500), rabbit anti-sV17 (1:10,000) and anti-sV23 (1:20,000), and HRP-conjugated secondary anti-mouse and anti-rabbit antibodies (Jackson ImmunoResearch; 1:10,000 for anti-Psd and anti-sV17, 1:40,000 for anti-sV23). The Glyko enzymatic deglycosylation kit (GK80110) with prO-LINK Extender (GK80115) was used according to kit instructions for removal of N-linked sugars and some sialylated O-linked sugars and polylactosamine, and treated and untreated samples were compared on 13% SDS-PAGE gels by blotting with Palisade antibody.
Electron microscopy
Ovaries were fixed in 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 16 hours at 4° C; washed with 0.1M sodium cacodylate buffer 3 × 15 minutes at room temperature; post-fixed in 4% osmium tetroxide for 1 hour; rinsed in distilled water, dehydrated in a graded series of ethanol and cleared with 3 changes of propylene oxide; then infiltrated and polymerized in EMbed 812 with Araldite 502 resin (Electron Microscopy Sciences) overnight in a 60° C oven. Ultra thin sections were cut and stained with uranyl acetate and lead citrate. The sections were examined and photographed using a JEOL 1010 (Fig. 5) or JEM 1230 (Fig. 6) transmission electron microscope (JEOL USA); the first of these microscopes used a film camera while the second was equipped with a 16 megapixel Ultrascan 4000 CCD camera (Gatan Inc.).
RESULTS
Comparison of Palisade primary sequence to sV23 and sV17
The palisade (psd) gene (CG9050) lies at 26A on chromosome 2, within a gene cluster that also includes the genes for two well-characterized major vitelline membrane components, sV23 and sV17 (Popodi et al., 1988). Previous studies have shown that psd mRNA is co-expressed with sV23 and sV17, although at 40- to 80-fold lower levels, and that the Palisade protein is found in enriched eggshell preparations (Fakhouri et al., 2006; Popodi et al., 1988). These findings are all consistent with Palisade being a component of the vitelline membrane.
Comparison of primary sequence features (Fig. 1B) illustrates that Palisade differs in several respects from the major vitelline membrane proteins (Waring, 2000, and references therein). It is more than twice as large as the largest of these proteins, sV23, and lacks homology to the 38 amino acid VM domain found in all 4 major VM proteins. It is an acidic protein (pI 4.42), similar to VM32E (pI 4.98) but distinct from sV17, sV23, and VM34C (pIs 7.75 to 8.34), suggesting differences in its general physical properties from these proteins. Apart from its signal sequence, Palisade can be divided into three domains. At the N-terminus is a 164 amino acid domain containing 10 clustered asparagines separated only by a pair of cysteines; its mixed alpha and beta structure, 6 cysteines, and relative hydrophilicity suggest this could be a globular domain. At the C-terminus is a 125 amino acid domain enriched in serines and threonines that may be O-glycosylated (supported by NetOGlyc prediction of 15 potential mucin-like O-glycosylation sites in Palisade, 11 of which are within this domain) (Julenius et. al., 2005), and containing 2 cysteines. Between these domains is a 77 amino acid predicted alpha-helical domain containing 7 copies of an imperfect 10 amino acid repeat with the consensus sequence, PAAPAYEAPA. This domain provides the one significant similarity to VM proteins, with organization similar to that found in a central domain of sV23 containing 7 copies of an imperfect 8 amino acid repeat with the consensus sequence, PAAPAYSA; small degenerate motifs enriched in PAYS are also seen immediately N-terminal to the VM domain in VM32E and VM34C but not sV17 (Waring, 2000).
Palisade biogenesis and localization in the vitelline membrane
To determine if Palisade is a genuine component of the vitelline membrane, we raised antibodies against its N-terminal domain for Western blot and immunostaining studies. Tissues derived from RNAi knockdown and homologous recombination knockout strains developed in the course of these studies were used to verify the specificity of the antibodies in these assays. Western blot analysis demonstrated that the Palisade protein is present in Stage 10 egg chambers when the vitelline membrane is being synthesized (Fig. 2A, D) and persists throughout oogenesis although is somewhat reduced in quantity in Stage 14 egg chambers compared to Stage 10 egg chambers. Interestingly, while Palisade is predicted to be a 41 kDa protein, it migrates instead as an apparent 70 kDa protein, with a doublet visible on some blots. This migration was not altered by enzymatic deglycosylation (see Methods); however, some O-linked structures, e.g., N-acetylgalactosamine found in mucins, cannot be removed enzymatically. On optimal blots (Fig. 2D), the Stage 14 form of Palisade appears to migrate more slowly than the Stage 10 form, although it is not clear what modification occurring in the extracellular space would cause this small shift.
Fig. 2. Palisade biogenesis and verification of RNAi knockdown and null allele by Western blotting.
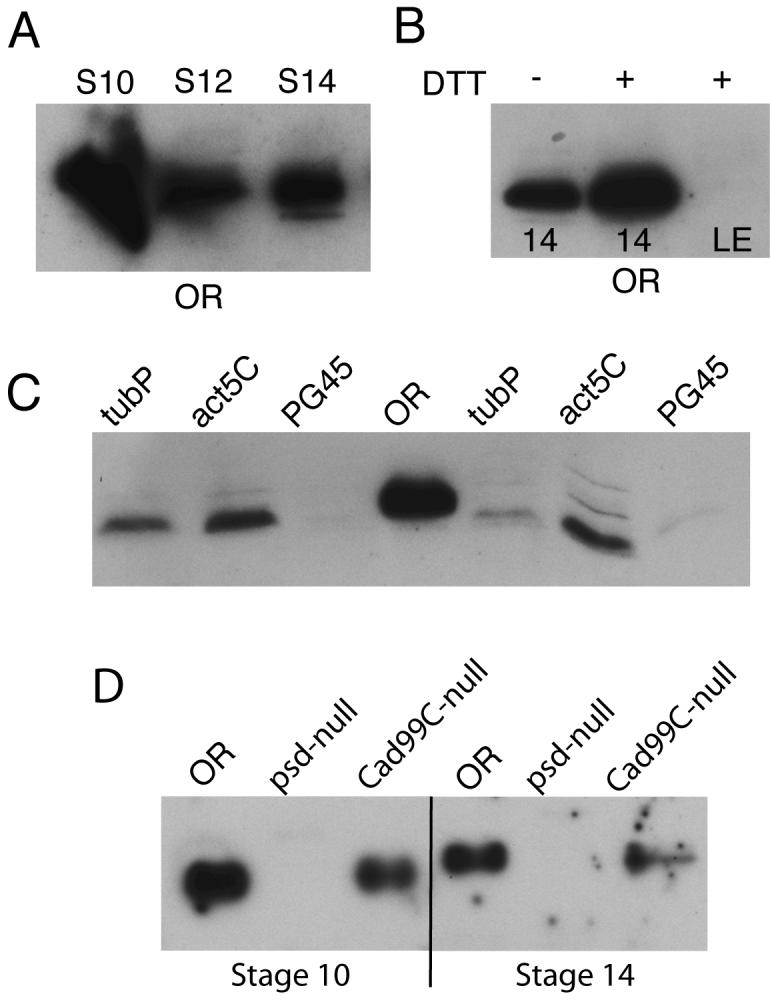
Equal numbers of egg chambers were loaded in each well; all samples were prepared under reducing conditions unless otherwise indicated. (A) Wild-type (OR) Stage 10, 12, and 14 egg chambers, showing that Palisade is synthesized in Stage 10 and persists throughout oogenesis. (B) OR Stage 14 egg chambers (labeled 14) or laid eggs (LE; chorions on) were extracted with or without 100 mM DTT as indicated. (C) Stage 14 egg chambers from psd knockdown driven by tubP, act5C, and PG45 Gal4 were compared to OR expression to demonstrate relative effectiveness of the psd knockdown by these drivers; two samples of each psd knockdown condition are shown to illustrate limited variability of expression among groups of egg chambers. (D) Comparison of Palisade levels and migration in OR, homozygous psdMF56 null, and homozygous Cad99C21-5 null Stage 10 and Stage 14 egg chambers.
