Abstract
We report the cloning and characterization of Pichia pastoris PEX19 by complementation of a peroxisome-deficient mutant strain. Import of peroxisomal targeting signal 1- and 2-containing peroxisomal matrix proteins is defective in pex19 mutants. PEX19 encodes a hydrophilic 299-amino acid protein with sequence similarity to Saccharomyces cerevisiae Pex19p and human and Chinese hamster PxF, all farnesylated proteins, as well as hypothetical proteins from Caenorhabditis elegans and Schizosaccharomyces pombe. The farnesylation consensus is conserved in PpPex19p but dispensable for function and appears unmodified under the conditions tested. Pex19p localizes predominantly to the cytosolic fraction. Biochemical and two-hybrid analyses confirmed that Pex19p interacts with Pex3p, as seen in S. cerevisiae, but unexpectedly also with Pex10p. Two-hybrid analysis demonstrated that the amino-terminal 42 amino acids of Pex19p interact with the carboxyl-terminal 335 amino acids of Pex3p. In addition, the extreme carboxyl terminus of Pex19p (67 amino acids) is required for interaction with the amino-terminal 380 amino acids of Pex10p. Biochemical and immunofluorescence microscopy analyses of pex19Δ cells identified the membrane protein Pex3p in peroxisome remnants that were not previously observed in S. cerevisiae. These small vesicular and tubular (early) remnants are morphologically distinct from other Pppex mutant (late) remnants, suggesting that Pex19p functions at an early stage of peroxisome biogenesis.
INTRODUCTION
Peroxisomes are single-membrane–bound organelles found in virtually all eukaryotes (De Duve, 1996). Although their involvement in H2O2 metabolism is a general feature, specific metabolic functions vary depending on the organism and the milieu it encounters. β-oxidation (Lazarow and De Duve, 1976; Tolbert and Essner, 1981), plasmalogen biosynthesis (Hajra and Bishop, 1982), and cholesterol biosynthesis (Thompson and Krisans, 1990) are among the various activities residing in mammalian peroxisomes. The importance of these organelles in humans is stressed by the existence of a number of fatal genetic disorders in which the peroxisome is defective (Subramani, 1997). In cells of these patients, remnants containing peroxisomal membrane proteins (PMPs) are present, whereas some, or even all, peroxisomal matrix proteins accumulate in the cytosol (Lazarow and Moser, 1989).
In recent years, much progress has been made in elucidating the molecular requirements for peroxisome biogenesis, especially through the use of yeasts as model organisms. In Pichia pastoris, peroxisomes are required for metabolizing carbon sources such as methanol and oleate but are dispensable for growth on glucose, glycerol, and ethanol. This feature made it relatively easy to obtain yeast mutants defective in peroxisome biogenesis. These mutants, and those in other yeasts, have contributed to the cloning and characterization of at least 21 PEX genes, encoding peroxins that are essential for peroxisome biogenesis (Distel et al., 1996; Götte et al., 1998; Purdue et al., 1998; Subramani, 1998; Titorenko et al., 1998). Screening the human Expressed Sequence Tags databases has revealed 13 human orthologues to the yeast PEX genes (Subramani, 1997; Fransen et al., 1998). Eight of these genes have been shown to complement the peroxisome-related defects of cells from human disease complementation groups (Subramani, 1997). Clearly, peroxisome biogenesis is a conserved process in eukaryotes, and yeasts are preeminently suited to study this process at the molecular level.
Despite the growing number of PEX genes, few details are known about their individual functions. Eight peroxins, Pex5p, Pex7p, Pex13p, Pex14p, Pex17p, Pex18p, Pex20p, and Pex21p, have been primarily implicated in the process of peroxisomal matrix protein import (Purdue et al., 1998; Subramani, 1998; Titorenko et al., 1998). Peroxisomal matrix proteins are synthesized in the cytosol and posttranslationally imported into the organelle (Lazarow and Fujiki, 1985). Two peroxisomal targeting signals (PTSs) have been identified; PTS1, a carboxyl-terminal tripeptide (SKL and conserved derivatives) (Gould et al., 1987, 1989; Elgersma et al., 1996), is most commonly found in matrix proteins; PTS2, an amino-terminal nonapeptide (Osumi et al., 1991; Swinkels et al., 1991), is found in a smaller subset of proteins. Pex5p and Pex7p are the receptors for PTS1 and PTS2, respectively (Rehling et al., 1996; Subramani, 1998). Three peroxisomal membrane-bound proteins, Pex13p, Pex14p, and Pex17p, interact to form a docking site on the organelle for these predominantly cytosolic signal receptors (Huhse et al., 1998; Subramani, 1998). The mechanism of protein translocation across the peroxisome membrane, however, is essentially unknown.
Most of the peroxins characterized to date are PMPs. Information about the sorting of this class of proteins is only rudimentary. A sequence involved in sorting of PMPs has been identified for a few proteins. The presence of a stretch of basic amino acids (four in a sequence of six) is the most common feature of these sequences (Elgersma et al., 1997). Although many PMPs seem to be synthesized in the cytosol and imported directly to the peroxisome, some were recently proposed to be targeted to the peroxisome via the endoplasmic reticulum (ER) (Titorenko and Rachubinski, 1998). This implies that a sorting machinery exists between the ER and the peroxisome, which was not envisioned in the original “multiplication-via-division model” for peroxisome biogenesis (Lazarow and Fujiki, 1985).
Here we report the cloning of the PEX19 gene and characterization of the gene product in P. pastoris. Previously, PEX19 has been cloned and characterized from Saccharomyces cerevisiae (Götte et al., 1998), Chinese hamster (PxF) (James et al., 1994), and human cells (HK33/PxF) (Braun et al., 1994; Kammerer et al., 1997) and shown to be modified by farnesylation, but no evidence for this modification was seen in P. pastoris. Consistent between S. cerevisiae and P. pastoris is the accumulation of lumenal peroxisomal proteins in the cytosol of pex19 mutants, an effect that has not been observed in mammalian cells because of lack of PEX19-defective cell lines. Biochemical localization of Pex19p/PxF in all organisms has suggested that it is mainly cytosolic, but immunocytochemical evidence suggests that it associates with the peroxisome membrane (James et al., 1994; Kammerer et al., 1997; Götte et al., 1998). Our results confirm a previously reported protein–protein interaction with Pex3p and also identify a novel one with Pex10p. In S. cerevisiae, no peroxisome remnant structures were observed in the pex19 mutants (Götte et al., 1998), but we present evidence that novel, morphologically distinct remnants are present in P. pastoris. The combination of novel cell biological, biochemical, and protein–protein interaction data allows us to augment models of Pex19p function beyond those reported previously.
MATERIALS AND METHODS
Strains and Growth Conditions
Strains used in this study are listed in Table 1 and were grown at 30°C. P. pastoris was grown in rich medium (YPD; 1% yeast extract, 2% Bacto-peptone, and 2% glucose) or synthetic medium (0.67% yeast nitrogen base) supplemented with 2% glucose (YND) or 0.5% (vol/vol) methanol (YNM). If needed, media were supplemented with appropriate amino acids to a final concentration of 50 μg/ml. Bacto-agar (2% wt/vol) was added for solid media. For oleate inductions, P. pastoris strains were grown in batch in mineral medium containing 0.2% (vol/vol) oleate and 0.02% (vol/vol) Tween 40 (MMOT) (Veenhuis et al., 1979).
Table 1.
P. pastoris strain list
| Strain | Relevant genotype | Source |
|---|---|---|
| PPY12 | his4, arg4 | Gould et al., 1992 |
| SMD1163 | his4 pep4 prb | Invitrogen |
| STW3 | PPY12 pTW2 | This study |
| STW4 | his4 arg4 pex19–112 | This study |
| SKF13 | PPY12 pex19Δ1::ZEO | This study |
| SKF14 | SMD1163 pex19Δ1::ZEO | This study |
| STW2 | PPY12 his4::pTW66 | Wiemer et al., 1996 |
| AK11 | SKF13 his4::pTW66 | This study |
| WSP50 | SKF14 his4::pWSP50 | This study |
| WSP51 | SKF14 his4::pWSP51 | This study |
| WSP52 | SKF14 his4::pWSP52 | This study |
| SSH4 | SMD1163 pex10Δ1::ZEO | This study |
| SSH18 | SSH4 his4::pSH38 | This study |
| PPY12Δ2 | PPY12 pex2Δ::ARG4 | Jim Cregg |
| PPY12Δ5 | PPY12 pex5Δ::ARG4 | McCollum et al., 1993 |
For the induction experiments (see Figures 3 and 8A) cells were precultured in MMG media (2% dextrose) and in the midlogarithmic phase shifted to MMM media (0.5% methanol) or MMOT media. PPY4 and SKF13 cultures were inoculated at an A600 of 0.2 and 0.5, respectively. At the indicated times, cells were harvested, and total cellular extracts were prepared as described previously (Monosov et al., 1996). Two to 20 μg of total protein were loaded per lane.
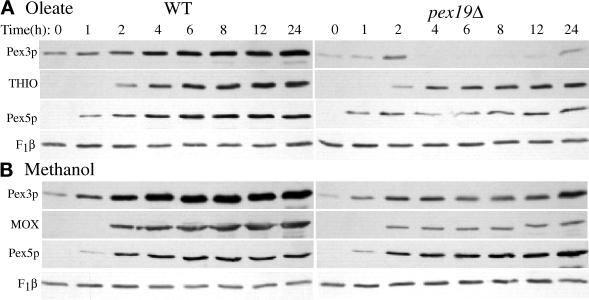
Figure 3.
