Abstract
At present little is known about the developmental mechanisms that give rise to inhibitory γ-aminobutyric acidergic interneurons of the neocortex or the timing of their subtype specification. As such, we performed a gene expression microarray analysis on cortical interneuron precursors isolated through their expression of a Dlx5/6Cre-IRES-EGFP transgene. We purified these precursors from the embryonic mouse neocortex at E13.5 and E15.5 by sorting of enhanced green fluorescent protein-expressing cells. We identified novel transcription factors, neuropeptides, and cell surface genes whose expression is highly enriched in embryonic cortical interneuron precursors. Our identification of many of the genes known to be selectively enriched within cortical interneurons validated the efficacy of our approach. Surprisingly, we find that subpopulations of migrating cortical interneurons express genes encoding for proteins characteristic of mature interneuron subtypes as early as E13.5. These results provide support for the idea that many of the genes characteristic of specific cortical interneuron subtypes are evident prior to their functional integration into cortical microcircuitry. They suggest interneurons are already relegated to specific genetic subtypes shortly after they become postmitotic. Moreover, our work has revealed that many of the genes expressed in cortical interneuron precursors have been independently linked to neurological disorders in both mice and humans
Keywords: cortical interneurons, subtype specification, transcriptional code
Introduction
The mammalian neocortex consists of 2 basic neuronal cell types: excitatory projection neurons, and inhibitory interneurons. The latter are typically γ-aminobutyric acidergic (GABAergic) and mediate local inhibition of other cortical neurons (Houser et al. 1983; DeFelipe and Farinas 1992). Although inhibitory interneurons comprise only about 20% of the total neuronal population in the cortex, they are crucial components of cortical circuits and their dysfunction is thought to contribute to a variety of neurological disorders such as epilepsy, autism, and schizophrenia (Baraban and Tallent 2004; Levitt et al. 2004; Cossart et al. 2005; Woo and Lu 2006). The exceptional heterogeneity in their morphological, biochemical, and functional properties (DeFelipe 1997; Kawaguchi and Kondo 2002; Markram et al. 2004; Somogyi and Klausberger 2005) likely reflects the complexity of regulating excitation in the cerebral cortex. The timing and molecular basis for the emergence of this diversity is largely unknown.
Two extreme theories have been posited to explain the emergence of cortical pyramidal cells specification and arealization. The tabula rasa view of this process is that the mature characteristics of cortical subtypes are imposed upon them by their neocortical environment, through cues such as neurotrophins and their synaptic partners (Marty et al. 1997; Kalisman et al. 2005; Sugino et al. 2006). The alternative point of view was invoked by the “protomap hypothesis” (Rakic 1988), which has lead to the idea that combinatorial gene expression during neurogenesis (Jessell 2000) initiates an intrinsic program that ultimately results in the emergence of specific neuronal subtypes. Within the neocortex, evidence for the latter point of view came from landmark studies where the fate of pyramidal cells was challenged through heterotopic transplantation. Through this work, it was found that the laminar fate of cortical pyramidal cells was determined shortly after these cells underwent their final round of DNA replication (McConnell 1988; McConnell and Kaznowski 1991). More recently genetic studies have discovered some of the molecular effectors that mediate these events; including the identification of specific transcription factors that control pyramidal cell fate and areal identity (Hamasaki et al. 2004; Chen, Schaevitz, et al. 2005; Chen, Rasin, et al. 2005; Molyneaux et al. 2005; Cholfin and Rubenstein 2007). The “protomap” and “tabula rasa” theories have been traditionally confined to pyramidal cells. We, however, feel that given the large contribution that cortical interneurons make to this structure the time has come to intergrate this critical class of cells into the existing framework. Our present study takes the first steps in initiating such an effort by attempting to understand how developmental events shape the generation of cortical interneuron diversity. Moreover, as with studies on pyramidal cells, this effort holds the promise of elucidating the genetic basis of developmental neurological disorders whose etiology we suggest results from a failure in this specification process.
To tackle this problem, it was necessary to devise a means to prospectively isolate cortical interneurons prior to their integration into the cortical plate. Cortical interneurons originate embryonically from the ganglionic eminences in the ventral telencephalon and migrate tangentially to populate the neocortex (Anderson, Eisenstat, et al. 1997; Wichterle et al. 2001; Nery et al. 2002). The ventral eminences are marked by the expression of specific transcription factors that are known to be essential for interneuron development. In particular, members of the Dlx gene family, which are expressed throughout the subpallial subventricular zone, have been shown to be critical for interneuron specification (Anderson, Qiu, et al. 1997; Pleasure et al. 2000; Petryniak et al. 2007). Mice containing compound Dlx1/Dlx2 mutations have a severe reduction in tangential migration of interneurons from the ventral eminences to the neocortex, resulting in a massive loss of neocortical GABAergic cells at birth (Anderson, Eisenstat, et al. 1997). Similarly, null mutations in Mash1, a proneural gene expressed in the ventral eminences (Casarosa et al. 1999), and Nkx2.1 (Sussel et al. 1999), a transcription factor expressed in the medial ganglionic eminence show a pronounced reduction of cortical interneurons and a concomitant reduction in Dlx5 expression.
These previous results suggest that the Dlx5 and Dlx6 genes provide an attractive means for the identification of precursors (i.e., immature but postmitotic cells) destined to give rise to cortical interneurons. To this end, we chose to utilize a Dlx5/6Cre-IRES-EGFP transgene (Stenman et al. 2003) to label and purify cortical interneuron precursors at embryonic ages. With this approach, we have identified genes that are enriched and/or highly expressed in embryonic interneuron precursors, many of which are involved in diverse biological functions such as transcription, cellular interaction, neurotransmission and network communication. These results suggest that similar to excitatory neurons (McConnell 1988; Rakic 1988; Chen, Schaevitz, et al. 2005; Chen, Rasin, et al. 2005; Molyneaux et al. 2005; Cholfin and Rubenstein 2007), interneuron specification is initiated prior to their integration into cortical circuitry. Moreover, we find that many of the genes expressed in cortical interneuron precursors are linked to specific neurological disorders.
Materials and Methods
Mouse Lines and Genotyping
All animal handling and maintenance were performed according to the regulations of the Institutional Animal Care and Use Committee of the NYU School of Medicine. The Dlx5/6Cre-IRES-EGFP (Stenman et al. 2003) and Z/EG (Novak et al. 2000) transgenic lines were maintained in the Swiss Webster background, and genotyped as previously described (Stenman et al. 2003; Novak et al. 2000).