The major vitelline membrane proteins are completely cross-linked by disulfide bonds in Stages 12-14 of oogenesis, then are further tyrosine cross-linked and insoluble upon egg-laying. Palisade showed less evidence of disulfide cross-linking during oogenesis. In Stage 14 egg chambers, a significant fraction of the total releasable Palisade could be extracted in the absence of reducing agents (Fig. 2B). This pattern could indicate that direct disulfide cross-linking of Palisade into the matrix occurs at a reduced efficiency compared to the major vitelline membrane proteins, or that Palisade is not disulfide cross-linked but is simply trapped within a network of disulfide-bonded proteins and more readily released when this network is disrupted. Upon egg-laying, however, Palisade can no longer be solubilized (Fig. 2B), consistent with it undergoing tyrosine-based cross-linking together with other eggshell proteins. In contrast to sV23 and sV17, which undergo proteolytic processing following their deposition in mid-oogenesis (Pascucci et al., 1996), Palisade appeared not to undergo any proteolytic processing.
For our subsequent functional studies of Palisade, the efficiency of RNAi knockdown could be monitored phenotypically (next section) or by detection of Palisade on blots of equal numbers of Stage 14 egg chambers (Fig. 2C). As previously described for transgenic RNAi inhibition of windbeutel in follicle cells (Zhu and Stein, 2004), only strong constitutive Gal4 drivers were capable of providing effective knockdown of Palisade protein levels, with tubulinP more effective than actin5C. An even stronger constitutive driver for follicle cell-expressed RNAi studies, PG45, has been characterized by Xianjun Zhu and David Stein (personal communication), in which there is a Gal4 transgene inserted in the myc gene promoter (Bourbon et al., 2002). The PG45 Gal4 driver gave nearly complete knockdown of Palisade protein expression when combined with a single copy of the UAS-psd-RNAi transgene. The psd null allele (psdMF56) generated by ends-out targeting was verified in part by the absence of Palisade protein in egg chambers from homozygous null females shown by Western blotting (Fig. 2D; see Methods for additional verification).
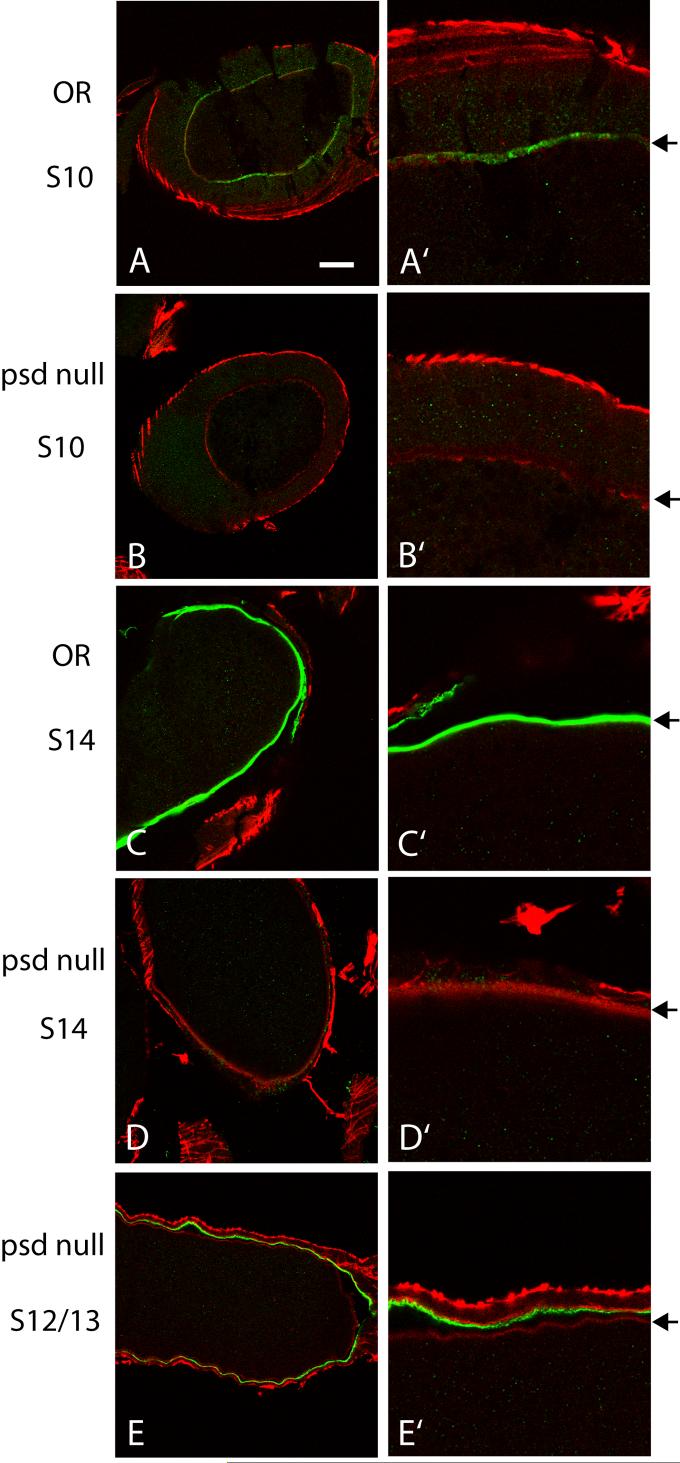
By immunostaining of sectioned OR ovaries, Palisade can be detected within the perivitelline space of Stage 10 egg chambers during synthesis of the vitelline membrane (Fig. 3A, A’). This staining is quite weak even with Alexa-488 fluorescent secondary antibody, and had been missed in earlier characterization using FITC secondary antibody (Suppl. Fig. 1A), but was verified as specific for Palisade by comparison to psd null egg chambers (Fig. 3B, B’). Only in Stage 13 and 14 OR egg chambers is there strong staining of the vitelline membrane lying immediately adjacent to the oocyte (Fig. 3C, C’) that is absent in Stage 14 psd null egg chambers (Fig. 3D, D’). The contrast between these immunostaining results and the Western blots showing that Palisade is more abundant in Stage 10 than in Stage 14 (Fig. 2A, D), documented with affinity-purified polyclonal antibodies from two mice, suggests that there may be masking of the N-terminal epitopes in Palisade that reduces detection of the native protein within the perivitelline space until late in oogenesis. There also may be some Palisade localized to the chorion, detected in the same late-stage egg chambers where the eggshell layers have separated (Fig. 3C, C’ and strongly in Supplemental Fig. 1B); however, some psd null egg chambers (typically Stage 12-13) show weak chorion staining (Fig. 3E, E’), suggesting that there are cross-reacting epitopes in this layer that do not represent authentic Palisade.
Fig. 3. Palisade is deposited in the vitelline membrane.
Panels at right (A’-E’) are 3X zoom images of sections of the corresponding egg chambers shown at left (A-E). The right-hand panels are oriented so that the oocyte is at the bottom, and the position of the vitelline membrane is marked with arrows. Palisade is detected weakly in Stage 10 (A), and strongly in Stage 13-14 (C), within the vitelline membrane, with antibody specificity verified by staining of psd null egg chambers (B, D, E). Weak, non-specific chorion staining is seen in some Stage 12-13 psd null egg chambers (E). Bar equals 30 μm in A-E and 10 μm in A’-E’.
Palisade is required for vitelline membrane function during embryogenesis
We first characterized the phenotype of embryos derived from mothers expressing the UAS-psd-RNAi construct, and will describe this in some detail as the strong hypomorphic condition allowed detection of some features missed in the psd null where only very rare embryos show any nuclear divisions and there are never hatching larvae. PG45/UAS-psd-RNAi expression resulted in early embryonic arrest with less than 1% of embryos hatching as larvae, while the weaker Act5C/UAS-psd-RNAi combination gave about 5% hatching larvae. The presence of occasional normal embryos most likely reflects variability in the knockdown of psd in individual egg chambers based on immunostaining results, and is consistent with previously reported escapers in windbeutel RNAi experiments (Zhu and Stein, 2004). In all subsequently described RNAi experiments, the PG45/UAS-psd-RNAi females were used and their tissues are referred to as psd knockdown ovaries and eggs. Similar terminology is applied to tissues derived from the psd null females.