Steady-state levels of peroxisomal proteins during oleate and methanol growth of wild-type and pex19Δ cells. Glucose-grown wild-type and pex19Δ cells (PPY12 and SKF13) were shifted to oleate (A) or methanol media (B), and samples were collected at the indicated times and processed as described in MATERIALS AND METHODS for SDS-PAGE and immunoblotting with the indicated antibodies.
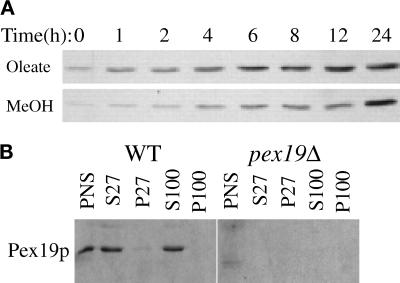
Figure 8.
Induction and subcellular fractionation of Pex19p. (A) Glucose-grown wild-type cells (PPY12) were shifted to oleate and methanol media, and samples were collected at the indicated times and processed as described for Figure 3 for immunoblotting with anti-Pex19p antibodies. (B) Oleate-grown spheroplasts of wild-type and pex19Δ cells (SMD1163 and SKF14) were lysed and subjected to sequential differential centrifugation. Equivalent amounts of the PNS, 27,000 × g supernatant (S27), 27,000 × g pellet (P27), 100,000 × g supernatant (S100), and 100,000 × g pellet (P100) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-Pex19p antibodies.
Molecular Biological Techniques
All plasmids used in this study are listed in Table 2. All DNA-oligonucleotide primers used are listed in Table 3.
Table 2.
Plasmids used in this study
| Name | Relevant elements | Reference |
|---|---|---|
| pTW2 | PAOX1BLE-PTS1 ScARG4 PARS2 | This study |
| pTW66 | PGAPDHPTS2-GFP HIS4 | Wiemer et al., 1996 |
| p112Sal | pSG560 2.2 kB-SalI-PEX19 | This study |
| pTW121 | pSG560 1.4 kB PCR PEX19 | This study |
| pKNSD169 | pex19Δ1::ZEO | This study |
| pTW128 | pQE9 HIS6-PEX19 | This study |
| pWSP50 | pIB1 PEX19 HIS4 | This study |
| pWSP51 | pIB1 pex19C296S HIS4 | This study |
| pWSP52 | pIB1 pex19C296ter HIS4 | This study |
| pSH5 | pex10Δ1::ZEO | This study |
| pSH38 | p21–43 NH-PEX10 | This study |
| Two-hybrid plasmids | ||
| pKNSD123 | pKNSD52 PEX19 | This study |
| pKNSD124 | pKNSD55 PEX19 | This study |
| pKNSD127 | pKNSD52 PEX19[1–232] | This study |
| pKNSD125 | pKNSD52 PEX19[1–75] | This study |
| pKNSD135 | pKNSD52 PEX19[1–42] | This study |
| pKNSD98 | pKNSD55 PEX3 | This study |
| pKNSD132 | pKNSD55 PEX3[1–110] | This study |
| pKNSD130 | pKNSD55 PEX3[110–445] | This study |
| pKNSD121 | pKNSD55 PEX10 | This study |
| pKNSD173 | pKNSD55 PEX10[1–380] | This study |
| pKNSD133 | pKNSD55 PEX10[1–217] | This study |
| pKNSD134 | pKNSD55 PEX10[193–217] | This study |
| pKNSD136 | pKNSD55 PEX10[193–419] | This study |
| pKNSD171 | pKNSD55 PEX10[260–419] | This study |
| pKNSD172 | pKNSD55 PEX10[260–380] | This study |
| pKNSD174 | pKNSD55 PEX10[217–380] | This study |
Table 3.
Primers
| Name | DNA sequence (5′ → 3′) |
|---|---|
| KNF16 | CCCGATTCTCAAAATACAACCAAGAAGTACC |
| KNF21 | CCCGGATCCATGCCCCCATCTGAAGAGATC |
| KNF32 | CCCGGATCCAAGAGGTTTTTCTCCATGAGG |
| KNF33 | CCCGAATTCCGTAGTTCCCATTTGGTG |
| KNF39 | CCCGGATCCTTACTAGGAGGGTTGATTG |
| KNF43 | CGCGGCCGCAAATTGGATAAGAACC |
| KNF44 | CCCGAATCATCTAAGATTGGCAGATATG |
| KNF45 | TCTCGAGTCTAGAGATATCTGAGGTAATGCAGGGAAC |
| KNF46 | CCCGCGGCCGCAATCAAAAGTACCCATCTC |
| KNF47 | CGCGGCCGCGGATAACAATTTCACAC |
| KNF48 | CCCGGAATTCATTTCGGAGACAGTTCGTTG |
| KNF49 | AAGATCTGATATCTGACCAGTAATCATCCTGC |
| KNF50 | CCCGCGGCCGCCAGACTGTTAACGTCAG |
| TW49 | AAGTCATCAGATGCCAG |
| TW59 | GTGTGGAATTGTGAGCGG |
| TW61 | GGATCCATGAGTAGCGAGGACGAACTAG |
| TW63 | GGATCCCTGCTGTTTACAAGTCTCATC |
| PEX10B | CCAGGATCCCAGAATTCAATCAAAAGTACC |
| PEX10C | GCCGGATCCATGCCCCCATCTGAAG |
| PEX19up | GCGGAGCTCAATTTCACACAGGTAAACAGC |
| PEX19dn | GCGCTGATCACTGCTGTTTACAAGTCTC |
| SAAX.19 | GCGCTGATCACTGCTGTTTACTAGTCTCATCC |
| ΔC4.19 | GCGCTGATCAAGTCTCATCCAATTCCTTGC |
Escherichia coli strains JM109, DH5α, and XL1blue were routinely used for propagation and amplification of plasmids and grown by standard methods. Enzyme digestions, cloning, plasmid isolation, PCRs, and Southern blotting were performed according to standard techniques (Sambrook et al., 1989). All reagents were handled according to the manufacturer’s instructions. DNA sequencing was performed by the method of Sanger et al. (1977).
P. pastoris transformations, mating, sporulation, and random spore analysis were performed as described (Gould et al., 1992).
Isolation of Peroxisome Assembly (pex) Mutants
Isolation of P. pastoris pex mutant strains was based on the procedure originally described for S. cerevisiae (Elgersma et al., 1993), and also for the P. pastoris screen for pex mutants defective in the import of PTS2-containing proteins (Elgersma et al., 1998). The Tn5-BLE-PTS1 gene from pEL43 (Elgersma et al., 1993) was excised as a BamHI fragment and inserted into the BamHI site of pAM2F plasmid (a gift from J. Heyman, University of California, San Diego, CA) creating pTW2. Plasmid pAM2F is based on the plasmid pSG464 (Gould et al., 1992) and contains a S. cerevisiae ARG4 gene, an autonomous-replicating sequence for P. pastoris, a 5′ AOX promoter fragment, a 3′ AOX transcription termination fragment, and a multiple-cloning site in between the 5′ and 3′ AOX fragments. This placed BLE-PTS1 under control of the methanol-inducible AOX1 promoter (Ellis et al., 1985). This plasmid was transformed into PPY12, yielding strain STW3. A 100-ml culture of strain STW3 was grown in YPD to an A600 of 1.0 and subjected to N-methyl-N-nitro-N-nitrosoguanidine mutagenic treatment as described (Elgersma et al., 1998). The enrichment in 25 μg/ml phleomycin was as described previously (Elgersma et al., 1998). Cells were then collected, washed, resuspended in 100 ml of YPD, and incubated for 2 h at 30°C. Subsequently, various dilutions were plated onto YPD agar. Cells were grown for 2 d and replica plated onto YNM plates. Methanol nonutilizers, putative pex mutants, were selected for further characterization.
Cloning and Sequencing of PEX19
A genomic library constructed in plasmid pYM8 (Liu et al., 1995) was used to transform mutant strain pex19-112. Ten clones, p112-1 to p112-10, were identified, which complemented strain pex19-112 for growth on methanol. Restriction analysis revealed four distinct, overlapping, genomic inserts. The complementing fragment was narrowed to a 2.2-kb SalI fragment by subcloning into pSG560 (p112Sal). Approximately 1.4 kb of the SalI fragment was sequenced (GenBank AF133271), which revealed a 900-bp ORF. The 1.4-kb fragment was amplified by PCR (Primers TW49 and TW59), cloned into pCRII (Invitrogen, Carlsbad, CA) (pTW119), excised with BamHI and XhoI, and cloned directly into pSG560 (Gould et al., 1992), yielding pTW121. This plasmid complemented pex19 deletion strains for growth on methanol and oleate media, confirming that this region comprised the PEX19 ORF and the required regulatory elements.