Cortex Dissection and Fluorescent Activated Cell Sorting
The cortex was identified by its anatomical position and morphology. The cortex of E13.5 and E15.5 Dlx5/6Cre-IRES-EGFP embryos were dissected in cold Dulbecco's Modified Eagle's Medium (DMEM) and treated with 0.25% trypsin (Worthington, Lakewood, NJ) and DNase I (0.1%; Sigma, St. Louis, MO) at 37 °C for 5 min. Dissociated cells from 6 to 8 pooled embryos were used for fluorescent activated cell sorting (FACS) according to the respective brightness of enhanced green fluorescent protein (EGFP). For each sorting, we collected cells not expressing EGFP (EGFP− cells) and cells expressing EGFP (EGFP+ cells).
RNA Isolation and Microarray Hybridization
Total RNAs from FACS purified cells was prepared by the TRIzol method (Invitrogen, Eugene, OR). Purified RNA (200 ng) was amplified and biotinylated using MessageAmp II-Biotin Kit (Ambion, Austin, TX), and hybridized to microarrays MOE430A (Affymetrix, Santa Clara, CA). This procedure was repeated in triplicate for each sample to produce 3 independent data sets per RNA sample.
Microarray Expression Analysis
The success of the amplification and hybridization was assessed by all the parameters recommended by Affymetrix. We performed array triplicates for each one of our 4 populations (E13.5 EGFP+, E13.5 EGFP−, E15.5 EGFP+, and E15.5 EGFP−). In order to select for the genes that were specifically enriched in cortical interneuron precursors, we performed comparative analysis of the interneuron (EGFP+) and noninterneuron populations (EGFP−) for each of the time points (E13.5 and E15.5). Within each pairwise comparison, our statistical analysis and validation of the candidates is based on analysis of variance (P ≤ 0.001), t-test (P ≤ 0.001), and template matching within at least 2 of the arrays. Data processing was performed using MAS 5.0 software (Affymetrix) (Genomics Core Facility, MSKCC, NY). Signal value assigns a relative measure of abundance to the transcript. The detection algorithm uses probe pair intensities to generate a detection P value and assign a Present (P), or Absent (A) call. To determine the fold change, EGFP+ versus EGFP− GeneChip probe arrays were compared against each other in order to detect and quantify changes in gene expression. The analysis compares the difference of values of each probe pair in the baseline array (EGFP−) to its matching probe pair on the experiment array (EGFP+). The fold enrichment number is computed using a 1-step Tukey's Biweight method. We select the genes that have a Fold Enrichment ≥ 2 and P ≤ 0.001 for all of the triplicates.
Immunohistochemistry
Brains were fixed by transcardiac perfusion followed by 1 hour of postfixation on ice with 4% formaldehyde/phosphate buffered saline (PBS) solution. Brains were rinsed with PBS and cryoprotected by using 25% sucrose/PBS solution overnight at 4 °C. Cryosections were prepared at 12 μm (for E13.5 and E15.5) or 20 μm (for P30) thickness. Cryostat tissue sections were stained with the following primary antibodies: rabbit anti-green fluorescent protein (GFP) (1:2000; Chemicon), rat anti-GFP (1:1000; Nacalai Tesque, Kyoto, Japan), rat anti-somatostatin (SST) (1:500; Chemicon), rabbit anti-NPY (1:500; Immunostar), mouse anti-parvalbumin (PV) (1:1000; Sigma), rabbit anti-vasointestinal polypeptide (VIP) (1:500; Incstar), and mouse anti-calretinin (CR) (1:1500; Chemicon), rabbit α-GABA (1:1000; Sigma) rat anti-platelet-derived growth factor receptor (PDGFRα) (1:500; BD PharMingen, Franklin Lakes, NJ). Secondary antibodies, raised in donkey, used at 1:1000 (Alexa 594 and Alexa 488) were obtained from Invitrogen. Standard immunohistochemical staining procedures were used. Nuclear counterstaining was performed with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) solution in PBS for 20 min. Fluorescent images were captured using a cooled CCD camera (Princeton Scientific Instruments, Trenton, NJ) using MetaMorph software (Universal Imaging, Downingtown, PA). Molecular expression profiles of interneurons were characterized at postnatal day 30 (P30) in somatosensory barrel cortex. The basic strategy is as described previously (Butt et al. 2005). One brain was used to quantify the percentage each one of the interneuron subtypes PV (n = 325), SST (n = 309), CR (n = 362), NPY (n = 312), and VIP (n = 293).
In Situ Hybridization
Messenger RNA (mRNA) in situ hybridizations was performed as described previously (Wilkinson and Nieto 1993). RNA antisense probes were prepared by PCR and labeled with digoxigenin. Most RNA probes were generated by linearization of IMAGE clones from Open Biosystems (OB) with their respective enzymes, and RNA polymerases transcription was used to obtain antisense probe. Ssbp2 (OB clone ID: 3486988, linearization: EcoRV, transcription: T7 RNA polymerase); Carhsp1 (OB clone ID: 4166353, KpnI, T7 RNA polymerases); Nxph1 (OB clone ID: 7930348, NcoI, T7 RNA polymerase); Viaat (OB clone ID: 5686843, XmaI, T3 RNA polymerase); Gria1 (OB clone ID: 6842391, EcoRI, T3 RNA polymerase); Kcc2 (OB clone ID: 6838880, EcoR1, T3 RNA polymerase); Nkcc1 (OB clone ID: 4824556, HincIII , T3 RNA polymerase); Cacnb4 (OB clone ID: 4501980, KpnI, T7 RNA polymerase); Cck (OB clone ID: 1400830, XhoI, T3 RNA polymerase); Npy (Phage image clone: 482891, EcoRI, T3 RNA polymerase); Sst (Phage EST clone: 480070, EcoRI, T3 RNA polymerase). Some RNA probes were produced by PCR using specific primers: Satb1 (5′ primer: ACACAGCTC TGCTGCCCAAGCC; 3′ primer GACCAGCTGAGGACTG ATCGG); Pbx3 (5′ primer: CGCGG ATCCAAGCAGGACATCGGCGACATCC; 3′ primer CCCAAGCTTTGCAGCATAGA GATTGGCCTCC); Pou3f4 (5′ primer: CGCGGATCCAC TGG GTGACCAGTCTTAGCG; 3′ primer CCCAAGCTTATCTCCTGCGCTGCAGGCTTGG); Neud4 (5′ primer: TCTGGATGG AGAAGACCCACCG; 3′ primer TCGGGACAC CCAGTCTTCTTGG), Cux2 (5′ primer: TCCTCCAGCTACTCCGGACAGC; 3′ primer GGAGTATGTGTCCAGCTCTGG). Images were obtained by bright-field photography on a Zeiss Axioskop using Spot Advanced software.