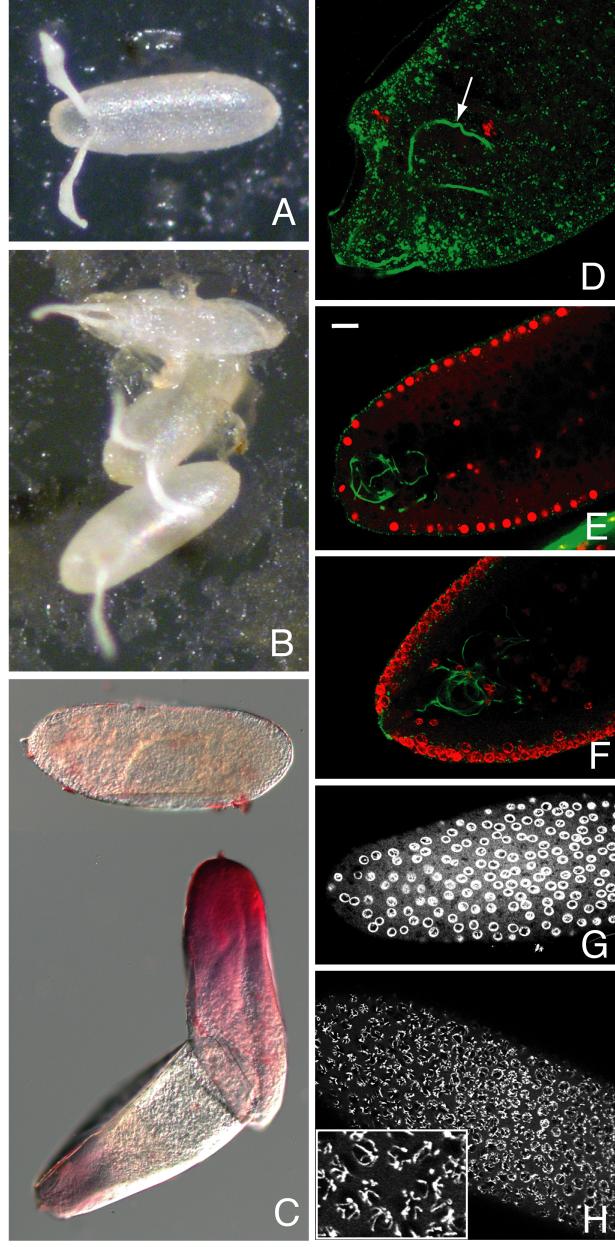
All eggs laid by psd knockdown females had grossly normal chorion morphology (cf. Fig. 4 A, B). Up to 10% of laid eggs were collapsed at or soon after deposition, while the remainder had normal chorion turgor and did not collapse over time. Following bleach dechorionation, most of the mutant eggs were permeable to neutral red dye and the vitelline membranes were difficult to remove with methanol, characteristics of mutations affecting vitelline membrane integrity (Degelmann et al., 1990; LeMosy and Hashimoto, 2000). The neutral red permeability of these eggs showed a strong preference for anterior staining near the micropyle, the structure through which the sperm enters (Fig. 4C). Fertilization did not appear to be compromised, however, as sperm could be detected in almost all embryos examined, including 7 of 10 most severely affected that showed no nuclear divisions after an hour pre-incubation on an egg-laying plate prior to fixation (Fig. 4D). The majority of psd knockdown embryos arrest prior to cellularization, although a small number (<10%) proceed to cellularization and all but the rare escapers have arrested at or before germband extension. This is an intermediate phenotype among mutations affecting vitelline membrane integrity, compared to the sV23 null mutant, lacking a major eggshell protein, that results in 100% collapsed, unfertilized eggs (Savant and Waring, 1989), and the Nudel protease mutants, affecting only tyrosine-based cross-linking, that result in embryos proceeding normally through early development but undergoing a dorsalized gastrulation (Hong and Hashimoto, 1996; LeMosy and Hashimoto, 2000). Of eggs laid by psd null females, 20-30% are collapsed at or soon after oviposition, and the others are very fragile to handling, but do not usually collapse unless in a dry environment and have apparently normal chorions as seen for the psd knockdown eggs.
Fig. 4. Vitelline membrane integrity and embryonic development are compromised by psd knockdown.
(A) OR and (B) psd knockdown laid eggs showing occasional collapse of the latter (top egg in B). (C) Most psd knockdown eggs are permeable to the dye neutral red following bleach dechorionation; top dye-impermeable egg was imaged in a different field of the same slide. (D-H) Embryos stained with propidium iodide to label DNA (red in D-F, white in G and H), and with DROP1.1 antibody to label the sperm tail (green in D-F only). (D) Severely affected psd knockdown embryo showing only oocyte and sperm DNA is still fertilized (sperm tail marked with arrow). (E) Sagittal section of wild-type syncytial blastoderm stage embryo showing even propidium iodide staining throughout nuclei. (F) Sagittal section of syncytial blastoderm stage psd knockdown embryo showing ring-like propidium iodide staining at periphery of nuclei. (G-H) Surface views of syncytial blastoderm stage psd knockdown embryos fixed after 0-4 hour collections to minimize effects of degradation after developmental arrest. These show either clear ring-like staining of DNA (G) or visibly separate chromosomes that are also at the nuclear periphery (H, enlarged in inset). Bar equals 20 μm in E-H, 10 μm in D.
A striking nuclear phenotype of abnormally condensed chromatin was visualized in the great majority of psd knockdown embryos by propidium iodide staining, and generally affected all of the nuclei in the embryo (cf. Fig. 4E and Fig. 4F-H). In some cases, individual chromosomes could be discerned at the periphery of the nucleus (Fig. 4H), but more commonly, chromatin appeared as a continuous ring around the periphery of the nucleus (Fig. 4F, G). There were also frequent empty spots and irregularities in depth of cortical nuclei in syncytial blastoderm embryos (Fig. 4F), and occasional examples of chromatin strands extending between adjacent nuclei during the earlier cleavage divisions (not shown), suggestive of cell cycle abnormalities. The dependence of these embryonic nuclear phenotypes on the psd knockdown and not on presence of PG45 insertion into the myc locus was verified by examination of psd knockdown embryos generated using the tubP and Act5C Gal4 drivers (not shown); also, in the rare psd null embryos that undergo early nuclear divisions, the peripheral chromatin phenotype could be seen (Supplemental Fig. 2). The condensed chromatin phenotype, particularly that of individual condensed chromosomes lining the periphery of the nucleus (Fig. 4H and inset), resembles that described for wild-type embryos subjected to anoxic conditions (Foe and Alberts, 1985), as well as the chromatin margination that occurs during apoptosis of mammalian cells (Sheridan et al., 1981). Apoptosis is not normally observed prior to gastrulation in Drosophila but can be induced by UV irradiation (Zhou and Steller, 2003), indicating that at least some apoptotic signaling components are present in early embryos. However, TUNEL, activated caspase-3, and anti-cleaved lamin staining each failed to demonstrate apoptosis in the psd knockdown embryos except for TUNEL labeling of many nuclei in a few late-arresting embryos (not shown), suggesting that apoptosis is not the principal endpoint of the developmental arrest in these embryos.