Two-Hybrid Analysis
Cloning vectors, tester strains, and screening by two-hybrid analysis, as well as two-hybrid vectors containing full-length PEX3, have been described (Faber et al., 1998). A full-length clone of PEX10 was amplified by PCR (primers KNF16 and KNF21) and inserted as a BamHI–EcoRI DNA fragment in pKNSD55, creating pKNSD121. A full-length clone of PEX19 was generated by cloning the BamHI fragment from pTW119 (see above) into pKNSD52 cut with the same enzyme, creating pKNSD123. Fragments of PEX3 and PEX19 were introduced in pKNSD55 and pKNSD52, making use of matching restriction sites present in the ORF of these genes and the vectors: PEX3[1–110] generated with BamHI creates pKNSD132; PEX3[110–445] generated with BamHI creates pKNSD130; PEX19[1–42] generated with BglII creates pKNSD135; PEX19[1–75] generated with PstI creates pKNSD125; and PEX19[1–232] generated with EcoRI creates pKNSD127. Fragments of PEX10 were cloned into pKNSD55 as follows. PEX10[1–217] was amplified by PCR (primers KNF21 and KNF33), cut with BamHI and EcoRI, and inserted into pKNSD55 cut with the same, creating pKNSD133. PEX10[193–217] was amplified by PCR (primers KNF32 and KNF33), cut with BamHI and EcoRI, and inserted into pKNSD55 cut with the same, creating pKNSD134. PEX10[193–419] was amplified by PCR (primers KNF16 and KNF32), cut with BamHI and EcoRI, and inserted into pKNSD55 cut with the same, creating pKNSD136. PEX10[1–380] was generated by cutting pKNSD121 with SalI and religating to create pKNSD173. PEX10[217–419] was amplified by PCR (primers KNF16 and KNF39), cut with BamHI and EcoRI, and inserted into pKNSD55 cut with the same, creating pKNSD170. PEX10[260–419] was generated by cutting pKNSD170 with EcoRV and EcoRI and the 490-bp fragment cloned into pKNSD53 cut with BamHI (blunted by Klenow) and EcoRI, creating pKNSD171. PEX10[260–380] was generated by cutting pKNSD170 with SalI and EcoRI and the 490-bp fragment cloned into pKNSD53 cut with BamHI, blunted by Klenow polymerase, and SalI, creating pKNSD172. PEX10[217–380] was generated by cutting pKNSD170 with SalI and religating to create pKNSD174.
Construction of pex19Δ Strains
The 5′ and 3′ flanking regions, 300 and 250 bp, respectively, of the PEX19 ORF were amplified by PCR (primers KNF47, KNF48, KNF49, and KNF50) and inserted into pBluescript II KS+ (Stratagene, La Jolla, CA), cut with XbaI (blunted by Klenow polymerase) and NotI for the 3′ region and with Asp718I (blunted by Klenow polymerase) and EcoRI for the 5′ region. This created a NotI site at both the extreme 5′ and 3′ ends, resulting in plasmid pKNSD168. The ZEO gene was cut from plasmid pPICZa (Invitrogen) as a 0.9-kb BamHI–StuI DNA fragment and cloned between the 5′ and 3′ PEX19 fragments in plasmid pKNSD168, digested by BamHI and EcoRV, resulting in pKNSD169. The disruption cassette was subsequently excised from pKNSD169 cut with NotI and used to transform PPY12, SMD1163, and STW2. Transformants were selected on YPD plates containing 100 μg/ml Zeocin (Invitrogen, Carlsbad, CA), and the expected genomic alteration in pex19 deletion strains, which were unable to grow on methanol, was confirmed by Southern blot analysis (our unpublished data).
Construction of the Farnesylation Mutants
Plasmid pWSP50 was generated by cloning the PCR (primers PEX19up and PEX19dn) product into pCR-Blunt (Invitrogen), excising with BamHI and EcoRI, and ligating to pIB1 (Sears et al., 1998), a HIS4 integrating vector, cut with the same enzymes. pWSP51 and pWSP52 were cloned using the same method as for pWSP50 using unique downstream primers (SAAX.19 and ΔC4.19). The pex19C296S mutation was confirmed by cutting at the unique restriction site (SpeI) that was introduced along with the codon change. These plasmids were introduced into SKF14 after linearization with SalI.
Preparation of Rabbit Anti-PpPex19p Antibodies
The PEX19 ORF was amplified by PCR (primers TW61 and TW63), introducing BamHI sites immediately 5′ and 3′ of the ORF, and cloned as a BamHI fragment into pQE9 (Qiagen, Chatsworth, CA) yielding plasmid pTW128. Amino-terminally HIS6-tagged PpPex19p was purified from E. coli using the manufacturer’s procedures for denatured proteins. Quantitative amounts of HIS6-PpPex19p were eluted from SDS-polyacrylamide gels and used to immunize rabbits (Bob Sergeant, Ramona, CA).
Biochemical Techniques
Crude cell-free extracts for Figure 9 were made as described previously (Babst et al., 1997). SDS-PAGE (Laemmli, 1970) and Western blot analyses (Towbin et al., 1979) were performed as described. Goat anti-rabbit HRP or goat anti-rabbit alkaline phosphatase (Bio-Rad, Hercules, CA) were used as secondary antibodies, which were detected by ECL (Amersham, Arlington Heights, IL) or bromochloroindoyl phosphate/nitro blue tetrazolium (Kirkland & Perry, Gaithersburg, MD) according to the manufacturers’ protocols. Primary antibodies and the dilutions used were α-Pex19p (1:4000), α-Pex3p (1:10,000), α-ScTHIO (1:10,000), α-CAT (1:10,000), α-MOX (1:10,000), α-Pex5p (1:10,000), α-Sc-glucose-6-phospate dehydrogenase (1:2000), and mouse-α-NH (1:5).
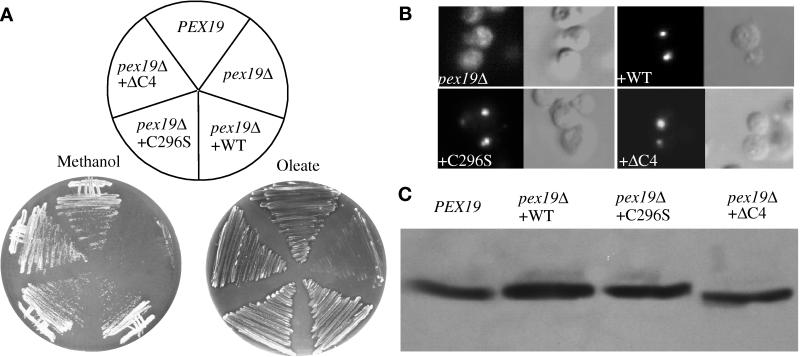
Figure 9.
The putative farnesylation site is not essential for Pex19p function. (A) Cells were grown on plates supplemented with methanol (left panel) or oleate (right panel) as sole carbon source. (B) Indirect immunofluorescence for catalase in methanol-grown pex19Δ cells and pex19Δ cells transformed with the indicated Pex19p-expressing plasmids. (C) Western blot analysis of total extracts of methanol-grown cells probed with Pex3p antibodies. PEX19, wild-type (SMD1163); pex19Δ, deletion mutant (SKF14); pex19Δ + WT, deletion mutant transformed with the wild-type PEX19 (WSP50); pex19Δ + C296S, deletion mutant transformed with the pex19C296S allele (WSP51); pex19Δ + ΔC4, deletion mutant transformed with the pex19C296ter allele (WSP52).
Immunoprecipitation was performed from 5 A600 units of cells in Tris-buffered saline and 0.5% Triton X-100 with 2 μl of anti-Pex19p per reaction as described previously (Stack et al., 1993).
Subcellular Fractionation Experiments
Differential centrifugation of oleate-grown cells was performed as described (Faber et al., 1998), except that 100 A600 units of cells were lysed in 5 ml of lysis buffer. Fractionations of methanol-grown cells were performed as with the oleate-grown cells, except that a HEPES-KOAc buffer (0.2 M sorbitol, 50 mM KOAc, 2 mM EDTA, and 20 mM HEPES, pH 6.8) was used as the lysis buffer. For floatation, all sucrose stocks contained the HEPES-KOAc buffer lacking the sorbitol, and the gradients were prepared as follows: 0.5 ml of postnuclear supernatant (PNS) from the methanol-grown cells was mixed with 1.5 ml of 80% (wt/vol) sucrose in a 5-ml ultracentrifuge tube; this was layered with 1.5 ml of 55% (wt/vol) sucrose and then 1.5 ml of 35% (wt/vol) sucrose. The gradients were centrifuged in a Beckman (Palo Alto, CA) SW50.1 rotor for 20 h at 40,000 rpm; 950-μl fractions were collected from the top and adjusted to 5% trichloroacetic acid (TCA). The material pelleted on the bottom of the tube was resuspended in 1 ml of 5% TCA and transferred to an Eppendorf tube. The TCA precipitations were incubated on ice for >20 min and washed once with 5% TCA and three times with cold acetone. The acetone was evaporated, and the pellets were resuspended in 100 μl of SDS-PAGE sample buffer.
Construction of a Strain Expressing NH-Pex10p
A pex10Δ construct was made as follows. The 3′ region of PEX10 was amplified by PCR (primers KNF45 and KNF46), cut with NotI, and ligated to pBluescript II KS+ cut with XbaI (filled in with Klenow) and NotI, creating pKNSD164. The 5′ region of PEX10 was amplified by PCR (primers KNF43 and KNF44), cut with NotI, and ligated to pKNSD164 cut with Asp718I (filled in with Klenow) and EcoRI, creating pKNSD167. The ZEO gene was released from pPICZa (Invitrogen) with StuI and BamHI and ligated into pKNSD167 cut with SmaI and BamHI, creating pSH5. pSH5 was cut with NotI and transformed into strain SMD1163 to create the pex10Δ strain SSH4, which was confirmed by genomic PCR and unable to grow on methanol- or oleate-containing medium. PEX10 was amplified by PCR (primers PEX10B and PEX10C), cut with BamHI and EcoRI, and inserted into p21-43 cut with the same, creating pSSH38. p21-43 was a gift of Ype Elgersma (University of California, San Diego, CA) and has the acyl-coenzyme A oxidase promoter upstream of a BglII–BamHI fragment encoding the NH epitope (Elgersma et al., 1996), with a downstream EcoRI site, and the HIS4 gene for selection in P. pastoris. pSSH18 was cut with StuI and integrated into SMD1163 creating strain SSH18.