Results
Genetic Fate Mapping of Dlx5/6 Expressing Cells
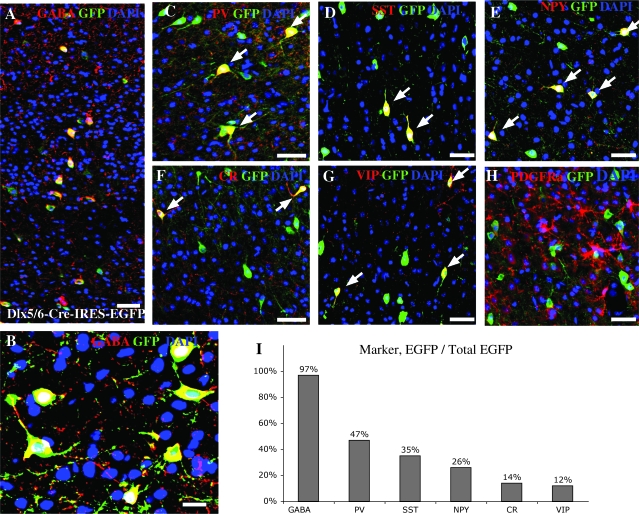
To confirm that the Dlx5/6Cre-IRES-EGFP driver we used (Stenman et al. 2003) is efficiently and selectively expressed within cortical interneuron precursors, we genetically fate mapped Dlx5/6 expressing cells within the adult cortex. We accomplished this by crossing the Dlx5/6Cre-IRES-EGFP transgenic mouse line (Stenman et al. 2003) to the Z/EG reporter line (Novak et al. 2000), whereby CRE activity removes a floxed-STOP cassette at the Z/EG locus, resulting in permanent EGFP expression. Previous work has suggested that Dlx genes are expressed in both interneuron and oligodendrocyte precursors (He et al. 2001; Marshall and Goldman, 2002), however colocalization immunological analysis of the cortex of Dlx5/6Cre-IRES-EGFP; Z/EG animals at P30 demonstrated that in this context we exclusively label cortical GABAergic interneurons (Fig. 1A,B,I), and not PDGFRα expressing oligodendrocytes (Fig. 1H). Moreover, with this strategy, we observed all the immunological characterized interneuron subtypes at the previously described frequencies, namely PV (47%), SST (35%), and CR (14%) (Wonders and Anderson 2006). We also quantified the percentage of 2 other interneuron markers, observing that 26% of the interneurons express NPY and 12% express VIP (Fig. 1C-I) (Martinez-Guijarro et al. 1998).
Figure 1.
Genetic fate mapping of Dlx5/6 expressing cells. Representative examples of sections for Dlx5/6 fate mapping. EGFP, DAPI, and various immunohistochemical markers on coronal sections of P30 brains. (A, B) show colocalization of GABA and EGFP in the vast majority of fate mapped cells. (C–G) Partial colocalization of EGFP and the interneuron subtype markers PV, SST, NPY, CR, and VIP signals. (H) No colocalization of GFP and PDGFRα signals was never observed. The frequency of cortical interneuron markers at P30 in mouse somatosensory barrel cortex PV, SST, NPY, CR, and VIP are calculated by comparing the number of cells that are double positive for EGFP and the markers examined as a ratio of the total number of EGFP cells (I). The histogram (I) summarizes the cell fate mapping in the Dlx5/6Cre-IRES-EGFP mice. Virtually all cells seen in the cortex are GABAergic. Notably the frequency and the distribution of various cell expressing immunomarkers shown is precisely consistent with that reported previously. Scale bars: (A) 35 μm, (B) 5 μm, (C–H) 35 μm.
Isolation of Embryonic Cortical Interneuron Precursors Using the Dlx5/6Cre-IRES-EGFP Transgenic Mouse Line
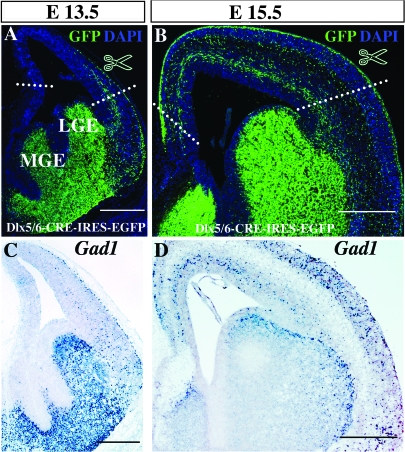
In order to identify novel genes that contribute to the embryonic generation and maturation of cortical interneuron subtypes, we have undertaken a genome-wide gene expression profile of interneuron precursors isolated from 2 different embryonic ages (E13.5 and E15.5). These ages span the early stages of interneuron precursor migration from the ventral telencephalon into the neocortex. We took advantage of a Dlx5/6Cre-IRES-EGFP transgenic mouse line, whose expression of EGFP from a minimal promoter activated by the Dlx5/6 enhancer allowed us to purify interneuron precursors that have migrated into the neocortex at embryonic stages. To obtain samples tissues, we dissected the cortices of transgenic embryos using the landmarks indicated in Figure 2A and B. By restricting our analysis to Dlx5/6 precursors in the cortex, we avoided the Dlx5/6 lineages that contribute to ventral structures, such as the striatum and olfactory bulb. Due to the robust expression of EGFP in interneuron precursors in these transgenic mice, it was possible to FACS cortical interneuron precursors (EGFP+ cells) from noninterneurons (EGFP− cells). Specifically, we collected 2.5% of the isolated EGFP+ E13.5 population, and 6.5% of the E15.5 EGFP+ cells (data not shown). We also collected non-EGFP expressing cells (EGFP− cells) from each time point for comparison. We detected no contamination in either the EGF+ or EGFP− populations (data not shown).
Figure 2.