Loss or reduction of Palisade leads to early disorganization of perivitelline space structures
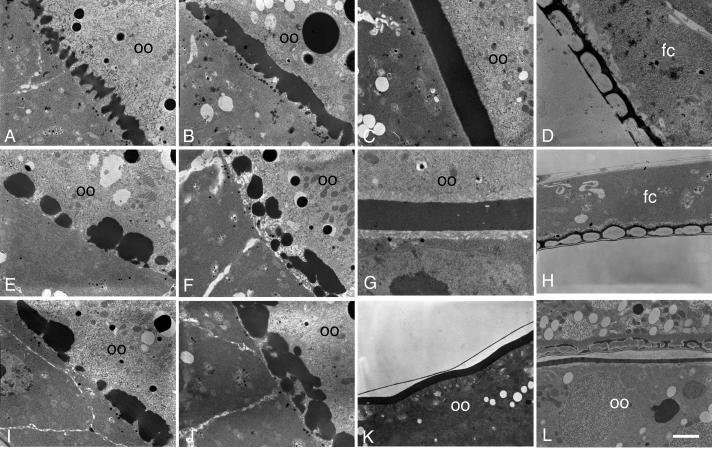
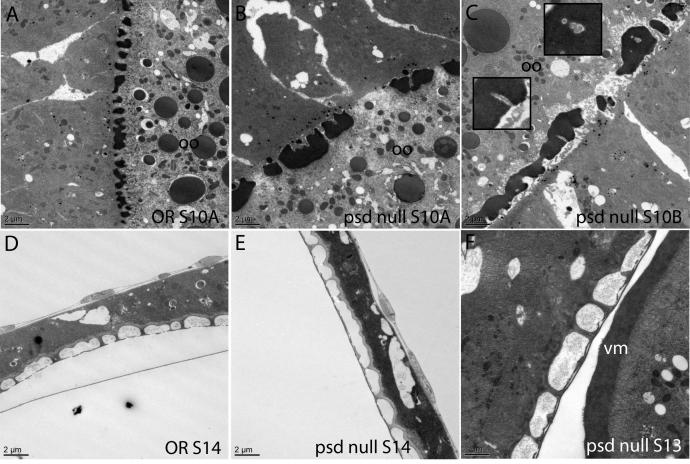
To address the underlying basis of the requirement for Palisade in vitelline membrane integrity, we first examined the biogenesis of this structure by electron microscopy. In wild-type egg chambers (Mahowald and Kambysellis, 1980; Trougakos et al., 2001), small, uniformly sized vitelline bodies are initially seen in Stages 9 and 10A with some lateral connections seen between them (Fig. 5A). These structures coalesce in Stage 10B as follicle cell microvilli retract from their contacts with the oocyte (Fig. 5B), and by Stage 11 have formed a continuous and homogeneous vitelline membrane (Fig. 5C), which will thin over the remainder of oogenesis (not shown). In psd knockdown egg chambers, defects are seen in the initial organization of the vitelline bodies. Instead of an array of evenly spaced and sized vitelline bodies, the psd knockdown vitelline bodies are irregular in size, sometimes very large or small, and unevenly spaced, giving rise to a “boulders and gaps” appearance in Stage 10A (Fig. 5E, I). These structures gradually coalesce with approximately normal timing (Fig. 5F, J), giving rise to a continuous vitelline membrane that can range from normal thickness to about half the thickness of wild type (cf. Fig. 5C, G where 5G is taken at twice magnification of 5C). Small holes are readily seen in the plane of the psd knockdown vitelline membrane at Stage 11 (Fig. 5G) and can still be detected after the vitelline membrane has thinned in late oogenesis (Fig. 5K); however, no large or full-thickness gaps were detected after coalescence was completed. The development of the inner chorionic layer (Fig. 5H, K) and endochorion structures (Fig. 5H) appears normal in the psd knockdown compared to wild-type (Fig. 5D); in contrast, the sV23 null shows collapse of an initially formed endochorion into the vitelline membrane in late oogenesis (Pascucci et al., 1996) (Fig. 5L).
Fig. 5. Electron microscopy reveals early defects in vitelline membrane morphogenesis in the psd knockdown.
(A-D) OR egg chambers. (A) Stage 10A, uniform array of vitelline bodies in the perivitelline space between the oocyte (oo) and follicle cells. (B) Stage 10B, coalescence of vitelline bodies. (C) Stage 11, uniform vitelline membrane. (D) Late-stage egg chamber, showing the endochorion, consisting of a thin floor and thick roof separated by pillars and spaces containing only light flocculent material. Follicle cells are indicated (fc), and the chorion has separated from the oocyte and vitelline membrane, which are out of the field of view. (E-K) psd knockdown egg chambers. (E, I) Stage 10A, irregularly sized and distributed vitelline bodies. and abnormally large gaps between them. (F, J) Late Stage 10A or Stage 10B, coalescence of vitelline bodies. (G) Stage 11, continuous vitelline membrane that can be thinner than in wild type, and that has small holes but no full-thickness gaps. (H, K) Late-stage egg chambers show a mature vitelline membrane with normal thickness but persistent holes (K), and normal inner chorionic layer (thin line above vitelline membrane in K) and endochorion (H). In a sV23-null mature egg chamber (L), an initially normal appearing endochorion has collapsed, with dense flocculent material in the endochorionic spaces and with electron-dense material scattered throughout a thin vitelline membrane (Pascucci et al., 1996). Bar equals 2 μm in all images except D, 750 nm, and G, 1 μm.
Similar results were seen with the psd null egg chambers (Fig. 6), establishing that the early disorganization of vitelline membrane assembly is due to the loss of Palisade rather than an RNAi artifact (cf. Fig. 6B and 6A). In several egg chambers in this newer study, we noted apparent trapping of microvilli as the vitelline membrane is coalescing from the vitelline bodies (Fig. 6C), also consistent with disorganization of the assembly process. The mature vitelline membrane was continuous without breaks in psd null egg chambers (Fig. 6F) and the chorion architecture appeared to be intact (Fig. 6D-F).
Fig. 6. Electron microscopy of the psd null confirms disorganized vitelline membrane assembly and intact chorion architecture.
Comparison of OR (A) and psd null (B) Stage 10A egg chambers illustrates the “boulders and gaps” appearance of the vitelline bodies in the psd null. During coalescence of the vitelline membrane in Stage 10B, there appeared to be trapping of microvilli within the vitelline membrane in the psd null (C; insets show enlarged images). The psd null egg chambers showed normal architecture of the chorion layers (E, F) as compared to OR (D), and the final vitelline membrane structure did not contain gaps. Bars equal 2 μm in all images except F, 1 μm.
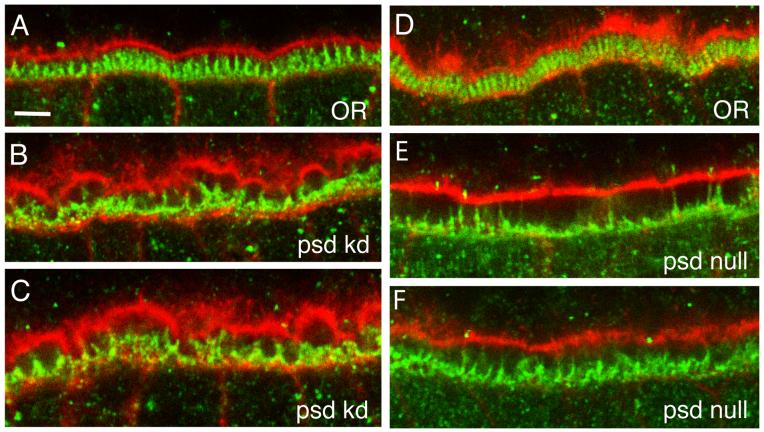
The disruption of the palisade-like arrangement of vitelline bodies in the psd knockdown and null is similar to the phenotype described for the absence of Cad99C, a component of follicle cell microvilli. In the absence of Cad99C, follicle cell microvilli are disorganized and often flattened against the surface of the follicle cells rather than making contact with oocyte microvilli, and the vitelline bodies appear to be somewhat irregular in size (D’Alterio et al., 2005; Schlichting et al., 2006). After coalescence of the vitelline bodies, irregularities in the vitelline membrane including full-thickness gaps readily detectable by light microscopy persist in the Cad99C null, in contrast to the more complete recovery of vitelline membrane morphology in later stages seen here for the psd null. To determine if microvillar organization is disrupted in the absence of Palisade, we examined the morphology of the follicle cell microvilli using a Cad99C antibody that stains these structures. In Stage 10A, when the follicle cell microvilli have reached their maximal length in wild type (Fig. 7A, D), the psd knockdown (Fig. 7B, C) and psd null (Fig. 7E, F) both showed grossly disorganized microvilli, with near absence of upright microvilli in some psd null egg chambers (Fig. 7E). In both psd knockdown and null conditions, there was a widening of the perivitelline space between the oocyte and the follicle cell epithelium, while in the psd knockdown but not the null there were also irregularities and a scalloped appearance of the cortical F-actin cytoskeleton of the oocyte. A similar widening of the perivitelline space can be noted in some images of Cad99C null follicle cells (Fig. 6D-F in Schlichting et al. (2006), and Fig. 5B in D’Alterio et al. (2005)). Thus, loss of Palisade, a minor protein of the vitelline membrane, results in the gross disorganization of both vitelline bodies and follicle cell microvilli within the perivitelline space during the phase when the vitelline membrane proteins are being secreted and assembled within this space.