Fluorescence and Electron Microscopy
Preparation and analysis of cells expressing green fluorescent protein (GFP) constructs were as described (Monosov et al., 1996). Samples for immunofluorescence were prepared from methanol-induced cells spheroplasted as described for biochemical fractionation and then fixed and prepared as described previously (Babst et al., 1998). Pex3p and catalase antibodies were used at dilutions of 1:10,000. Microscopy for Figure 5 is as described (Odorizzi et al., 1998). Immunofluorescence was visualized on a Delta Vision (Issaquah, WA) optical sectioning microscope (Pogliano et al., 1999) for Figure 6. Cells for electron microscopy were prepared as described previously (Sakai et al., 1998).
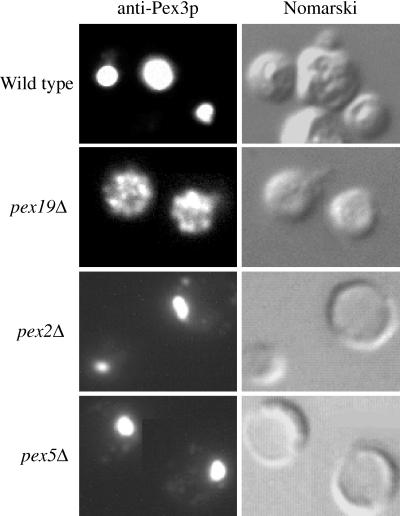
Figure 5.
Immunofluorescence microscopy of Pex3p-containing compartments in wild-type and pex mutant cells. Methanol-grown wild-type (PPY12), pex19Δ (SKF13), pex2Δ (PPY12Δ2), and pex5Δ (PPY12Δ5) spheroplasts were indirectly labeled with the anti-Pex3p antibody and visualized by fluorescence microscopy (Pex3p) and Nomarski optics.
Figure 6.
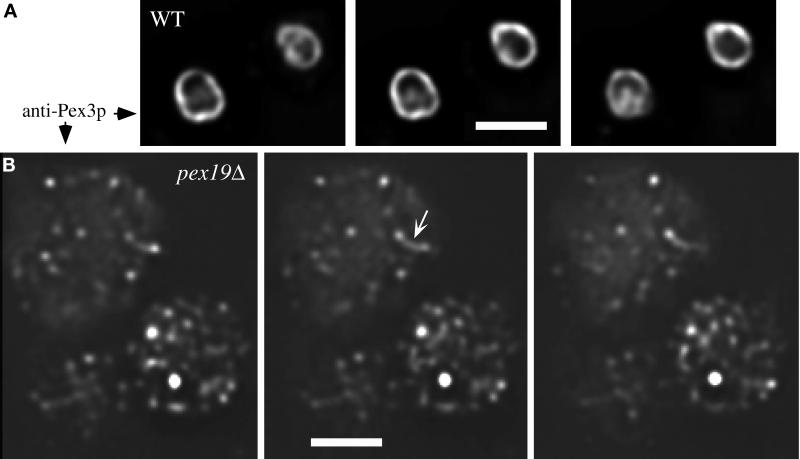
Deconvolution microscopy of Pex3p immunofluorescence from wild-type and pex19Δ cells. Pex3p-containing compartments from wild-type (PPY12) (A) and pex19Δ (SKF13) (B) cells were visualized by immunofluorescence staining with Pex3p antibodies on a Delta Vision optical sectioning microscope. Three consecutive 0.2-μm sections are shown from each strain. Bar, 2 μm.
RESULTS
A Positive Selection Yields New pex Mutants
To characterize novel PEX genes encoding proteins required for peroxisomal protein import, we applied a positive-screening method, originally described for S. cerevisiae (Elgersma et al., 1993), in P. pastoris. This procedure uses a drug resistance protein fused to a PTS to select mutants; in wild-type cells the drug resistance protein hybrid is imported into the peroxisome, and the cells are sensitive to the drug. Import mutants accumulate the resistance protein hybrid in the cytosol and thus become resistant to the drug. To this end, the gene encoding the bleomycin-resistance protein BLE from the Tn5 transposon was fused with a carboxyl-terminal PTS1 encoding sequence (-SKL) and placed under control of the P. pastoris alcohol oxidase 1 promoter. After N-methyl-N-nitro-N-nitrosoguanidine mutagenesis of the strain carrying this construct and enrichment of mutants (see MATERIALS AND METHODS) that were defective for the import of the reporter protein into the peroxisome, several hundred candidate colonies were obtained. Twenty-two of these strains were unable to grow on oleate or methanol as the sole carbon source but grew well on media containing glucose, glycerol, or ethanol, indicating the specificity of the peroxisome defect. These strains were crossed with available pex mutants and assigned to complementation groups. Most mutants fell into known complementation groups (Table 4). However, one mutant (112) did not belong to any of the existing P. pastoris pex complementation groups and accumulated both PTS1- and PTS2-containing GFP fusion proteins in the cytosol (our unpublished data). Accordingly, this mutant was chosen for further analysis.
Table 4.
Complementation groups of new P. pastoris pex mutants
| Complementation group | No. of mutants found |
|---|---|
| PEX1 | 1 |
| PEX2 | 2 |
| PEX3 | 2 |
| PEX4 | 0 |
| PEX5 | 4 |
| PEX6 | 2 |
| PEX8 | 4 |
| PEX10 | 0 |
| PEX12 | 6 |
| PEX13 | 0 |
| Unknown | 1 |
Cloning of P. pastoris PEX19
Complementation of the methanol growth defect of mutant 112 by a genomic DNA library led to the identification of four distinct clones that contained overlapping genomic inserts (our unpublished data). The minimal complementing region was narrowed to a 2.2-kb SalI fragment (our unpublished data). Sequence analysis revealed an ORF of 897 nucleotides (GenBank accession number AF133271), encoding a 299-amino acid protein (Figure 1) with a predicted molecular mass of 33.7 kDa. This protein is predicted to be negatively charged (pI 3.88) and hydrophilic, lacking recognizable hydrophobic transmembrane domains. Comparison of the protein sequence with those in databases revealed five proteins with significant homology, namely Chinese hamster PxF (James et al., 1994), human PxF (Kammerer et al., 1997) (originally called HK33; Braun et al., 1994), S. cerevisiae Pex19p (Götte et al., 1998), and uncharacterized ORFs from Schizosaccharomyces pombe and Caenorhabditis elegans (Figure 1). The similarity among these proteins suggests that the complementing fragment encodes the P. pastoris PEX19. All previously described PxF/Pex19p proteins have been shown to be farnesylated at a carboxy;-terminal sequence, CaaX (C, cysteine; a, aliphatic; X, any amino acid). Notably, Pex19p contains a CKQQ residue at the carboxyl terminus that is fully conserved between S. cerevisiae and P. pastoris.
Figure 1.
Sequence alignment of Pex19p orthologues. The amino acid sequences were aligned using the ClustalW program. Identical residues (black) and similar residues (gray) in at least half of the orthologues are shaded. Pp, P. pastoris Pex19p; Sc, S. cerevisiae Pex19p; Hs, Homo sapiens HK33/PxP; Cg, Cricetulus griseus PxP; Sp, S. pombe hypothetical protein C17C9.14; Ce, C. elegans hypothetical protein F54F2.8. Similarity rules: G = A = S, A = V, V = I = L = M, I = L = M = F = Y = W, K = R = H, D = E = Q = N, and S = T = Q = N. Dashes represent gaps.
Peroxisome Protein Synthesis and Localization Defects in the pex19Δ Mutant
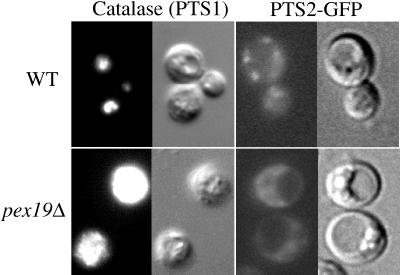
P. pastoris strains deleted for PEX19 were constructed as described in MATERIALS AND METHODS. These pex19Δ mutants were unable to grow on methanol or oleate as sole carbon sources (see below) and mislocalized catalase, a PTS1-containing peroxisome matrix protein, and PTS2-GFP proteins in the cytosol (Figure 2), indicating a defect in the import of both PTS1- and PTS2-containing peroxisome matrix proteins. In contrast, wild-type cells showed the characteristic punctate peroxisomal staining pattern (Figure 2).
Figure 2.
pex19Δ mutants are defective for the import of catalase (PTS1) and PTS2-GFP. Wild-type and pex19Δ cells (SMD1163 and SKF14) grown in methanol media were prepared for anti-catalase immunofluorescence, and PTS2-GFP expressing wild-type and pex19Δ cells (STW2 and AK11) grown in oleate media were visualized using a fluorescence microscope (anti-catalase, GFP) and Nomarski optics as described in MATERIALS AND METHODS.