Isolation of cortical interneuron precursors using the transgenic Dlx5/6Cre-IRES-EGFP mouse. EGFP immunohistochemistry on coronal sections of E13.5 and E15.5 brains. (A, B) The cortical region indicated was dissected, dissociated and FACS sorted at both ages (A) E13.5 and (B) E15.5. (C, D) In situ hybridization on similar cryosections using the Gad1 antisense probe. Note that within the cortex the distribution of the EGFP precursors shown in (A) and (B) closely resembles the pattern of Gad1 expression seen in (C) and (D). Scale bars: (A–D) 250 μm.
Microarray Analysis of Gene Expression in Cortical Interneuron Precursors
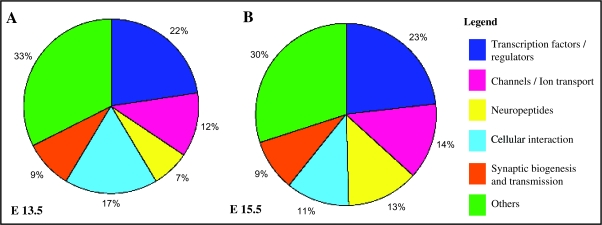
Validating our technical approach, we found that many of the genes known to be expressed in cortical interneurons (i.e., Gad1, Dlx5, Dlx1, Dlx2, Lhx6) are highly enriched in our data set (Fig. 2; Supplemental Tables 1 and 4). By contrast and as expected, we did not detect the expression of genes restricted to interneuron progenitors (such as Nkx2.1 and Nkx6.2). As these genes shut off very quickly within the ventral progenitor regions, they are no longer present in the interneuron precursors that have migrated into the embryonic neocortex. Similarly, we did not detect the expression of genes expressed in pyramidal cells (such as Ngn2, and Pax6), nor markers of oligodendrocyte lineages (such as Sox10 and Pdgfrα). In Figure 3, we show a summary of the classes of proteins encoded by the genes whose expression was upregulated by at least 2-fold in the EGFP+ population identified in this screen. With the exception of the cellular interaction and neuropeptide classes, there were not substantial differences between the 2 ages. The total number of genes with at least 2-fold upregulated expression at E13.5 was 416 and at E15.5 was 371.
Figure 3.
Classes of genes enriched in the interneuron population. The proportion of different classes of genes with assigned function, which are enriched in the interneuron population by at least 2-fold at E13.5 (total number genes: 416) (A) and E15.5 (total number genes: 371) (B). In this study we validated the expression of some interneuron enriched genes encoding transcription factors, neuropeptides, channels, and ion transport, and finally genes encoding cellular interaction molecules.
Transcription Factors: New Candidates for Interneuron Maturation and Subtype Determination
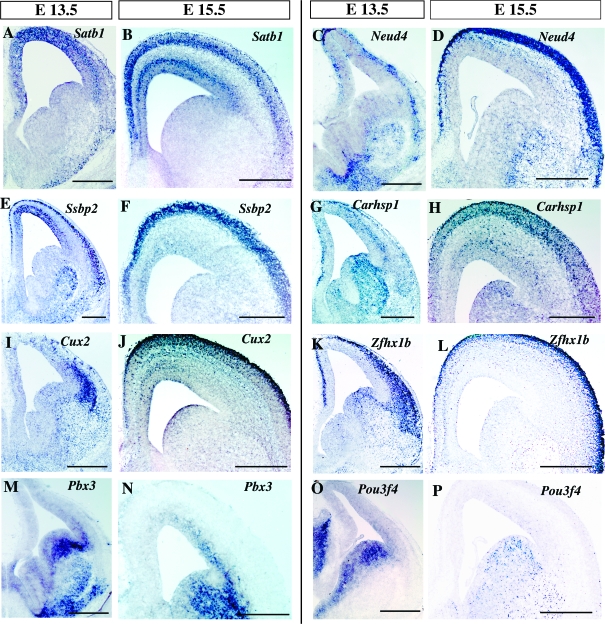
In our analysis, we identified a number of transcription factor mRNAs that were enriched in interneuron precursors (Supplementary Table 1). Some of these genes such as Mafb (Cobos et al. 2006; Wonders and Anderson 2006) are known to be expressed in cortical interneuron precursors , whereas others have yet to be characterized. Among the genes encoding for transcription factors identified, the expression of a subset of them was restricted to postmitotic interneurons (Satb1, Neud4, and Ssbp2) (Fig. 4A–F), whereas the expression of others such as Carhsp1, Cux2, Zfhx1b, Pbx3, and Pou3f4 was detected both within the ganglionic eminences, as well as migrating interneurons (Fig. 4G–P).
Figure 4.
Expression of genes encoding transcription factors. (A–F) Expression patterns of genes encoding transcription factors enriched in interneuron precursors at E13.5 and E15.5 revealed by in situ hybridization. Satb1, Neud4, and Ssbp2 were broadly expressed in postmitotic interneurons at both ages. (G–L) Carhsp1, Cux2, and Zfxh1 were strongly expressed in migrating interneurons and detected within the ganglionic eminences. (M–P) Pbx3 and Pou3f4 were more strongly expressed in the ganglionic eminences and were downregulated in interneurons as they migrated toward the cortex. Scale bars: (A–F) 250 μm.
Migrating Interneurons Express Subtype-Specific Neuropeptides
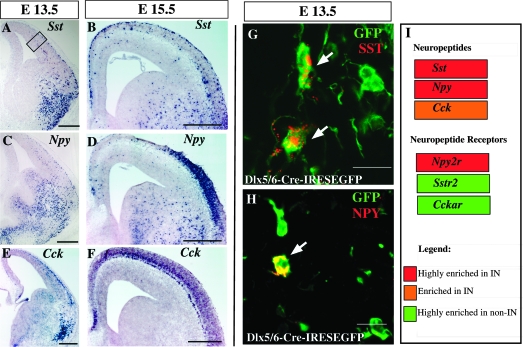
Neuropeptides and calcium-binding proteins provide some of the most robust markers for mature cortical interneuron subtypes. Surprisingly we found that some of these markers are already expressed in interneuron precursors by E13.5. Our microarray and in situ hybridization data revealed that the genes encoding the neuropeptides Sst, Npy, and Tac1 (Substance P) are exclusively expressed in cortical interneurons at both E13.5 and E15.5 (Fig. 5A–F Supplementary Table 2). We observed that many of the interneuron precursors invading the cortex already express SST and NPY (Fig. 5G,H). Similarly, Cck is enriched in interneuron precursors but expressed to a lesser extent in other cortical cells (Fig. 5I, Supplementary Table 2). Interestingly the genes encoding for the SST and CCK receptors (Sstr2 and Cckar, respectively) have a complementary expression pattern in the noninterneuron populations (Fig. 5, Supplementary Table 2), whereas Npy2r, the gene encoding the receptor for NPY, is only expressed in interneurons (Fig. 5I, Supplementary Table 2).
