Abstract
A third DNA base pair, which is synthesized efficiently and selectively, would have wide ranging applications from synthetic organisms to nucleic acids biotechnology. Hydrophobic unnatural nucleobases offer a promising route to such a pair, but are often limited by inefficient extension, defined as synthesis immediately following the unnatural pair. Here, we describe a simple screen which enables the characterization of large numbers of previously uncharacterized hetero base pairs. From this screen, we identified a class of unnatural base pairs which are extended more efficiently than any unnatural base pair reported to date. Screening, when complemented by further kinetic analysis, can improve the understanding of the determinants of efficient extension as well as identify viable hetero base pairs.
The four natural nucleotides of DNA form two base pairs that are capable of encoding complex genetic information as well as functional nucleic acids. Expansion of the genetic alphabet to include a third base pair, formed between two identical or different unnatural nucleotides, referred to as self pairs and heteropairs, respectively, would expand the informational and functional potential of DNA. While the pairing of the natural nucleobases is mediated by hydrogen-bonding (H-bonding) complementarity, it has become clear that, like proteins, hydrophobic forces are also capable of mediating these interactions.1 Indeed, a variety of nucleotides bearing predominantly hydrophobic nucleobase analogs form pairs that are both stable in duplex DNA and synthesized by the exonuclease deficient Klenow fragment of DNA Pol I (Kf), by incorporation of an unnatural dNTP opposite the unnatural base in the template, with reasonable efficiency and selectivity.1b,2 However, extension of the newly synthesized unnatural base pair, by incorporation of the next correct dNTP, has consistently limited the enzymatic synthesis of unnatural DNA. Only recently have efforts begun to focus on the determinants of efficient extension,1b,3 however, they remain largely unknown. This has limited the rational design of viable unnatural base pairs.
We have synthesized over 80 unnatural nucleotides bearing predominantly hydrophobic nucleobase analogs and systematically evaluated the synthesis and extension of the corresponding self pairs. However, because these nucleotides may be combined to form more than 6400 heteropairs, their synthetic evaluation requires a more efficient screening strategy. In an attempt to identify more efficiently extended heteropairs, as well as to begin to understand the determinants of extension, we developed and performed a simple screen of the unnatural nucleotides to identify those that formed heteropairs which Kf efficiently extends.
Each nucleobase was incorporated at the 3′-end of a 24-mer primer oligonucleotide, as well as at position 24 of a 45-mer template oligonucleotide. Hybridization of these two oligonucleotides forms a polymerase substrate with a primer that terminates at the X:Y heteropair, where X and Y are the unnatural nucleotides in the primer and template, respectively. In all cases, the next nucleobase in the template is dG; incorporation of dCTP results in correct extension of the unnatural heteropair. Sixty primers were organized into ten interrelated groups based on their shape and functionality. In each experiment, a single group, consisting of a pool of 4–10 radiolabeled primers, was annealed to a common template strand, producing heteropairs differentiated by the nucleotide X at the primer terminus. Kf (0.3 nM) and dCTP (400 μM) were added in reaction buffer and allowed to incubate with the unnatural DNA for 5 minutes before quenching with EDTA. The amount of the primer pool extended by the addition of dCTP was quantified by PAGE and phosphorimaging. While the detailed analysis of the results will be published elsewhere, it was immediately apparent that one group of primers, the bicyclic 5/6 series,4 including BFr, BTp, BTz, and IN, was extended efficiently when paired with 4-methyl pyridone (4MP) in the template (Figure 1). We were thus interested in the more detailed characterization of this class of unnatural heteropair.
Figure 1.
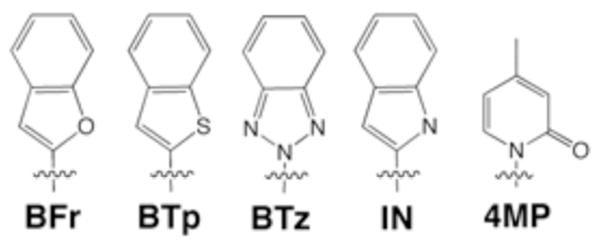
Unnatural Nucleobases
To quantitate the efficiency with which Kf extends the BFr:4MP, BTp:4MP, BTz:4MP, and IN:4MP heteropairs, we employed steady-state kinetics (Table 1). In this context, with the bicyclic nucleobase analog in the primer and 4MP in the template, the second order rate constants range between 3 × 105 and 1 × 106 M−1 min−1. The most efficiently extended pairs, BFr:4MP, BTz:4MP, and IN:4MP, are extended at rates that are two- to four-fold higher than the most efficiently extended unnatural base pair reported to date, and the BTp:4MP heteropair was extended only slightly less efficiently. The efficient extension rates are predominantly due to low KM values, which range from 4 to 12 μM and, remarkably, are all within a factor of ten of that typical for the extension of a natural base pair (3.5 μM).2 The KM values for the corresponding self pairs are all significantly higher, ranging between 85 and 294 μM, indicating that the tight binding of dCTP is specific to the heteropairs (Supporting Information). The data suggest that in this sequence context, the heteropair forms a specific primer terminus that tightly binds the incoming dNTP.