Fig. 7. Loss of Palisade results in disruption of follicle cell microvillar organization during vitelline membrane synthesis.
Shown are close-up views of the perivitelline space from six different stage 10A egg chambers from OR (A, D), psd knockdown (B, C), and psd null (E, F) females. The cortical actin cytoskeleton of the oocyte (top portion of each panel) and follicle cell epithelium (bottom portion) is stained with Texas Red-phalloidin (red), while the follicle cell microvilli are stained with a Cad99C antibody (green). Bar equals 5 μm.
Loss of Palisade or Cad99C results in uptake of sV17 but not sV23 by the oocyte
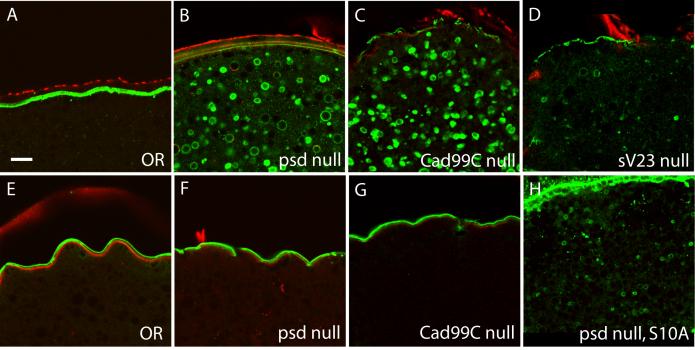
To look in more detail at the consequences of loss of Palisade on the assembly of the vitelline membrane, we examined the localization of two major vitelline membrane proteins, sV17 and sV23 (Fig. 8). In wild-type egg chambers, sV17 is localized exclusively in the vitelline membrane after its secretion by follicle cells (Fig. 8A). In contrast, the psd null egg chambers showed extensive redistribution of sV17 epitopes to vesicles within the oocyte. This was first detectable in some but not all Stage 10 egg chambers (Fig. 8H), and by later stages of oogenesis (Stage 12-14), all egg chambers showed very reduced sV17 staining in the vitelline membrane and striking vesicular staining within the oocyte (Fig. 8B).
Fig. 8. Loss of Palisade or Cad99C results in relocalization of sV17, but not sV23, into the oocyte.
(A-D) Staining of sectioned Stage 14 egg chambers with anti-sV17 antibody (green). (A) OR (B) psd null (C) Cad99C null (D) sV23 null. (E-G) Staining of sectioned Stage 14 egg chambers with anti-sV23 antibody (green). (E) OR (F) psd null (G) Cad99C null. (H) Staining of a sectioned psd null Stage 10A egg chamber with anti-sV17 antibody. Counterstain in all panels is Texas Red-phalloidin (red). Bar equals 10 μm.
This redistribution did not affect all vitelline membrane proteins, however, as the sV23 antibody consistently stained only the vitelline bodies and vitelline membrane throughout the latter portion of oogenesis in both psd null and wild-type egg chambers (cf. Fig. 8E, F); this staining represents both sV23 and a cross-reacting material that appears to be another vitelline membrane protein still present in sV23 null egg chambers (Manogaran and Waring, 2004; Pascucci et al., 1996). Thus, there is no detectable mislocalization of sV23 in the psd null. Similar results were observed for the psd knockdown egg chambers (Supplemental Fig. 3).
For comparison to other vitelline membrane mutants, we also examined the localization of sV17 and sV23 in Cad99C null egg chambers, and the localization of sV17 in sV23 null egg chambers. Similarly to the psd null, the Cad99C null egg chambers showed striking vesicular localization of sV17 within the oocyte in all Stage 12-14 egg chambers (Fig. 8C), although they tended to retain stronger vitelline membrane staining than seen in the psd null and did not show staining within the oocyte in Stage 10 (data not shown). Also similar to the psd null, Cad99C null egg chambers did not show an abnormal distribution of the sV23 protein (Fig. 8G). In contrast, the sV23 null showed much weaker oocyte vesicle staining with the sV17 antibody in Stage 12-14 egg chambers (Fig. 8D and Suppl. Fig. 3G) than seen in the Cad99C and psd nulls, and no oocyte staining in Stage 10 egg chambers. Occasional degenerating egg chambers from the sV23 null did show bright staining of large and irregularly shaped structures within the oocyte that may represent uptake of pieces of the vitelline membrane (Supplemental Fig. 3H), however, extensive sV17 uptake did not appear to be a characteristic feature of the sV23 null as it is for the psd and Cad99C nulls.
Loss of Palisade results in incomplete processing and cross-linking of sV17 and sV23
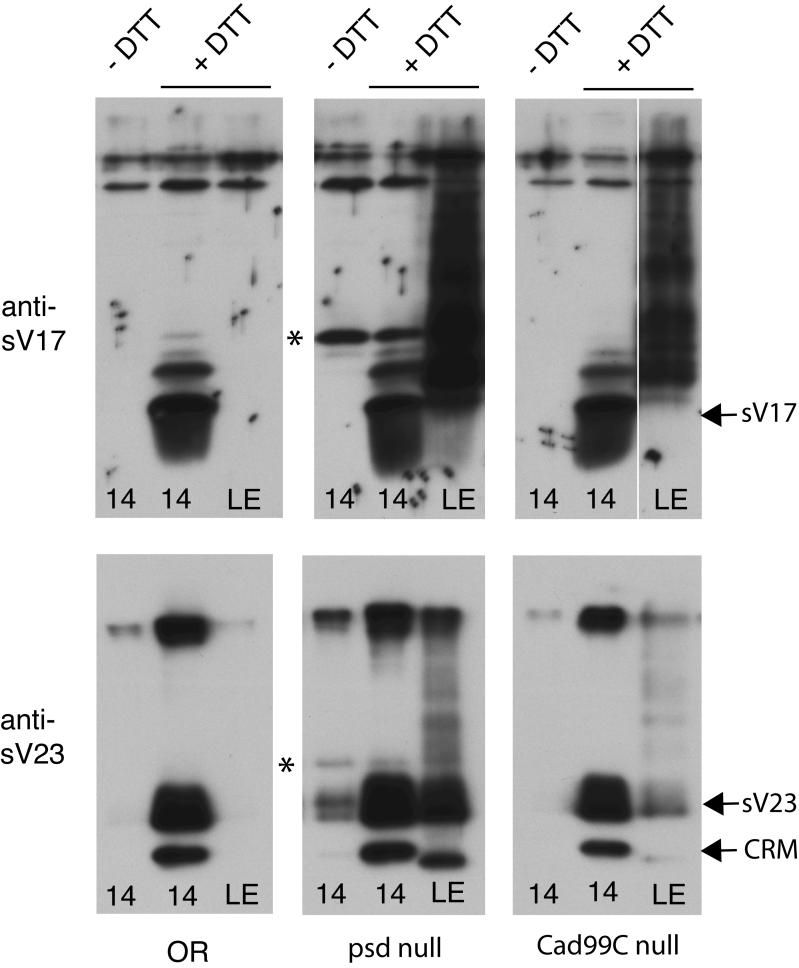
To address at a molecular level how Palisade may function in the assembly of the vitelline membrane, we compared the cross-linking of sV17 and sV23 in Stage 14 egg chambers and laid eggs from wild type, psd null, and Cad99C null females. The total releasable sV17 or sV23 present in late oogenesis is determined by solubilization in the presence of 100 mM DTT (center lanes of each panel in Fig. 9), as the vitelline membrane proteins are normally cross-linked by disulfide bonds and completely insoluble in the absence of reducing agents from Stage 12 onward. Comparing these samples among the three genotypes, we observed no decrease in total amount of sV17 or sV23 in the absence of Palisade or Cad99C, indicating that stability of these proteins is not compromised. While the majority of the sV17 and sV23 proteins were properly processed, the psd null samples alone demonstrated persistence of incompletely processed forms (marked by asterisks) that were either very minor species (sV17) or absent (sV23) in the wild type by Stage 14.