Figure 5.
Expression of neuropeptides. (A–F) In situ hybridization for Sst, Npy, and Cck and double immunostaining for (G) EGFP/ SST, and (H) EGFP/ NPY of Dlx5/6Cre-IRES-EGFP mouse shows that subpopulations of cortical interneuron precursors express SST and NPY (arrows). Panels (G) and (H) corresponds to the location of the box in (A). (I) Microarray fold enrichment (color code) in interneuron precursors for the neuropeptides Sst, Npy, Cck, and neuropeptide receptors Npy2r, Sstr2, and Cckar2. Scale bars: (A–F) 250 μm, (G–H) 10 μm.
Cellular Interaction Genes Enriched in Migratory Interneurons
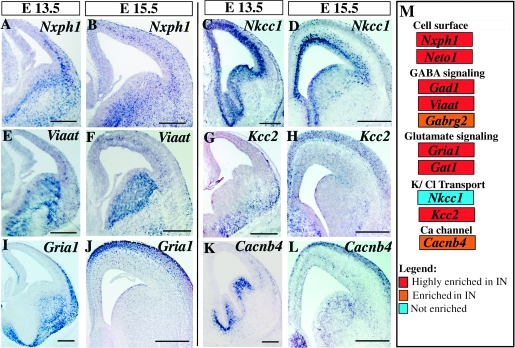
Cell surface molecules are involved in essential neurological processes, such as migration and synapse formation. In Supplementary Table 3 we present the genes involved in cellular interactions that are enriched in interneuron precursors. With the exception of Cxcr4, the genes we identified have not previously been implicated in cortical interneuron development. One of the highly expressed interneuron genes is Nxph1 (Fig. 6A,B), which encodes a secreted peptide that binds to α-neurexins. Although known to be expressed in the adult murine brain (Petrenko et al. 1996), its specific expression in interneurons during development was not recognized (Fig. 6A,B). We also detected that Neto I expression, a gene encoding a member of the neuropilin family is confined to interneurons. In contrast, we have detected that Nrp1 expression, another member of the same family, while present in interneurons, is more strongly expressed in noninterneuron populations (Supplementary Table 3). In addition, we identified a number of additional genes encoding cell surface proteins, such as Chl1, Dscaml, and Nlgn2 with enriched expression within cortical interneuron precursors (Supplementary Table 3).
Figure 6.
Expression of genes encoding cellular interaction molecules. In situ hybridization for Nxph1, Nkcc1 (C, D), Viaat (E, F), Kcc2 (G, H), Gria1 (I, J), and Cacnb4. (I) Microarray fold enrichment (color code) in the interneuron precursor population for select genes involved in cell surface interactions, GABA signaling, glutamate signaling, and ionic regulation. Scale bars: (A–L) 250 μm.
Migrating Interneurons Express Channels and Synaptic Machinery
Not surprisingly, many of the genes we identified are involved in GABA signaling and are enriched at GABAergic synapses. As expected the expression of Viaat, the gene encoding the GABA vesicular transporter (VGAT) was confined to the interneuron population. However we were surprised that its expression was observed as early as E13.5. Similarly, the GABA-A transporter (Gat1), and the GABA-A receptor (Gabrg2) genes were already expressed in embryonic interneuron precursors (Fig. 6M, Supplementary Table 4). Although the expression of the K/Cl cotransporter Nkcc1 was present at similar levels in both interneuron precursors and other cortical cells, Kcc2 expression was highly enriched in interneuron precursors (Fig. 6C,D,G,H,M, Supplementary Table 4). Conversely, there was an enrichment in the expression of genes involved in glutamate signaling and glutamatergic-synapses in the interneuron population. For instance, we detected the exclusive expression of the glutamate receptor gene Grik1 (also known as Glur5), as well as the enriched expression of both the ionotropic glutamate receptor gene AMPA1 (Gria1), and the gene encoding the glutamate receptor interacting protein 1 (Grip1) in interneuron precursors (Fig. 6I,J,M, Supplementary Table 4). Finally, we also found genes encoding multiple calcium, sodium, and potassium voltage-gated channels to be strongly expressed within interneuron precursor populations (Fig. 6K,L,M, Supplementary Table 4).
Migrating Interneurons Express Genes Implicated in Neurological Disorders
A variety of genes with enriched expression in cortical interneuron precursors have been linked to different neurological disorders. In Table 1 we show which of these genes have been implicated in specific human disorders, as well as the consequence of the dysfunction of these genes in mouse models. The expression data for these genes is listed in Supplementary Tables 1, 3, 4, and 5.
Table 1.