Table 1.
Steady State Rate Constants of Unnatural Terminus Extension by Insertion of dCTPa
| 5′ - dTAATACGACTCACTATAGGGAGA (X) | ||||
|---|---|---|---|---|
| 3′ - dATTATGCTGAGTGATATCCCTCT (Y) GCTAGGTTACGGCAGGATCGC
| ||||
| X | Y | kcat (min−1) | KM (μM) | kcat/KM(M−1min−1) |
| BFr | 4MP | 5.4 ± 1.2 | 6.1 ± 2.1 | 8.8 × 105 |
| BTp | 4MP | 3.3 ± 1.0 | 11.5 ± 1.3 | 2.9 × 105 |
| BTz | 4MP | 5.4 ± 1.7 | 7.6 ± 2.2 | 7.1 × 105 |
| IN | 4MP | 5.3 ± 1.2 | 3.9 ± 0.9 | 1.4 × 106 |
| 4MP | BFr | 30.9 ± 6.5 | 1 | 1.4 × 105 |
| 4MP | BTp | 19.8 ± 5.1 | 205 ± 39 | 9.7 × 104 |
| 4MP | BTz | 14.9 ± 3.3 | 89 ± 23 | 1.7 × 105 |
| 4MP | IN | 15.8 ± 4.1 | 402 ± 108 | 3.9 × 104 |
See supporting information for experimental details.
Viable heterobase pair candidates must be efficiently extended in both sequence contexts. With 4MP in the primer and the bicyclic analog in the template, each heteropair is also extended efficiently, with second order rate constants ranging from 4 × 104 to 2 × 105 M−1 min−1 (Table 1). In stark contrast to the data discussed above, efficient extension of the unnatural heteropairs in this context results primarily from large kcat values, which range from 15 to 31 min−1. Remarkably, these kcat’s are all within 10-fold of that for the extension of a natural base pair (163 min−1).2 With the exception of the BTz self pair, which is extended at a rate of 2 × 105 M−1 min−1, the rates in either context are significantly faster than any of the rates of extension of the corresponding self pairs, which range from 7 × 102 to 1 × 104 M−1 min−1. This again indicates that efficient extension is generally specific for the heteropairs. In addition, the data suggest that in this sequence context, the heteropair forms a primer terminus that interacts favorably with the incoming dCTP in the developing transition state.
The efficient extension of the heteropairs must not come at the expense of fidelity. To characterize the fidelity with which each heteropair is extended, we examined extension of all eight unnatural termini with dATP, dGTP, and dTTP. Importantly, the rates of misincorporation of these nucleobases opposite dGTP are all below detectable rates (kcat/KM < 103 M−1 min−1). Thus, the unnatural heteropairs are extended with high efficiency and fidelity.
Replicative DNA polymerases, such as Kf, interact with up to five base pairs upstream of the primer terminus, and it has been demonstrated that modified nucleobases at these upstream positions may have an effect on replication.5 To determine if the heteropairs compromise full length synthesis, we examined synthesis in the presence of all dNTPs (Figure 2). Kf (3.7 nM), dNTPs (50 μM), and primer/template (40 nM) were incubated for five minutes in the presence or absence of dCTP. After five minutes of incubation, full length synthesis was the predominant product in the presence of dCTP, but only unextended primer was apparent in the absence of dCTP. While the presence of a small amount of unextended unnatural termini indicates that the reaction is less efficient than wild type, the data clearly demonstrate that full length synthesis is both efficient and selective.
Figure 2.