Figure 9. Loss of Palisade results in defects in processing and cross-linking of sV17 and sV23, while loss of Cad99C has more limited effects on cross-linking.
Equal numbers of Stage 14 egg chambers (14) or laid eggs (LE) were loaded in each lane, following extraction by boiling in Laemmli sample buffer with (+) or without (-) 100 mM DTT as indicated above the top row of panels. Blotting was with either anti-sV17 (top panels) or anti-sV23 (bottom panels), and positions of the major Stage 14 processed forms of sV17, sV23, and a cross-reacting material (CRM) are indicated at right. In wild-type (OR), sV17 and sV23 are only soluble in the Stage 14 egg chambers extracted with DTT, while psd null and Cad99C null show solubility of a ladder of partially cross-linked forms of these proteins in laid eggs. The psd null also shows persistence of incompletely processed forms in Stage 14 egg chambers (marked with asterisks), which together with a small amount of correctly processed sV23 fail to be disulfide cross-linked during oogenesis.
In the absence of DTT (lefthand lanes of each panel in Fig. 9), sV17 and sV23 should be completely insoluble in Stage 14 egg chambers. This is the case for the OR and Cad99C null samples, while in the psd null, we observed apparently complete solubility of the incompletely processed forms of each protein as well as a small proportion of the correctly processed sV23.
Following ovulation, the vitelline membrane proteins undergo further non-disulfide cross-linking and normally become completely insoluble in the DTT-containing Laemmli sample buffer that effectively solubilized these proteins in Stage 14 (see righthand lanes of each panel in Fig. 9). In contrast to wild type, the psd null showed persistent and nearly complete solubility of both sV17 and sV23 in laid eggs, though the presence of a ladder of larger species in these lanes indicates that some non-disulfide cross-linking has occurred in the absence of Palisade. The Cad99C null also showed persistent solubility of both sV17 and sV23 in laid eggs, but the defect in sV23 cross-linking was mild compared to that in sV17 cross-linking.
DISCUSSION
Using a combination of RNAi knockdown and homologous recombination knockout approaches, we have shown that Palisade is an integral component of the vitelline membrane that is essential for multiple aspects of vitelline membrane biogenesis and function. The psd knockdown and psd null mutants give very similar results in most assays, but each also provides information that could not be obtained by using only the other approach. The psd null exhibits normal chorion structure, indicating that psd is not essential for chorion formation even if some Palisade is normally deposited in this eggshell layer. Comparison of the knockdown and null suggests that non-specific RNAi effects may contribute to the appearance of oocyte F-actin cytoskeletal defects (Fig. 7) and possibly to a greater defect in sV23 disulfide cross-linking than seen in the psd null (cf. Fig. 9 and Supplemental Fig. 4). The psd knockdown demonstrates a hypomorphic phenotype affecting embryonic nuclei that was confirmed to be specific by careful examination of the psd null, and also provides confirmation of unpublished work from the Stein lab that the PG45 Gal4 driver gives the best available knockdown in follicle cells.
Palisade is required early in the assembly of the vitelline membrane, as judged by three criteria: the morphologic abnormalities in the vitelline bodies and follicle cell microvilli in Stage 10 of oogenesis; the early and continuing uptake of sV17-related epitopes into the oocyte beginning in Stage 10; and the partial defect in proteolytic processing of sV17 and sV23. These abnormalities could result from a primary defect either in interactions among the vitelline membrane proteins or in matrix assembly on a potential microvillar scaffold, either of which in turn could cause the later-observed defects in disulfide- and non-disulfide cross-linking of sV17 and sV23 (as described for the N-terminal domain of sV23 by Manogaran and Waring, 2004). It is not yet understood how the major VM proteins are assembled into the homogeneous vitelline membrane, or how the microvilli on follicle cells and the oocyte are involved in this assembly, so it is difficult at this time to propose a model or straightforward experiments to determine the mechanism by which Palisade functions. Two main questions relevant to mechanism are raised by the novel observations we have made, however, which may influence future work. Why is sV17 taken up into the oocyte, while sV23 appears not to be? What do the similarities and differences between the psd null and Cad99C null effects on vitelline membrane assembly suggest about a relationship between Palisade and Cad99C?
Immunostaining experiments suggest that there is striking uptake of sV17 into the oocyte in psd null and Cad99C null egg chambers, but no uptake of sV23. Less interesting possibilities to explain this difference between these proteins are that sV17 is more sensitive than sV23 to any perturbations in vitelline membrane assembly, or that the polyclonal sV17 antibody is particularly reactive with a fragment or conformation of sV17 that is highly represented in the endocytosed fraction of this protein. These possibilities are diminished by, in the first case, the finding that sV23 null egg chambers do not show this characteristic uptake of sV17, and, in the second case, the observation that, at least in the Cad99C null egg chambers, the overall complement of sV17 processed forms appears identical to that of wild-type egg chambers. Alternatively, if taken at face value, our data suggest that sV17 requires Palisade and Cad99C, but not sV23, for stable incorporation into the vitelline membrane, while sV23 does not require Palisade or Cad99C for its stable incorporation into this matrix. In the case of the Cad99C null, sV17 has undergone apparently complete disulfide cross-linking (Fig. 9), suggesting that the sV17 represented within oocyte vesicles has formed a large disulfide-bonded network independently of sV23. Consistent with a model in which sV17 and sV23 initially undergo separate assembly processes, Gail Waring’s lab has found that in wild-type Stage 10 egg chambers sV17 and sV23 are found in distinct small disulfide-bonded complexes that could represent homo-oligomers or complexes with other matrix components (G. Waring, personal communication). In the absence of Palisade or Cad99C, the sV17 network may never form proper linkages with other vitelline membrane proteins to stabilize it in the matrix.
The psd and Cad99C nulls demonstrate an overlapping spectrum of phenotypes. We initially were struck by the similarity of disorganization of the vitelline bodies and follicle cell microvilli in Stage 10 of oogenesis in these mutants, although they later diverge in that only the Cad99C null vitelline membranes retain obvious full-thickness breaks that are not repaired later in oogenesis (D’Alterio et al., 2005; Schlichting et al., 2006). Both mutants show greater effects on sV17 than on sV23, including the similar movement into the oocyte and a greater defect in non-disulfide cross-linking of sV17. The psd null has additional effects on the major vitelline membrane proteins, including a modest defect in proteolytic processing of both sV17 and sV23 and failure of these incompletely processed forms to be incorporated into a disulfide-bonded network, and a greater defect than Cad99C in the non-disulfide cross-linking of sV23. These results suggest that Palisade and Cad99C could both act in a similar process, such as coordinating the assembly of the vitelline bodies on a microvillar scaffold, with the structural abnormalities of that scaffold being more profound in the absence of Cad99C and with Palisade being additionally required for interactions among the major vitelline membrane proteins. At this time, we do not have clear evidence that Palisade and Cad99C directly interact or function in a common process. Flies hemizygous for both psd and Cad99C null alleles do not exhibit any decrease in hatch rate (not shown), suggesting there is no significant defect in the vitelline membrane when both Palisade and Cad99C are present at half their normal levels. While there is no difference in the amount of Cad99C in psd null Stage 10 egg chambers compared to wild type (not shown), we have seen modest but variable decreases in the amount of Palisade in Cad99C null Stage 10 or 14 egg chambers compared to wild type (Fig. 2D). This result suggests that Palisade may be less stable in the absence of Cad99C but does not demonstrate a direct interaction. Nonetheless, our results suggest that Palisade may be an important molecule to examine in tests of models proposed by researchers of Cad99C (D’Alterio et al., 2005; Schlichting et al., 2006) that the vitelline bodies and follicle cell microvilli make important functional contacts within the perivitelline space that stabilize the microvillar structure and/or modulate the biogenesis of the vitelline membrane.