To characterize protein synthesis and stability defects in the pex19Δ mutant, wild-type and pex19Δ cells were grown in peroxisome-inducing conditions, and the steady-state levels of peroxisomal proteins was determined by SDS-PAGE and immunoblotting. In oleate-grown wild-type cells, a characteristic induction pattern is observed for Pex3p, Pex5p, and thiolase (Figure 3A). In contrast, the pex19Δ cells contained lower levels of Pex3p, a PMP, after 4 h of induction when compared with wild-type cells (Figure 3A). The initial levels of Pex3p appeared normal during the first 2 h of induction, suggesting that the defect at later time points is not due to lower induction of Pex3p synthesis in oleate media. Pex5p and thiolase were present at near wild-type levels in pex19Δ mutants throughout the oleate induction, suggesting that induction of peroxisomal proteins is normal in pex19Δ mutants. The internal loading control, mitochondrial F1β-ATPase, was similar in all samples. In methanol-grown cells no significant difference was observed between wild-type and pex19Δ cells for the levels of Pex3p, alcohol oxidase (MOX), and Pex5p (Figure 3B). These results suggest that there is a defect in the biogenesis of Pex3p, which could lead to protein instability, in oleate-grown pex19Δ cells.
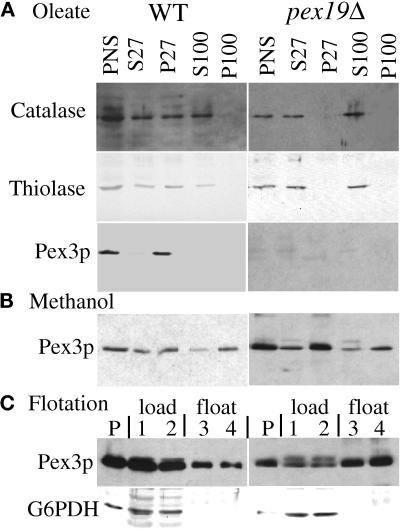
Differential centrifugation of a lysate prepared from oleate-induced wild-type and pex19Δ cells (see MATERIALS AND METHODS) confirmed the peroxisome protein import defect of pex19Δ cells (Figure 4). PTS1- and PTS2-containing proteins catalase and thiolase were detected in the lysate or PNS after a low-speed spin (Figure 4A). Upon centrifugation at 27,000 × g, the pellet fraction from wild-type cells contained peroxisome-associated catalase, thiolase, and the membrane protein Pex3p (P27). Some of the catalase and thiolase leaked from the peroxisome during the biochemical manipulations, consistent with previous reports (Kalish et al., 1996; Waterham et al., 1996; Elgersma et al., 1998; Faber et al., 1998), and could not be further pelleted even at 100,000 × g. In contrast, in pex19Δ cells thiolase and catalase were found exclusively in the supernatant fractions (Figure 4A, S27 and S100). The PMP Pex3p could not be detected in oleate-grown cells (Figure 4A; also see Figure 3A). These data clearly demonstrate a defect for the import of lumenal peroxisomal proteins containing PTS1 or PTS2 sequences in pex19Δ mutants.
Figure 4.
Mislocalization of peroxisomal proteins by pex19Δ mutants. Oleate-grown (A) and methanol-grown (B) spheroplasts of wild-type and pex19Δ cells (SKF14 and SMD1163) were lysed and subjected to sequential differential centrifugation. Equivalent amount of the PNS, 27,000 × g supernatant (S27), 27,000 × g pellet (P27), 100,000 × g supernatant (S100), and 100,000 × g pellet (P100) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the indicated antibodies. (C) The PNS from methanol-grown cells was adjusted to 60% sucrose, layered with 55 and 35% sucrose, and centrifuged to allow floatation of membranes into the lighter fractions as described in MATERIALS AND METHODS. P, pelleted material at the bottom of the tube. Fractions are numbered from the bottom to the top and were collected as described in MATERIALS AND METHODS. Fractions 1 and 2 are the 60% sucrose load zone; fractions 3 and 4 are the 55% sucrose float zone. The top fraction contained no protein.
To follow the fate of Pex3p in the absence of Pex19p, a fractionation experiment was performed with methanol-grown cells. We observed the majority of Pex3p in the 27,000 × g and 100,000 × g pellet fractions (P27 and P100) of both wild-type and pex19Δ cells (Figure 4B). The S100 and P100 pools of Pex3p are likely due to the increased rupture of methanol-grown peroxisomes, compared with oleate-grown peroxisomes, during the fractionation procedure, a phenomena that has been observed previously with lumenal peroxisomal proteins (Waterham et al., 1996). To determine whether the Pex3p that pellets in the pex19Δ cells is membrane associated, or simply in large protein aggregates, we loaded a PNS fraction at the bottom of a sucrose density gradient to see whether Pex3p could float to lighter-density fractions, a behavior that is consistent with membrane association. A large portion of Pex3p from both strains floated from the load fractions (Figure 4C, lanes 1 and 2), which contain 60% sucrose, to the float fractions (Figure 4C, lanes 3 and 4), which contain 55% sucrose. Very little detectable Pex3p was found in the very top fraction of the gradient containing 35% sucrose, and some of the Pex3p was pelleted to the bottom of the tube (Figure 4C, lanes P). The cytosolic protein glucose-6-phosphate dehydrogenase did not float into fractions that contain 55% sucrose (Figure 4C) as expected. This analysis suggests that at least a portion of Pex3p is localized to membranous structures in pex19Δ cells grown in methanol medium.
Novel Peroxisome Remnants in pex19Δ Cells
Methanol-induced wild-type and pex mutant cells were analyzed for the presence of peroxisomal remnants by using indirect immunofluorescence to visualize Pex3p-containing structures. In wild-type cells the characteristic large cluster of peroxisomes was observed, which contain Pex3p (Figure 5). In pex2Δ and pex5Δ mutants, a large cluster of peroxisome remnants, or ghosts, was observed (Figure 5). Similar structures were observed in all other pex mutants examined, including pex1, 4, 6, 7, 8, 10, 12, and 13 (Gould et al., 1992, 1996; McCollum et al., 1993; Spong and Subramani, 1993; Heyman et al., 1994; Kalish et al., 1995, 1996; Liu et al., 1995; Waterham et al., 1996; Elgersma et al., 1998; Snyder, unpublished results). In contrast, pex19Δ cells did not contain the large cluster of organelle remnants, but instead Pex3p was found in small bright dots dispersed throughout the cell (Figure 5). To further elucidate the morphology of the Pex3p-containing structures in pex19Δ cells, Pex3p immunofluorescence was visualized in a deconvolution microscope, which provides greater resolution, allowing the visualization of peroxisome membrane staining by Pex3p antibodies in wild-type cells (Figure 6A). In contrast, the Pex3p-containing structures in pex19Δ cells appeared as very small spheres and tubules (Figure 6B). The arrowhead in Figure 6B points to one such tubule that seems to be connecting two of the small spherical structures. These results and those presented above support the conclusion that Pex3p is localized to small spherical and tubular membranous compartments in the absence of Pex19p, and that these structures are smaller and more dispersed than the typical peroxisome ghosts seen in other P. pastoris pex mutants.
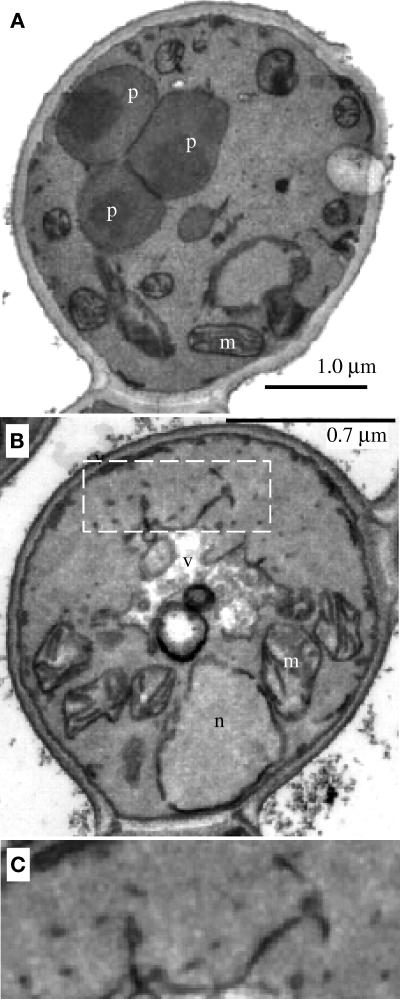
Analysis of the methanol-induced pex19Δ cells by electron microscopy revealed structures that were absent in wild-type cells (Figure 7, compare A and B). These structures are small vesicles and tubules (Figure 7C), which likely correspond to the Pex3p-containing structures seen in the immunofluorescence experiment. Immunoelectron microscopy revealed clusters of gold particles when stained with Pex3p antisera; however, no membranous structures were visualized around these clusters because of insufficient contrast or resolution (our unpublished data). The data presented above clearly indicate the presence of Pex3p-containing peroxisome remnants in pex19Δ cells, but our inability to clearly visualize such structures by electron microscopy demonstrates the difficulties with identification of certain types of peroxisome remnants by this technique.
Figure 7.
Electron microscopy of wild-type and pex19Δ cells. Methanol-grown wild-type (PPY12) (A) and pex19Δ (SKF13) (B) cells were prepared as described in MATERIALS AND METHODS. (C) Enlargement of the area highlighted by dashed white box in B. n, nucleus; m, mitochondria; v, vacuole; p, peroxisome.