Genes expressed in developing interneurons linked to neurological disorders
| Gene function | Gene | Human |
Mouse |
||
| Disease | Ref. | Endophenotype | Ref. | ||
| Transcription factors/regulators | Npas3 | Schizophrenia | (Pickard et al. 2005; Pickard et al. 2006) | Impaired prepulse inhibition; Impaired social recognition. | (Erbel-Sieler et al. 2004) |
| Hdac11 | Autism | (Szatmari et al. 2007) | NA | ||
| Cited2 | Autism | (Szatmari et al. 2007) | NA | ||
| Meis1 | Autism | (Szatmari et al. 2007) | NA | ||
| Zfhx1b | Autism; Mental retardation; Epilepsy | (Inlow and Restifo 2004; Dastot-Le Moal et al. 2007; Hoffer et al. 2007) | NA | ||
| Arx | Epilepsy; mental retardation; lissencephaly | (Inlow and Restifo 2004; Kato et al. 2007) | Deficit in interneuron migration. | (Colombo et al. 2007) | |
| Cell surface | Ncam1 | Schizophrenia; Bipolar disorder | (Atz et al. 2007) | Abnormal interneurons; Impaired sensory gating and fear conditioning; Deficient prepulse inhibition; Impaired startle response. | (Pillai-Nair et al. 2005) (Plappert et al. 2005, 2006) |
| Chl1 | Mental retardation | (Frints et al. 2003) | Impaired prepulse inhibition; Impaired acoustic startle response | (Irintchev et al. 2004) | |
| Cntnap4 | Autism | (Sebat et al. 2007) | NA | ||
| Sema3a | Autism | (Szatmari et al. 2007) | NA | ||
| Channels/neurotransmission | Cacnb4 | Autism; epilepsy | (Escayg, De Waard, et al. 2000; Szatmari et al. 2007) | Ataxia; lethargy; seizures. | (Fletcher and Frankel 1999) (Burgess and Noebels 1999a, 1999b) |
| Cacng2 | Schizophrenia; epilepsy | (Sutrala et al. 2007) | Epileptic seizures | (Letts et al. 1998) | |
| Scn1a | Mental retardation; epilepsy | (Escayg, MacDonald, et al. 2000; Escayg et al. 2001; Inlow and Restifo 2004) | Epileptic seizures | (Ogiwara et al. 2007) | |
| Abat | Mental retardation; Autism; Schizophrenia | (Inlow and Restifo 2004: Barnby et al. 2005; Zhang et al. 2005) | NA | ||
| Kcnk2 | Autism | (Szatmari et al. 2007) | Kcnk2 deletion as an antidepression behavioral phenotype | (Heurteaux et al. 2006) | |
| Others | Dcx | Epilepsy; Mental retardation | (Reiner et al. 2006) | Deficient interneuron migration. | (Friocourt et al. 2007) |
| Man2a1 | Autism | (Szatmari et al. 2007) | NA | ||
| Nr2f2 | Autism | (Szatmari et al. 2007) | Defective tangential cell migration | (Tripodi et al. 2004) | |
| Shank3 | Autism | (Durand et al. 2007; Szatmari et al. 2007) | NA | ||
| Sez6l2 | Autism | (Szatmari et al. 2007) | NA | ||
| Dpp6 | Autism | (Szatmari et al. 2007) | NA | ||
| Centg2 | Autism | (Szatmari et al. 2007) (Wassink et al. 2005) | NA | ||
| Dtna | Autism | (Szatmari et al. 2007) | NA | ||
Discussion
Through a whole genome analysis of mRNAs expressed in embryonic cortical interneuron precursors, we have identified genes that are likely involved in the maturation of this population. Moreover the proteins encoded by these genes likely have functional significance in establishing cortical interneuron diversity. To our surprise, we found that as early as E13.5 cortical interneuron precursors express mature markers, such as neurotransmitters, channels, receptors, and synaptic machinery. This data suggests that as in pyramidal neurons (McConnell 1988; Rakic 1988; Chen, Schaevitz, et al. 2005; Chen, Rasin, et al. 2005; Molyneaux et al. 2005; Cholfin and Rubenstein 2007), interneuron subtype fate is restricted prior to their integration into cortex (Butt et al. 2005; Whichterle et al. 2001; Nery et al. 2001). Moreover, the observation that many of the genes identified are associated with developmental neurological disorders argue that disruption of cortical interneuron development contributes to the underlying etiology for a number of these diseases (Rodier 2000).
Transcription Factors
Acquisition of interneuron subtype identity, similar to the spinal cord, likely relies on the combinatorial expression of specific transcription factors (Flames et al. 2007). Moreover, the assignment of a particular neuron to a given subclass is likely the result of progressive restrictions beginning in the proliferative neuroblasts and subsequently refined (Jessell 2000; Fishell 2008). In our analysis, we have identified novel transcriptional activator/repressor pathways that likely direct the maturation of specific cortical interneuron subtypes. Although some are present in the majority of interneuron precursors, others exhibit a more restricted pattern of expression that may be indicative of a role in establishing subtype or regional specificity.
Of particular interest in cortical interneuron fate determination are global regulators of gene expression, such as Satb1, a gene that is known to be involved in chromatin remodeling and which modulates expression at loci (Alvarez et al. 2000; Cai et al. 2003, 2006) including the specific Dlx5/6 region (Horike et al. 2005). Similarly, both Ssbp2 and Neud4 appear to be broadly expressed in postmitotic interneurons, and are likely involved in general maturation. In addition, genes such as Cux2, Zfhx1b, Carhsp1 show robust expression in postmitotic neurons, but are also minimally expressed within the proliferative ventral eminences. Although Cux2 has been previously shown to be expressed in some migrating cortical interneuron precursors (Zimmer et al. 2004), the expression of Zfhx1b and Carhsp1 in interneurons was unknown. Our analysis also revealed a group of transcription factor genes including Pou3f (de Kok et al. 1995; Phippard et al. 1999) and Pbx3 that are mainly expressed in the ventral eminences. The Pbx genes encode a family of highly conserved homeodomain proteins, and it has been shown that PBX colocalizes in the ventral telencephalon with DLX early in development (Toresson et al. 2000). Given their restricted expression patterns, these genes represent good candidates for regulating the maturation, and diversification of cortical interneurons.
Migrating Interneuron Express Mature Subtype Interneuron Markers
Interneurons are typically subdivided into different classes according to their expression of calcium-binding proteins (PV, CR, and calbindin) and neuropeptides (SST, NPY, VIP, CCK, Substance P). Here we show that cortical interneuron precursors express some of these markers during embryonic development. Specifically, we detected the selective expression of genes encoding for Npy, Sst, Tac1 (Substance P), and Cck in the interneuron precursor population as early as E13.5, earlier than has been previously reported (De Belleroche et al. 1990; Morozov and Freund, 2003). Interestingly, in a complementary fashion, the expression of both the SST receptor gene (Sstr2), and the CCK receptor gene (Cckar) are enriched in noninterneuron populations. Although both of these receptors are expressed in mature pyramidal cells and are thought to modulate glutamatergic transmission (Gallopin et al. 2006), it was not previously appreciated that the genes encoding these receptors are expressed at embryonic stages. Furthermore, in contrast to CCK and SST, both NPY and the NPY receptor gene (Npy2r) were both exclusively expressed in immature interneurons and thus may function specifically within these caudal ganglionic eminence–derived subclasses (Butt et al. 2005). In addition to their role in functional signaling, these signaling peptides, as elsewhere in the central nervous system (Komuro and Rakic 1998), may also be utilized in directing the migration of this population.