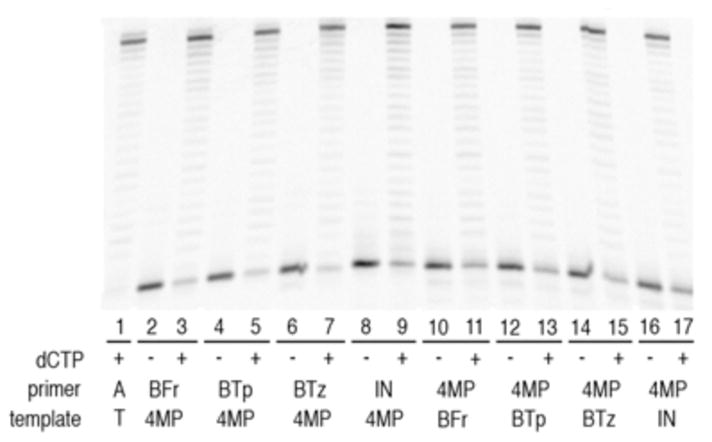
Full-length extension of DNA termini. All reactions contain 50 μM dATP, dGTP, dTTP. Odd lanes also contain 50 μM dCTP. Sequences are identical to Table 1. See supporting information for full experimental details.
The origins of the efficient extension of the bicyclic-pyridone heteropairs likely lie in specific interactions between the unnatural base pair, the polymerase, and the incoming dNTP. For the efficient extension of a natural Watson-Crick base pair, Kf appears to require the precise assembly of an arginine fork - formed between the primer terminus minor groove H-bond acceptor, residue Arg668 of Kf, and the incoming dNTP ribosyl oxygen.6 With the bicyclic analogs in the primer, the larger aromatic surface area may facilitate dNTP binding via favorable stacking interactions. However, this strong interaction may induce the formation of a slightly distorted primer terminus-Kf-dNTP complex, resulting in the slightly compromised kcat observed. With 4MP at the primer terminus, its pyrimidine-like structure may result in a more natural like primer terminus structure, including the correct alignment of the nucleophilic 3′-OH and minor groove H-bond acceptor. However, compared to the bicyclic analogs at the primer terminus, the relatively reduced surface area of 4MP may underlie its weaker dCTP binding. Nonetheless, the base pairs are reasonably well extended in either context, while the self pairs are not, suggesting that the corresponding nucleobase analogs are complementary and truly form a base pair.
Recent systematic efforts to understand the role of hydrophobicity, aromatic surface area, and electronic contributions have begun to shed light on the determinants of polymerase-mediated insertion of unnatural dNTPs.1b,7 However, the rate-limiting extension of the resulting base pairs has remained relatively less studied, and thus, less understood, limiting the rational design of viable unnatural base pairs. From a pool of thousands of novel nucleobase combinations, we identified one class of unnatural heteropairs that are extended by Kf with second order rate constants that are orders of magnitude higher than typically observed for unnatural base pairs. In fact, they represent the most efficiently extended unnatural base pairs identified to date. Thus, it is apparent that interbase interactions mediated by predominantly hydrophobic interactions are sufficient to support both base pair synthesis and extension. The fact that each of the four members of the bicyclic analog family formed efficiently extended heteropairs when paired opposite 4MP suggests that their recognition is general to this class of heteropair. Experiments designed at further understanding the efficient extension of this family of heteropairs should prove invaluable for future base pair design efforts, and may also yield derivatives that are even better recognized by DNA polymerases. Moreover, further progress will be stimulated by more sophisticated screens which might sort through libraries of heretopairs, based not only on their efficient extension, but also on their efficient and high fidelity synthesis. Such screens are currently under development.
Supplementary Material
Screen details and kinetic data.
Acknowledgments
This work was supported by the National Institutes of Health (NIGMS 60005).
References
- 1.(a) Moran S, Ren RX-F, Sumney S, IV, Kool ET. J Am Chem Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Henry AA, Olsen AG, Matsuda S, Yu C, Geierstanger BH, Romesberg FE. J Am Chem Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]; (c) Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. J Am Chem Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa AK, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg FE. J Am Chem Soc. 2000;122:3274–3287. [Google Scholar]
- 3.(a) Adelfinskaya O, Nashine VC, Bergstrom DE, Davisson VJ. J Am Chem Soc. 2005;127:16000–16001. doi: 10.1021/ja054226j. [DOI] [PubMed] [Google Scholar]; (b) Morales JC, Kool ET. J Am Chem Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuda S, Henry AA, Schultz PG, Romesberg FE. J Am Chem Soc. 2003;125:6134–6139. doi: 10.1021/ja034099w. [DOI] [PubMed] [Google Scholar]
- 5.Miller H, Grollman AP. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 6.Meyer AS, Blandino M, Spratt TE. J Biol Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 7.(a) Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels J, Kuchta RD. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang X, Lee I, Zhou X, Berdis AJ. J Am Chem Soc. 2006;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]; (c) Kim TW, Delaney JC, Essigmann JM, Kool ET. Proc Natl Acad Sci USA. 2005;102:15803–15. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screen details and kinetic data.