Characterization of Pex19p and Its Predominantly Cytosolic Location
Rabbit antibodies were raised against bacterially expressed HIS6-tagged Pex19p. These antibodies specifically recognized a single protein band of ∼45 kDa in wild-type cells, which was absent in the pex19Δ strain (see below). Like many Pex proteins, Pex19p was detectable in glucose-grown cells and was induced 5- to 10-fold during peroxisome-inducing conditions (Figure 8A). The observed molecular weight of Pex19p was significantly different from the calculated molecular mass of 33.7 kDa but is in agreement with the molecular mass observed for the bacterially expressed protein (our unpublished data) and is the same for oleate- and methanol-grown cells. Therefore, the reduced migration in SDS-PAGE seems to be an intrinsic feature of Pex19p. Noteably, only a single species of Pex19p was observed in either growth condition.
The subcellular localization of Pex19p was determined by the differential fractionation technique. Controls for this experiment are shown in Figure 4A, where peroxisome marker proteins fractionated in the usual manner. Centrifugation of the PNS at 27,000 × g left Pex19p predominantly (>95%) in the supernatant fraction (Figure 8B, S27). Minor amounts (<5%) of Pex19p were present in this organelle pellet fraction (Figure 8B, P27), consistent with this pool of Pex19p being localized to the peroxisome. Further centrifugation of the 27,000 × g supernatant at 100,000 × g did not pellet any of the Pex19p, suggesting that the protein is predominantly cytosolic (Figure 8B, S100 and P100). The absence of the Pex19p band in pex19Δ cells indicates the specificity of the antibody (Figure 8B).
The Putative Farnesylation Site Is Not Essential for Protein Function
All members of the PxF/Pex19p family examined so far are known to be farnesylated at a carboxyl-terminal CaaX motif (James et al., 1994; Kammerer et al., 1997; Götte et al., 1998). Moreover, in S. cerevisiae the farnesylation site was important for protein function but not absolutely required; a mutant form of Pex19p, with a serine replacing the cysteine four amino acids from the carboxyl terminus (ScPex19pC347S), can partially complement the deletion mutant for growth on oleate media and shows partial localization of peroxisomal markers (Götte et al., 1998). This SaaX-containing mutant protein is not a substrate for farnesylation. PpPex19p also contains the same carboxyl-terminal four amino acids as ScPex19p. To test whether this sequence is essential for protein function in P. pastoris, we deleted the four carboxyl-terminal amino acids (pex19C296ter) and also mutated the critical cysteine residue to serine (pex19C296S) and expressed these proteins from their own promoter, integrated in the chromosome of pex19Δ cells. Both mutant proteins complemented the pex19Δ phenotype for growth on methanol and oleate media (Figure 9A) and for the import of catalase into peroxisomes (Figure 9B). The mutant proteins were expressed at wild-type levels (Figure 9C). In addition, the Pex19pC296S displayed the same electrophoretic mobility as the wild-type protein (Figure 9C), and because the farnesylated form of the ScPex19p migrates faster in SDS-PAGE than the unmodified form (Götte et al., 1998), we conclude that PpPex19p is not farnesylated under the conditions tested.
Pex19p Interacts with Pex3p and Pex10p
The yeast two-hybrid system was used to assay for potential protein–protein interactions between Pex19p and other peroxins from a collection of PEX gene two-hybrid constructs. A combination of a Pex19p–activation domain fusion protein with Pex3p– or Pex10p–DNA-binding domain fusions, both PMPs, resulted in activation of transcription of the reporter genes (HIS3 and LacZ/β-galactosidase) (Figures 10 and 11). Unfortunately, the two-hybrid analysis could only be tested with PEX19 in the activation domain plasmid, because this protein alone confers activation of transcription of the reporter genes when expressed as a DNA-binding domain fusion protein in the absence of an activation domain fusion (our unpublished data).
Figure 10.
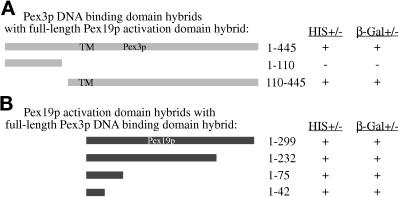
Two-hybrid analysis of Pex3p and Pex19p. The indicated Pex3p and Pex19p hybrid protein constructs were tested for trans-activation of the HIS3 gene, resulting in growth on media lacking histidine (+) or LacZ, resulting in the production of β-galactosidase assayed by blue color change (+) as described in MATERIALS AND METHODS. Numbers refer to amino acids from Pex3p (A) or Pex19p (B). TM, relative position of the putative transmembrane domain in Pex3p.
Figure 11.
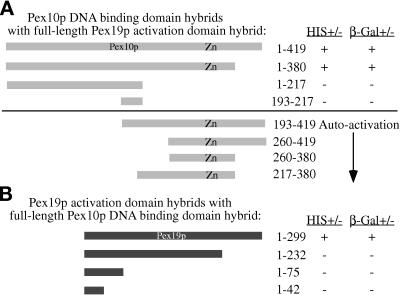
Two-hybrid analysis of Pex10p and Pex19p. The indicated Pex10p and Pex19p hybrid protein constructs were tested for trans-activation of the HIS3 gene, resulting in growth on media lacking histidine (+) or LacZ, resulting in the production of β-galactosidase assayed by blue color change (+) as described in MATERIALS AND METHODS. Numbers refer to amino acids from Pex10p (A) or Pex19p (B). Zn, relative position of the zinc-binding domain in Pex10p.
Mapping of the Protein–Protein Interaction Domains
The interacting domains of Pex3p and Pex19p were mapped by expressing subdomains of both proteins in the yeast two-hybrid system. Two subdomains of Pex3p, expressed as DNA-binding domain fusions, were tested for interaction with the full-length Pex19–activation domain protein. The amino-terminal domain of Pex3p (amino acids 1–110) did not activate transcription of the reporter genes when expressed with the full-length Pex19–activation domain fusion protein (Figure 10A). The carboxyl-terminal region, from amino acids 110–440, did result in activation of both reporter genes (Figure 10A) in the presence of the Pex19p–activation domain fusion protein. None of the Pex3p containing DNA-binding domain fusions autoactivated transcription from the reporter genes. The region of Pex19p responsible for the interaction with Pex3p was narrowed to the extreme amino terminus of Pex19p. Only 42 amino acids of the amino terminus of Pex19p were required to yield a positive interaction with the full-length Pex3p (Figure 10B). These results indicate that the amino terminus of Pex19p interacts with the carboxyl-terminal 335 amino acids of Pex3p in the two-hybrid system.
Pex10p domains were expressed in the two-hybrid system to further elucidate the site of interaction with Pex19p. DNA-binding domain fusions comprising many different regions of Pex10p were created with the goal of precisely mapping the interacting region; however, many of these fusion proteins led to the activation of transcription in the absence of an activation domain fusion (Figure 11A). The ability of these DNA-binding domain fusions to autoactivate transcription made the two-hybrid test impossible, because the DNA-binding domain fusions to Pex19p also autoactivate transcription. A few of the Pex10p fusions do not autoactivate and were therefore useful to narrow the region of interaction to a domain between amino acids 1 and 380 (Figure 11A). The shorter amino-terminal fusion (amino acids 1–217) and the internal fragment (amino acids 193–217) that did not activate were not useful for a positive evidence–based conclusion. Moreover, only the full-length Pex19p fusion interacted with the full-length Pex10p fusion in the two-hybrid test (Figure 11B). Taken together these results suggest that a region of Pex10p, excluding the carboxyl-terminal 40 amino acids, interacts with Pex19p in a manner that is dependent on the extreme carboxyl terminus of Pex19.
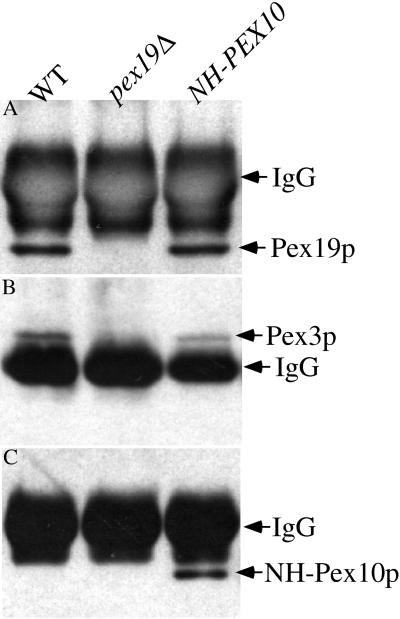
The interactions suggested by the two-hybrid system were confirmed by coimmunoprecipitation. We created a strain expressing an epitope-tagged version of Pex10p, NH-Pex10p, that fully complements a pex10Δ mutant (our unpublished data). Oleate-induced wild-type, pex19Δ, and NH-PEX10 cells were lysed, and Pex19p was immunoprecipitated from these lysates. The material from the immunoprecipitation was separated by SDS-PAGE and immunoblotted to identify other Pex proteins in a complex with Pex19p. Pex19p was easily detected in these immunoprecipitations from wild-type and NH-PEX10 strains but not from the pex19Δ cells (Figure 12A). Pex3p was also observed in the Pex19p immunoprecipitations from wild-type and NH-PEX10 strains but not from the pex19Δ cells (Figure 12B) as expected. We also observed coimmunoprecipitation of NH-Pex10p with Pex19p antisera (Figure 12C) in the NH-PEX10 strain. Pex19p immunoprecipitations did not contain detectable levels of Pex8p or Pex5p (our unpublished data), indicating the specificity of the result. These results demonstrate that Pex19p physically interacts with Pex3p and Pex10p.
Figure 12.