Embryos surrounded by a psd knockdown vitelline membrane generally arrest during the first 13 nuclear divisions prior to cellularization and show an unusual nuclear morphology of chromatin margination that somewhat resembles the phenotype of anoxic arrest (Foe and Alberts, 1985). Although detailed molecular analyses would be required to compare the physiology and cell biology of psd knockdown and anoxic embryos, one possibility is that the abnormally assembled vitelline membrane surrounding these embryos blocks the normal passage of oxygen into the perivitelline space. Studies in Manduca sexta have shown that subchorion layers of the insect eggshell, including inner chorionic layer, wax layer and vitelline membrane, provide significant resistance to the free delivery of oxygen to the embryo (Woods et al., 2005), so alterations in these layers might increase, as well as decrease, this resistance. Either insufficient oxygen delivery or excessive water loss across the psd knockdown vitelline membrane would be incompatible with the metabolic needs of the embryo, and eventually could result in developmental arrest and death via non-apoptotic mechanisms. Such an embryonic role for the vitelline membrane has not been explored previously because most described mutants affecting this structure, including the psd null, result in absent or very limited embryonic development (Claycomb et al., 2004; D’Alterio et al., 2005; Degelmann et al., 1990; Hong and Hashimoto, 1996; Konrad et al., 1993; Savant and Waring, 1989; Schlichting et al., 2006).
Supplementary Material
Supplemental Fig. 1. Comparison of Palisade localization in wild-type and psd knockdown egg chambers. Sectioned egg chambers were stained with Palisade antibody using an FITC-conjugated secondary antibody, and counterstained with Texas Red-phalloidin. Staining was not detected until late oogenesis (B, and egg chamber 2 in A) in wild-type ovaries (OR), but then was associated with both the vitelline membrane and chorion, seen where these structures separate (arrow in B). Staining could not be seen in Stages 9 and 10 (egg chamber 1 in A), although this is when the psd mRNA is expressed (Fakhouri et al., 2006). Staining was greatly diminished in egg chambers expressing the UAS-psd-RNAi construct (C, D; act5C Gal4 driver), with vitelline membrane staining reduced more than chorion staining (arrow in D). Bar equals 20 μm.
Supplemental Fig. 2. Presence of the peripheral chromatin phenotype in embryos derived from psd null females. Embryos were formaldehyde-fixed and then chopped into large pieces (typically ½ embryo) because the vitelline membrane could not be easily removed with methanol or by hand, to allow entry of Rnase A and propidium iodide; embryos treated in this way tended to show strong cytoplasmic staining with propidium iodide near the cut edges (toward left in these images). Shown are three confocal images from a single psd null embryo that showed the peripheral chromatin phenotype in all of its nuclei.
Supplemental Fig. 3. The psd knockdown egg chambers show specific relocalization of sV17 into the oocyte. (A, B) Staining of sectioned Stage 14 egg chambers with anti-sV23 antibody (green). (A) psd knockdown. (B) OR. (C-F) Staining of sectioned egg chambers with anti-sV17 antibody (green). (C) psd knockdown, Stage 10. (D) OR, Stage 10. (E) psd knockdown, Stage 14. (F) OR, Stage 14. Counterstain in all panels is Texas Red-phalloidin (red). Bar equals 20 μm.
Supplemental Fig. 4. The psd knockdown shows effects on processing and cross-linking of major vitelline membrane proteins similar but not identical to those seen for the psd null. Disulfide cross-linking was assayed by comparison of protein releasable from Stage 14 egg chambers (14) by boiling in Laemmli sample buffer containing 100 mM DTT (+ DTT, center lane in each panel), which is presumed to represent the total amount of that eggshell protein present, to protein releasable by boiling in the absence of DTT (- DTT, left lane in each panel). OR egg chambers show nearly complete disulfide cross-linking of sV23 and a cross-reacting material, CRM (A, left), and of sV17 (B, left), while a substantial fraction of sV23 was not disulfide cross-linked in psd knockdown egg chambers (A, right) but sV17 disulfide cross-linking was unaffected (B, right). Non-disulfide covalent cross-linking occurring at ovulation in wild-type eggs (laid eggs, LE) renders both sV23 and sV17 completely insoluble to conditions of boiling in Laemmli sample buffer + DTT (right lane in each panel). In contrast, both of these proteins remain completely DTT-soluble in the psd knockdown laid eggs. Compared to the psd null (Fig. 9), the incompletely processed forms of sV17 and sV23 are present (marked with asterisks) but not as prominently seen in the samples extracted without DTT, and there appears to be a greater defect in sV23 and CRM disulfide cross-linking.
Acknowledgments
We are indebted to David Stein and Xianjun Zhu for generously providing advice and unpublished data on follicle cell RNAi. We thank Dorothea Godt, David Gubb, Timothy Karr, Norbert Perrimon, David Stein, Gail Waring, the Bloomington Stock Center, the Drosophila Genomics Resource Center, and the ExPASy Proteomics Server for generously providing reagents, fly stocks, and resources, and Darren Baker, Katsuya Miyake, Libby Perry and Bob Smith of the MCG Cell Imaging and Electron Microscopy Core Facilities for advice or assistance. This work was supported by a Basil O’Connor Starter Scholar Research Award (5-FY02-258) from the March of Dimes Birth Defects Foundation, and by a National Institutes of Health RO1 (GM067738).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrenacci D, Cernilogar FM, Taddei C, Rotoli D, Cavaliere V, Graziani F, Gargiulo G. Specific domains drive VM32E protein distribution and integration in Drosophila eggshell layers. J. Cell Sci. 2001;114:2819–2829. doi: 10.1242/jcs.114.15.2819. [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Fabbri F, Andrenacci D, Taddei C, Gargiulo G. Drosophila vitelline membrane cross-linking requires the fs(1)Nasrat, fs(1)polehole and chorion genes activities. Dev. Genes Evol. 2001;211:573–580. doi: 10.1007/s00427-001-0192-1. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev. Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- D’Alterio C, Tran DDD, Au Yeung MWY, Hwang MSH, Li MA, Arana CJ, Mulligan VK, Kubesh M, Sharma P, Chase M, et al. Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J. Cell Biol. 2005;171:549–558. doi: 10.1083/jcb.200507072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degelmann A, Hardy PA, Mahowald AP. Genetic analysis of two female-sterile loci affecting eggshell integrity and embryonic pattern formation in Drosophila melanogaster. Genetics. 1990;126:427–434. doi: 10.1093/genetics/126.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri M, Elalayli M, Sherling D, Hall JD, Miller E, Sun X, Wells L, LeMosy EK. Minor proteins and enzymes of the Drosophila eggshell matrix. Dev. Biol. 2006;293:127–141. doi: 10.1016/j.ydbio.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J. Cell Biol. 1985;100:1623–1636. doi: 10.1083/jcb.100.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Hong CC, Hashimoto C. The maternal nudel protein of Drosophila has two distinct roles important for embryogenesis. Genetics. 1996;143:1653–1661. doi: 10.1093/genetics/143.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- Karr TL. Intracellular sperm/egg interactions in Drosophila: a three-dimensional structural analysis of a paternal product in the developing egg. Mech. Dev. 1991;34:101–111. doi: 10.1016/0925-4773(91)90047-a. [DOI] [PubMed] [Google Scholar]
- Konrad KD, Wang D, Marsh JL. Vitelline membrane biogenesis in Drosophila requires the activity of the a-methyl dopa hypersensitive gene (l(2)amd) in both the germline and follicle cells. Insect Mol. Biol. 1993;1:179–187. doi: 10.1111/j.1365-2583.1993.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Hashimoto C. The Nudel protease of Drosophila is required for eggshell biogenesis in addition to embryonic patterning. Dev. Biol. 2000;217:352–361. doi: 10.1006/dbio.1999.9562. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Kemler D, Hashimoto C. Role of Nudel protease activation in triggering dorsoventral polarization of the Drosophila embryo. Development. 1998;125:4045–4053. doi: 10.1242/dev.125.20.4045. [DOI] [PubMed] [Google Scholar]