Cellular Interactions
Cellular interactions play important roles during development, as well as in the maintenance and modification of synaptic functions in the adult. Cell adhesion molecules are known to play a role in interneuron migration (Metin et al. 2006). In this study, we have observed the expression of a number of genes encoding cell surface proteins, including Neto1, Kitl, Chl1, Dscaml Ncam, and Astn1, which were not previously known to be expressed in interneuron precursors. Neto1 shares some homology with the neuropilin family of cell surface receptor genes, and there are indications that like neuropilins, this receptor can bind to class 3 semaphorins (Kolodkin et al. 1997; Michishita et al. 2003). Class 3 semaphorins act as chemorepulsive signals for neuropilins, and mediate the directed migration of striatal and cortical interneurons, deriving from the ventral eminences (Marin et al. 2001).
Interestingly, our embryonic screen uncovered the expression of genes that are thought to be involved in the wiring of cortical circuits. For instance, we find that Nxph1, which encodes a secreted peptide that binds to α-neurexins, is highly expressed as early as E13.5. It has been shown that α-neurexins have a crucial role at both GABAergic and glutamatergic synapses, as α-neurexin knockout mice have reduced neurotransmitter release due to dysfunction of high-voltage activated Ca2+ channels (Missler et al. 2003; Beglopoulos et al. 2005). The observation that Nxph1 is specifically enriched in cortical interneuron precursors suggests a potential role in neurotransmitter release at very early stages of cortical development.
GABAergic Signaling and Synaptic Activity
During development, ambient GABA and glutamate release are known to control neuronal proliferation and migration (Cancedda et al. 2007; Manent and Represa 2007). Previous work on the development of hippocampal circuits has demonstrated that GABA synapses generally become functional before glutamatergic ones (Hennou et al. 2002). Consistent with this, by showing the strong expression of the VGAT gene (Viaat) by E13.5, and the preferential expression of genes encoding molecules involved in the synaptic machinery such as Synpr, Scamp, Vamp2, and Sv2a in developing interneurons, our data suggests that cortical interneurons are among the first neurons to participate in synaptogenesis in the neocortex. However, the lower expression of many of these genes in the noninterneuron population is also consistent with the fact that Cajal-Retzius cells (Mienville and Barker 1997), which are glutamatergic, also establish early synapses.
We find that both the GABAA transporter Gat1 and receptor gene Gabrg2 were enriched in interneuron populations by E13.5. In immature pyramidal cells, GABA is excitatory rather than inhibitory due to the higher intracellular concentration of Cl-, resulting from low perinatal expression of KCC2 (Gulyas et al. 2001; Ben-Ari 2002). However, during this same period, the Kcc2 gene is highly expressed and enriched in cortical interneuron precursors. This suggests that unlike immature pyramidal cells, interneuron precursors may exhibit an inhibitory response to GABA. In fact, this is the case in hippocampal interneurons, where during the first postnatal weeks GABAergic synapses onto interneurons have been shown to be inhibitory (Banke and McBain 2006).
Taken together our study identifies a plethora of novel genes involved in cortical interneuron development. Moreover, our work demonstrates that cortical interneurons express genes that confer physiological function much earlier than presently recognized. These observations suggest the integration of interneurons into cortical circuitry is initiated during embryogenesis. This in turn may provide an explanation for the growing body of evidence supporting a central role for cortical interneurons in the early activity-dependent remodeling of the cortex during the critical period (Hensch and Stryker 2004; Gandhi et al. 2005; Hensch 2005; Katagiri et al. 2007).
Interneuron Development and Neurological Disorders
Many of the genes enriched in migrating cortical interneurons have been implicated in a variety of human developmental and neurological diseases (Table 1). By cross-correlating genes known to be involved in such disorders with our data, we were able to identify disease-related genes encoding all classes of proteins that we found to be enriched in cortical interneuron precursors. These findings potentially implicate cortical interneurons as being causal in a broad swath of neurological disorders. In Table 1, we list some of the genes identified through this analysis.
Six of the genes enriched in cortical interneuronal precursors encoded transcription factors that were found to be to be linked to specific brain dysfunctions. Interestingly, some of these genes resulted in apparent brain abnormalities in genetically engineered mouse knockouts. Specifically, Npas3 and Npas1/3 compound mutants have behavioral deficits associated with mental disorders (Erbel-Sieler et al. 2004). Arx null mice have been implicated in autism and have an impairment in interneuron migration (Colombo et al. 2007).
A number of the genes encoding cell surface molecules were also associated with serious neurological diseases. For example, both Ncam1 and Chl1 have been linked to schizophrenia (Irintchev et al. 2004; Atz et al. 2007). Moreover, interneurons in transgenic mice lacking Ncam1 function have compromised synaptic connectivity and exhibit abnormal behavior (Pillai-Nair et al. 2005). Chl1 null mice have similar behavioral deficits, as well as axonal guidance (Montag-Sallaz et al. 2002; Irintchev et al. 2004). Similarly, disruption of genes encoding the specific channels that we found to be enriched in cortical interneuron precursors have also been associated with neurological disorders. Indeed, Cacnb4 mutations have been strongly linked to epilepsy in both mice and humans (Burgess and Noebels 1999a, 1999b; Fletcher and Frankel 1999; Escayg et al. 2000). In addition, a deficiency of 4-aminobutyrate animotransferase in humans, which is essential for catabolism of GABA, was found in our screen to be enriched in cortical interneuron precursors and has been implicated in mental retardation, autism, and schizophrenia (Inlow and Restifo 2004; Barnby et al. 2005; Zhang et al. 2005). Furthermore, neuroligins and their receptors neurexins have been implicated in autism (Ylisaukko-oja et al. 2005; Szatmari et al. 2007). Although not directly linked to this disorder, our microarray results show that both Nxph1 and Nlgn2 are enriched in cortical interneuron precursors, making these good candidates for autism related genes.
Although the association between neurological disorders and interneuron function is already substantial, our understanding of this connection is in its nascence. It seems likely that many of the genes identified in our screen will ultimately be causally implicated in the development, establishment and maintenance of interneurons in cortical circuitry. This in turn will no doubt help elucidate the functions of these genes in both normal and abnormal brain function.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
Research in the Fishell laboratory was supported by the Simons Foundation, the National Institutes of Health; National Institute of Mental Health; National Institute of Neurological Disorders and Stroke (R01 MH068469, R01NS039007). Science and Technology Foundation of Portugal grant to R. B. B.