Coimmunoprecipitation of Pex3p and NH-Pex10p with Pex19p. Pex19p was immunoprecipitated from cleared lysates of wild-type (SMD1163), pex19Δ (SKF14), or NH-PEX10 (SSH18) strains as described in MATERIALS AND METHODS. The material was resolved by SDS-PAGE and analyzed by immunoblotting with anti-Pex19p (A), anti-Pex3p (B), and anti-NH antibodies (C).
DISCUSSION
Pex19p has been cloned and characterized from two different yeasts (P. pastoris and S. cerevisiae), mammalian cells (Chinese hamster and human PxF), and homologous ORFs identified in C. elegans and S. pombe. We identified the P. pastoris PEX19 gene, characterized the mutant cells, analyzed the protein product, and defined protein–protein interactions. These results combined with information regarding Pex19p from other systems reinforce the convergence of model systems used to study peroxisomes. Despite the fact that the overall identity between the various Pex19p orthologues is low, several lines of evidence suggest that they belong to the same Pex group. First, all six proteins are approximately the same size, are hydrophilic, and contain a consensus sequence for farnesylation at the carboxyl terminus. In addition, there are regions of homology that are conserved between all the orthologues, and these regions are scattered throughout the proteins (Figure 1). Finally, similar protein–protein interactions have been observed for Pex19p from S. cerevisiae and P. pastoris.
Novel Peroxisome Remnants in pex19Δ Cells
Previous studies in S. cerevisiae had suggested that the pex19Δ strain lacks peroxisome remnants (Götte et al., 1998). This conclusion was based solely on observations using electron microscopy. However, our immunofluorescence studies (Figures 5 and 6), particularly with the greater resolution afforded by the deconvolution microscope (Figure 6), and the biochemical fractionation experiments (Figure 4, B and C) show unequivocally the presence of small spherical and tubular remnants containing Pex3p in the pex19Δ strain. It is noteworthy that these remnants could easily have been missed using conventional immunofluorescence or electron microscopy, because the structures are much smaller and morphologically distinct from the remnants observed in many other pex mutants or the large (∼1.3-μm-diameter) cluster of peroxisomes seen in wild-type cells (Figure 5). Consistent with earlier reports, no normal peroxisomes, or the usual peroxisome remnant structures observed in many other pex mutants (Gould et al., 1992, 1996; McCollum et al., 1993; Spong and Subramani, 1993; Heyman et al., 1994; Kalish et al., 1995, 1996; Liu et al., 1995; Waterham et al., 1996; Elgersma et al., 1998), were seen in Pppex19Δ cells by electron microscopy. However, structures resembling the spherical and tubular forms seen by deconvolution microscopy were observed in our electron micrographs (Figure 7C). It is quite possible that these unusual remnants were missed in S. cerevisiae because they represent novel structures not observed in other pex mutants. Others have also stressed the importance of immunofluorescence for the identification of remnants in mutants when they were not observed by electron microscopy (Purdue and Lazarow, 1995). In addition, when the pex19Δ allele was introduced into pex mutant strains that contained the normal, large peroxisome remnants, these double mutants contained only the type of remnants seen in pex19Δ mutants (Snyder, unpublished observation), suggesting that the pex19Δ remnants are formed before the remnants seen in other pex mutants (see below).
Farnesylation of PpPex19p Is Not Essential for Peroxisome Biogenesis
Despite the finding that the Chinese hamster, human, and S. cerevisiae Pex19/PxF proteins are modified by farnesylation at a conserved CaaX motif (James et al., 1994; Kammerer et al., 1997; Götte et al., 1998), this modification is unnecessary for PpPex19p function. ScPex19p migrates as a doublet in SDS-PAGE; the faster-migrating species is the farnesylated form, which comprises ∼60% of the steady-state level of Pex19p (Götte et al., 1998). We only detect a single band of Pex19p in P. pastoris (Figures 8A and 9C), suggesting that this modification is absent under the induction conditions tested, despite the fact that the carboxyl-terminal sequence CKQQ, which is farnesylated in S. cerevisiae, is fully conserved in PpPex19p. Furthermore, PpPex19p mutant proteins that cannot be modified by farnesylation, Pex19pC296ter and Pex19pC296S, fully complement Pppex19Δ mutant cells for growth on methanol or oleate (Figure 9A) and contain morphologically wild-type peroxisomes that import catalase (Figure 9B). In addition, the point mutant has the same mobility in SDS-PAGE as the wild-type protein (Figure 9C). These observations contrast results obtained for ScPex19p, for which farnesylation is clearly involved in protein function. What purpose the farnesylation serves in S. cerevisiae and how this requirement is bypassed in P. pastoris remain to be determined. Taken together, these data strongly suggest that PpPex19p is not farnesylated; however, we cannot rule out the possibility that farnesylation may be transient or occur at a different time of peroxisome induction other than the ones tested here.
Other observations also raise questions about the functional significance of farnesylation of Pex19p: 1) in S. cerevisiae, not all of the protein is modified, suggesting that modification is slow, or perhaps reversible; 2) the addition of a hydrophobic molecule such as farnesyl might be expected to act as a membrane anchor, but the majority of yeast Pex19p has been localized to the cytosol; and 3) in S. cerevisiae the Pex19pC347S allows partial complementation of pex19Δ cells, suggesting that farnesylation is not an absolute requirement for ScPex19p function. Farnesylation seems only to improve the efficiency of Pex19p function in S. cerevisiae and may be redundant in P. pastoris, either because of the extended carboxyl terminus of the PpPex19p relative to the other orthologues or some other activity.
The Subcellular Localization of Pex19p
Pex19p was found predominantly in the cytosolic fractions of P. pastoris (Figure 8B), S. cerevisiae (Götte et al., 1998), and Chinese hamster (James et al., 1994) cells. However, the mammalian orthologues and overexpressed myc-ScPex19p have been observed at the peroxisome membrane by electron microscopy (James et al., 1994; Kammerer et al., 1997; Götte et al., 1998). Perhaps the protein is loosely associated with peroxisomes, and this association is disrupted by biochemical manipulations. Moreover, a small fraction of total cellular Pex19p that associates with the peroxisome may be detected immunocytochemically, because of the high local concentration, but the majority of the protein could essentially go unnoticed in the cytosol because of low local concentrations. For now we must conclude that the majority of PpPex19p resides in a nonperoxisomal, cytosolic location, and that a small pool may transientley associate with peroxisomes, perhaps by binding to Pex3p (see below).
Interactions of Pex19p with Other Peroxins
We demonstrate that PpPex19p interacts with two PMPs, Pex3p and Pex10p (Figures 10–12). ScPex19p interacts with the ScPex3p (Götte et al., 1998), and our results confirm this interaction in P. pastoris. In addition, we show that the extreme amino terminus (amino acids 1–42) of PpPex19p interacts with the carboxyl-terminal domain (110–455) of Pex3p. PpPex3p contains a putative transmembrane domain at amino acids 149–174, suggesting that the carboxyl-terminal two-thirds of the protein are on one side of the peroxisomal membrane. Indeed, protease protection experiments suggest that the majority of the protein, and thus this carboxyl-terminal region of PpPex3p, is exposed to the cytosol (our unpublished data). This topology of PpPex3p is consistent with that reported for ScPex3p (Höhfeld et al., 1991). This result allows us to conclude that the cytosolic domain of Pex3p interacts with Pex19p. Consistent with these observations is the hypothesis that Pex19p, which is predominantly cytosolic, transiently interacts with the cytosolic domain of Pex3p, which acts like a Pex19p receptor (Götte et al., 1998). The minor amount of Pex19p found in the organelle pellet fraction might represent the pool that is transiently interacting with Pex3p at the peroxisome. The apparent instability of Pex3p in oleate-grown pex19Δ cells (Figure 3A) could be caused by its missing binding partner, Pex19p.
Interaction of the extreme carboxyl terminus of Pex19p with Pex10p suggests that Pex19p might have multiple functions. Because much of Pex10p, including its zinc-binding domain, is believed to be within the peroxisome lumen (Kalish et al., 1995), Pex19p likely interacts with Pex10p before its insertion in the membrane. In this regard, Pex19p may function to facilitate the targeting, import, or assembly of Pex10p at the peroxisome. On the other hand, Pex19p may interact with the small cytosolic domain of Pex10p (amino acids 193–217). We must point out that the topology model for human Pex10p (Warren et al., 1998) is opposite from that of PpPex10p; therefore caution must be used when incorporating the topology data into interaction models. Nonetheless, we believe that the interactions of Pex19p with the two PMPs must be transient, because the majority of Pex19p is not found on the peroxisome membrane. Additional studies will be required to determine the functional significance of the interaction of Pex19p with Pex3p and Pex10p.
Role of Pex19p in Peroxisome Biogenesis
A role for Pex19p in peroxisome biogenesis and the import of peroxisomal proteins has been clearly established for P. pastoris and S. cerevisiae. In both yeast mutants there is a strong defect for the import of both PTS1- and PTS2-containing peroxisomal matrix proteins. Because Pppex19Δ cells clearly contain novel peroxisome remnants that are smaller and morphologically distinct (termed early remnants) from those (late remnants) seen in most other pex mutants, we postulate that the structures accumulating in the absence of Pex19p are normal intermediates (and not aberrant structures) in peroxisome biogenesis. If this is true, we propose the following model for peroxisome biogenesis. Early remnants that accumulate in the pex19Δ strain progress by further protein and/or membrane addition to the late remnants seen in other pex mutants, and these, in turn, mature to normal peroxisomes by the import of matrix proteins. In this model, Pex19p has a direct role in the maturation of the early remnants to the late ones. We do not know whether the block in the pex19Δ strain is due to the inability to incorporate certain PMPs into the peroxisomal membrane or due to some other defect, such as lipid addition. However, because Pex3p is found in the early remnant, its incorporation into membranes could not be dependent on Pex19p. Rather, Pex3p may serve as the Pex19p receptor to transiently dock it at the peroxisome membrane. This would be consistent with the hypothesis that for the earliest stages of peroxisome biogenesis Pex3p functions concurrently with Pex19p. This notion is also supported by previous reports that pex3Δ cells have no remnants, although those results have to be reexamined in the light of the novel remnants reported here in the pex19Δ strain (Höhfeld et al., 1991; Baerends et al., 1996; Wiemer et al., 1996).