- Mahowald AP, Kambysellis MP. Oogenesis. In: Ashburner M, Wright TRF, editors. The Genetics and Biology of Drosophila. Academic Press; New York: 1980. pp. 141–224. [Google Scholar]
- Manogaran A, Waring GL. The N-terminal prodomain of sV23 is essential for the assembly of a functional vitelline membrane network in Drosophila. Dev. Biol. 2004;270:261–271. doi: 10.1016/j.ydbio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Margaritis LH, Kafatos FC, Petri WH. The eggshell of Drosophila melanogaster. I. Fine structure of the layers and regions of the wild-type eggshell. J. Cell Sci. 1980;43:1–35. doi: 10.1242/jcs.43.1.1. [DOI] [PubMed] [Google Scholar]
- Papassideri IS, Margaritis LH, Gulik-Krzywicki T. The egg-shell of Drosophila melanogaster. VI, Structural analysis of the wax layer in laid eggs. Tissue Cell. 1991;23:567–575. doi: 10.1016/0040-8166(91)90014-k. [DOI] [PubMed] [Google Scholar]
- Pascucci T, Perrino J, Mahowald AP, Waring GL. Eggshell assembly in Drosophila: processing and localization of vitelline membrane and chorion proteins. Dev. Biol. 1996;177:590–598. doi: 10.1006/dbio.1996.0188. [DOI] [PubMed] [Google Scholar]
- Petri WH, Mindrinos MN, Lombard MF. Independence of vitelline membrane and chorion cross-linking in the Drosophila melanogaster eggshell. Dev. Biol. 1979;83:23a. [Google Scholar]
- Petri WH, Wyman AR, Kafatos FC. Specific protein synthesis in cellular differentiation. III. The eggshell proteins of Drosophila melanogaster and their program of synthesis. Dev. Biol. 1976;49:185–199. doi: 10.1016/0012-1606(76)90266-9. [DOI] [PubMed] [Google Scholar]
- Popodi E, Minoo P, Burke T, Waring GL. Organization and expression of a second chromosome follicle cell gene cluster in Drosophila. Dev. Biol. 1988;127:248–256. doi: 10.1016/0012-1606(88)90312-0. [DOI] [PubMed] [Google Scholar]
- Reichhart JM, Ligoxygakis P, Naitza S, Woerfel G, Imler JL, Gubb D. Splice-activated UAS hairpin vector gives complete RNAi knockdown of single or double target transcripts in Drosophila melanogaster. Genesis. 2002;34:160–164. doi: 10.1002/gene.10122. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Savant SS, Waring GL. Molecular analysis and rescue of a vitelline membrane mutant in Drosophila. Dev. Biol. 1989;135:43–52. doi: 10.1016/0012-1606(89)90156-5. [DOI] [PubMed] [Google Scholar]
- Schlichting K, Wilsch-Brauninger M, Demontis F, Dahmann C. Cadherin Cad99C is required for normal microvilli morphology in Drosophila follicle cells. J. Cell Sci. 2006;119:1184–1195. doi: 10.1242/jcs.02831. [DOI] [PubMed] [Google Scholar]
- Sheridan JW, Bishop CJ, Simmons RJ. Biophysical and morphological correlates of kinetic change and death in a starved human melanoma cell line. J. Cell Sci. 1981;49:119–137. doi: 10.1242/jcs.49.1.119. [DOI] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation. In: Roberts DB, editor. Drosophila: a Practical Approach. IRL Press; Oxford: 1986. pp. 175–197. [Google Scholar]
- Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. The Drosophila embryonic patterning determinant torsolike is a component of the eggshell. Curr. Biol. 2003;13:1058–1063. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- Trougakos IP, Papassideri IS, Waring GL, Margaritis LH. Differential sorting of constitutively co-secreted proteins in the ovarian follicle cells of Drosophila. Eur. J. Cell Biol. 2001;80:271–284. doi: 10.1078/0171-9335-00163. [DOI] [PubMed] [Google Scholar]
- Turcotte CL, Hashimoto C. Evidence for a glycosaminoglycan on the Nudel protein important for dorsoventral patterning of the Drosophila embryo. Dev. Dyn. 2002;224:51–57. doi: 10.1002/dvdy.10081. [DOI] [PubMed] [Google Scholar]
- Tzarfaty-Majar V, Lopez-Alemany R, Feinstein Y, Gombau L, Goldshmidt O, Soriano E, Munoz-Canoves P, Klar A. Plasmin-mediated release of the guidance molecule F-spondin from the extracellular matrix. J. Biol. Chem. 2001;276:28233–28241. doi: 10.1074/jbc.M102585200. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. Intnl. Rev. Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Woods HA, Bonnecaze RT, Zrubek B. Oxygen and water flux across eggshells of Manduca sexta. J. Exp. Biol. 2005;206:1297–1308. doi: 10.1242/jeb.01525. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xie L, Huang J, Liu X, Zhang R. A novel matrix protein family participating in the prismatic layer framework formation of pearl oyster, Pinctada fucata. Biochem. Biophys. Res. Comm. 2006;344:735–740. doi: 10.1016/j.bbrc.2006.03.179. [DOI] [PubMed] [Google Scholar]
- Zhang G, Young BB, Ezura Y, Favata M, Soslowsky LJ, Chakravarti S, Birk DE. Development of tendon structure and function: regulation of collagen fibrillogenesis. J. Musculoskelet. Neuronal Interact. 2005;5:5–21. [PubMed] [Google Scholar]
- Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev. Cell. 2003;4:599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Zhu X, Stein D. RNAi-mediated inhibition of gene function in the follicle cell layer of the Drosophila ovary. Genesis. 2004;40:101–108. doi: 10.1002/gene.20070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Comparison of Palisade localization in wild-type and psd knockdown egg chambers. Sectioned egg chambers were stained with Palisade antibody using an FITC-conjugated secondary antibody, and counterstained with Texas Red-phalloidin. Staining was not detected until late oogenesis (B, and egg chamber 2 in A) in wild-type ovaries (OR), but then was associated with both the vitelline membrane and chorion, seen where these structures separate (arrow in B). Staining could not be seen in Stages 9 and 10 (egg chamber 1 in A), although this is when the psd mRNA is expressed (Fakhouri et al., 2006). Staining was greatly diminished in egg chambers expressing the UAS-psd-RNAi construct (C, D; act5C Gal4 driver), with vitelline membrane staining reduced more than chorion staining (arrow in D). Bar equals 20 μm.
Supplemental Fig. 2. Presence of the peripheral chromatin phenotype in embryos derived from psd null females. Embryos were formaldehyde-fixed and then chopped into large pieces (typically ½ embryo) because the vitelline membrane could not be easily removed with methanol or by hand, to allow entry of Rnase A and propidium iodide; embryos treated in this way tended to show strong cytoplasmic staining with propidium iodide near the cut edges (toward left in these images). Shown are three confocal images from a single psd null embryo that showed the peripheral chromatin phenotype in all of its nuclei.
Supplemental Fig. 3. The psd knockdown egg chambers show specific relocalization of sV17 into the oocyte. (A, B) Staining of sectioned Stage 14 egg chambers with anti-sV23 antibody (green). (A) psd knockdown. (B) OR. (C-F) Staining of sectioned egg chambers with anti-sV17 antibody (green). (C) psd knockdown, Stage 10. (D) OR, Stage 10. (E) psd knockdown, Stage 14. (F) OR, Stage 14. Counterstain in all panels is Texas Red-phalloidin (red). Bar equals 20 μm.
Supplemental Fig. 4. The psd knockdown shows effects on processing and cross-linking of major vitelline membrane proteins similar but not identical to those seen for the psd null. Disulfide cross-linking was assayed by comparison of protein releasable from Stage 14 egg chambers (14) by boiling in Laemmli sample buffer containing 100 mM DTT (+ DTT, center lane in each panel), which is presumed to represent the total amount of that eggshell protein present, to protein releasable by boiling in the absence of DTT (- DTT, left lane in each panel). OR egg chambers show nearly complete disulfide cross-linking of sV23 and a cross-reacting material, CRM (A, left), and of sV17 (B, left), while a substantial fraction of sV23 was not disulfide cross-linked in psd knockdown egg chambers (A, right) but sV17 disulfide cross-linking was unaffected (B, right). Non-disulfide covalent cross-linking occurring at ovulation in wild-type eggs (laid eggs, LE) renders both sV23 and sV17 completely insoluble to conditions of boiling in Laemmli sample buffer + DTT (right lane in each panel). In contrast, both of these proteins remain completely DTT-soluble in the psd knockdown laid eggs. Compared to the psd null (Fig. 9), the incompletely processed forms of sV17 and sV23 are present (marked with asterisks) but not as prominently seen in the samples extracted without DTT, and there appears to be a greater defect in sV23 and CRM disulfide cross-linking.