The functional significance of the Pex19p–Pex10p interaction is likely different from the Pex3p–Pex19p interaction. pex10 mutant cells contain late remnant membranes (Kalish et al., 1995), suggesting that Pex19p performs early functions, proposed above, that are epistatic to a later Pex10p function. One possibility is that the Pex19p–Pex10p interaction is necessary for the import and/or assembly of Pex10p into peroxisomes and that Pex10p has to be in the early remnants for them to mature into normal peroxsiomes.
An alternative to the biogenesis model presented above is the possibility that Pex19p coordinates growth and division of peroxisomes, and in its absence these are no longer coordinated, leading to the development of small membrane fragments by unchecked division. Growth and division of peroxisomes must be tightly controlled to maintain the normal size and morphology of the organelle in wild-type cells. In pex19Δ cells, the Pex3p-containing structures might be created by the inability to maintain a large organelle, not due to a defect in maturation from a preperoxisome to a peroxisome as suggested above. The fragmentation would be due to unregulated division in the absence of coordinated growth. This is consistent with the conclusion from Hansenula polymorpha studies that Pex3p is required for the maintenance of the peroxisome membrane (Baerends et al., 1996) and our hypothesis that Pex19p may function concurrently during this process. Additional support for a role of Pex10p in this process is again provided from H. polymorpha experiments, which suggest that Pex10p functions to regulate peroxisome division (Tan et al., 1995). With these observations in mind, it would be reasonable to suggest that the interacting proteins Pex19p, Pex3p, and Pex10p function to regulate peroxisome division. In this scenario, Pex19p and Pex3p would act like negative regulators of division, whereas Pex10p would perform a stimulatory role.
Future experiments will be required to address what, if any, remnant structures containing peroxisomal markers exist in the pex3Δ mutants and whether other PMPs localize to the same structures in pex3Δ and pex19Δ cells. Clearly we must distinguish between the possibilities that Pex19p mediates recruitment of membrane from an as yet unidentified source, or the fusion of early remnants into larger structures, and the assembly of membrane components to allow lumenal import to begin. In addition, it will be important to determine whether these types of remnant structures are a normal precursor in the biosynthesis of peroxisomes or the product of aberrant peroxisome division. Isolation of conditional alleles of PEX19 could help distinguish these possibilities.
The origin of the early remnants is an intriguing question that is relevant in light of recent suggestions that ER-derived vesicles play a role in peroxisome biogenesis (Titorenko and Rachubinski, 1998). It will also be interesting to determine the connection between the nonperoxisomal membranous structures containing the two AAA-ATPases, Pex1p and Pex6p (Faber et al., 1998), and the types of remnants described here.
ACKNOWLEDGMENTS
We thank Jim Cregg for providing strains and unpublished information about pex mutant collections and Su Hua for technical assistance. We are grateful to Kit Pogliano and Joe Pogliano for assistance with the deconvolution microscopy and Scott Emr for use of his microscope. We also appreciate the advice of Peter Rehling, Andy Wurmser, and Marcus Babst. This work was supported by fellowships from the American Cancer Society (to W.B.S.), the Human Frontier Science Program Organization (to K.N.F.), the European Molecular Biology Organization (to T.J.W.), the Swiss National Funds (to A.K.), and the Deutsche Forschungsgemeinshaft (to G.H.L.) and National Institutes of Health grant NIHDK-41737 and the March of Dimes (to S.S.). The nucleotide sequence of PpPEX19 has been submitted to GenBank with accession number AF133271.
Abbreviations used:
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- PMP
peroxisomal membrane protein
- PNS
postnuclear supernatant
- PTS
peroxisomal targeting signal
- TCA
trichloroacetic acid
REFERENCES
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei I, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- Braun A, Kammerer S, Weissenhorn W, Weiss EH, Cleve H. Sequence of a putative human housekeeping gene (HK33) localized on chromosome 1. Gene. 1994;146:291–295. doi: 10.1016/0378-1119(94)90308-5. [DOI] [PubMed] [Google Scholar]
- De Duve C. The peroxisome in retrospect. Ann NY Acad Sci. 1996;804:1–10. doi: 10.1111/j.1749-6632.1996.tb18603.x. [DOI] [PubMed] [Google Scholar]
- Distel B, et al. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Elgersma HM, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Van den Berg M, Tabak HF, Distel B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics. 1993;135:731–740. doi: 10.1093/genetics/135.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Sluijs P, Distel B, Tabak HF. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- Ellis SB, Brust PF, Koutz PJ, Waters AF, Harpold MM, Gingeras TR. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol Cell Biol. 1985;5:1111–1121. doi: 10.1128/mcb.5.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M, Terlecky SR, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau WH, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Hajra AK, Bishop JE. Glycerolipid biosynthesis in peroxisomes via the acyl dihydroxyacetone phosphate pathway. Ann NY Acad Sci. 1982;386:170–182. doi: 10.1111/j.1749-6632.1982.tb21415.x. [DOI] [PubMed] [Google Scholar]
- Heyman JA, Monosov E, Subramani S. Role of the PAS1 gene of Pichia pastoris in peroxisome biogenesis. J Cell Biol. 1994;127:1259–1273. doi: 10.1083/jcb.127.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau WH. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James GL, Goldstein JL, Pathak RK, Anderson RG, Brown MS. PxF, a prenylated protein of peroxisomes. J Biol Chem. 1994;269:14182–14190. [PubMed] [Google Scholar]
- Kalish JE, Keller GA, Morrell JC, Mihalik SJ, Smith B, Cregg JM, Gould SJ. Characterization of a novel component of the peroxisomal protein import apparatus using fluorescent peroxisomal proteins. EMBO J. 1996;15:3275–3285. [PMC free article] [PubMed] [Google Scholar]
- Kalish JE, Theda C, Morrell JC, Berg JM, Gould SJ. Formation of the peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol. 1995;15:6406–6419. doi: 10.1128/mcb.15.11.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer S, Arnold N, Gutensohn W, Mewes HW, Kunau WH, Hofler G, Roscher AA, Braun A. Genomic organization and molecular characterization of a gene encoding HsPXF, a human peroxisomal farnesylated protein. Genomics. 1997;45:200–210. doi: 10.1006/geno.1997.4914. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, De Duve C. A fatty acyl-CoA oxidizing system in rat liver peroxisomes; enhancement by clofibrate, a hypolipidemic drug. Proc Natl Acad Sci USA. 1976;73:2043–2046. doi: 10.1073/pnas.73.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Moser HW. Disorders of peroxisome biogenesis. In: Scriver CR, Beaudet AL, Sly WS, editors. The Metabolic Basis of Inherited Disease. D. Valle, New York: McGraw-Hill; 1989. pp. 1479–1509. [Google Scholar]
- Liu H, Tan X, Russell KA, Veenhuis M, Cregg JM. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J Biol Chem. 1995;270:10940–10951. doi: 10.1074/jbc.270.18.10940. [DOI] [PubMed] [Google Scholar]
- McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun Y-L, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Identification of peroxisomal membrane ghosts with an epitope-tagged integral membrane protein in yeast mutants lacking peroxisomes. Yeast. 1995;11:1045–1060. doi: 10.1002/yea.320111106. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Albertini M, Kunau WH. Protein import into peroxisomes: new developments. Ann NY Acad Sci. 1996;804:34–46. doi: 10.1111/j.1749-6632.1996.tb18606.x. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears IB, O’Connor J, Rossanese OW, Glick BS. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–790. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Spong AP, Subramani S. Cloning and characterization of PAS5: a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Cell Biol. 1993;123:535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. PEX genes on the rise. Nat Genet. 1997;15:331–333. doi: 10.1038/ng0497-331. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Waterham HR, Veenhuis M, Cregg JM. The Hansenula polymorpha PER8 gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Krisans SK. Rat liver peroxisomes catalyze the initial step in cholesterol synthesis. The condensation of acetyl-CoA units into acetoacetyl-CoA. J Biol Chem. 1990;265:5731–5735. [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. The endoplasmic reticulum plays an essential role in peroxisome biogenesis. Trends Biochem Sci. 1998;23:231–233. doi: 10.1016/s0968-0004(98)01226-2. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE, Essner E. Microbodies: peroxisomes and glyoxysomes. J Cell Biol. 1981;91:271s–283s. doi: 10.1083/jcb.91.3.271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis MJ, Keizer I, Harder W. Characterization of peroxisomes in glucose-grown Hansenula polymorpha and their development after the transfer of cells into methanol-containing media. Arch Microbiol. 1979;120:167–175. [Google Scholar]
- Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ. Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet. 1998;63:347–359. doi: 10.1086/301963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, de Vries Y, Russel KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Luers G, Faber KN, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]