Supplementary Material
Acknowledgments
We are thankful to members of the Fishell lab for helpful comments on this work. We are especially grateful to Elsa Rossignol for intellectual insights. We would like to thank Kenneth Campbell for providing us with the Dlx5/6Cre-IRES-EGFP transgenic line. We are thankful to Dr Quifu Ma, for giving us the necessary primers for RNA probe generation. We would like to in particular thank Melissa Mckenzie, as well as Elizabeth Maxwell, Alessandro de Camille, and Chika Sakamoto for excellent technical assistance with in situ hybridization and cryosectioning. Conflict of Interest: None declared.
References
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Atz ME, Rollins B, Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007;17:55–67. doi: 10.1097/YPG.0b013e328012d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, McBain CJ. GABAergic input onto CA3 hippocampal interneurons remains shunting throughout development. J Neurosci. 2006;26:11720–11725. doi: 10.1523/JNEUROSCI.2887-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraban SC, Tallent MK. Interneuron diversity series: interneuronal neuropeptides—endogenous regulators of neuronal excitability. Trends Neurosci. 2004;27:135–142. doi: 10.1016/j.tins.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglopoulos V, Montag-Sallaz M, Rohlmann A, Piechotta K, Ahmad M, Montag D, Missler M. Neurexophilin 3 is highly localized in cortical and cerebellar regions and is functionally important for sensorimotor gating and motor coordination. Mol Cell Biol. 2005;25:7278–7288. doi: 10.1128/MCB.25.16.7278-7288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y. Excitatory actions of GABA during development: the nature of the nurture. Nat Rev Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Burgess DL, Noebels JL. Single gene defects in mice: the role of voltage-dependent calcium channels in absence models. Epilepsy Res. 1999a;36:111–122. doi: 10.1016/s0920-1211(99)00045-5. [DOI] [PubMed] [Google Scholar]
- Burgess DL, Noebels JL. Voltage-dependent calcium channel mutations in neurological disease. Ann N Y Acad Sci. 1999b;868:199–212. doi: 10.1111/j.1749-6632.1999.tb11287.x. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cancedda L, Fiumelli H, Chen K, Poo MM. Excitatory GABA action is essential for morphological maturation of cortical neurons in vivo. J Neurosci. 2007;27:5224–5235. doi: 10.1523/JNEUROSCI.5169-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci USA. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl. 1):i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, Rubenstein JL, Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart R, Bernard C, Ben-Ari Y. Multiple facets of GABAergic neurons and synapses: multiple fates of GABA signalling in epilepsies. Trends Neurosci. 2005;28:108–115. doi: 10.1016/j.tins.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- De Belleroche J, Bandopadhyay R, King A, Malcolm AD, O'Brien K, Premi BP, Rashid A. Regional distribution of cholecystokinin messenger RNA in rat brain during development: quantitation and correlation with cholecystokinin immunoreactivity. Neuropeptides. 1990;15:201–212. doi: 10.1016/0143-4179(90)90014-p. [DOI] [PubMed] [Google Scholar]
- de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FP. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Farinas I. The pyramidal neuron of the cerebral cortex: morphological and chemical characteristics of the synaptic inputs. Prog Neurobiol. 1992;39:563–607. doi: 10.1016/0301-0082(92)90015-7. [DOI] [PubMed] [Google Scholar]
- Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA. 2004;101:13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, De Waard M, Lee DD, Bichet D, Wolf P, Mayer T, Johnston J, Baloh R, Sander T, Meisler MH. Coding and noncoding variation of the human calcium-channel beta4-subunit gene CACNB4 in patients with idiopathic generalized epilepsy and episodic ataxia. Am J Hum Genet. 2000;66:1531–1539. doi: 10.1086/302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, Heils A, MacDonald BT, Haug K, Sander T, Meisler MH. A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus–and prevalence of variants in patients with epilepsy. Am J Hum Genet. 2001;68:866–873. doi: 10.1086/319524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Fishell G. Perspectives on the developmental origins of cortical interneuron diversity. In: Bock G, Goode J, editors. Cortical development: genes and genetic abnormalities. Hoboken (NJ): Wiley; 2008. pp. 21–45. [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CF, Frankel WN. Ataxic mouse mutants and molecular mechanisms of absence epilepsy. Hum Mol Genet. 1999;8:1907–1912. doi: 10.1093/hmg/8.10.1907. [DOI] [PubMed] [Google Scholar]
- Frints SG, Marynen P, Hartmann D, Fryns JP, Steyaert J, Schachner M, Rolf B, Craessaerts K, Snellinx A, Hollanders K, et al. CALL interrupted in a patient with non-specific mental retardation: gene dosage-dependent alteration of murine brain development and behavior. Hum Mol Genet. 2003;12:1463–1474. doi: 10.1093/hmg/ddg165. [DOI] [PubMed] [Google Scholar]
- Friocourt G, Liu JS, Antypa M, Rakic S, Walsh CA, Parnavelas JG. Both doublecortin and doublecortin-like kinase play a role in cortical interneuron migration. J Neurosci. 2007;27:3875–3883. doi: 10.1523/JNEUROSCI.4530-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallopin T, Geoffroy H, Rossier J, Lambolez B. Cortical sources of CRF, NKB, and CCK and their effects on pyramidal cells in the neocortex. Cereb Cortex. 2006;16:1440–1452. doi: 10.1093/cercor/bhj081. [DOI] [PubMed] [Google Scholar]
- Gandhi SP, Cang J, Stryker MP. An eye-opening experience. Nat Neurosci. 2005;8:9–10. doi: 10.1038/nn0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas AI, Sik A, Payne JA, Kaila K, Freund TF. The KCl cotransporter, KCC2, is highly expressed in the vicinity of excitatory synapses in the rat hippocampus. Eur J Neurosci. 2001;13:2205–2217. doi: 10.1046/j.0953-816x.2001.01600.x. [DOI] [PubMed] [Google Scholar]
- Hamasaki T, Leingartner A, Ringstedt T, O'Leary DD. EMX2 regulates sizes and positioning of the primary sensory and motor areas in neocortex by direct specification of cortical progenitors. Neuron. 2004;43:359–372. doi: 10.1016/j.neuron.2004.07.016. [DOI] [PubMed] [Google Scholar]
- He W, Ingraham C, Rising L, Goderie S, Temple S. Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci. 2001;21:8854–8862. doi: 10.1523/JNEUROSCI.21-22-08854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennou S, Khalilov I, Diabira D, Ben-Ari Y, Gozlan H. Early sequential formation of functional GABA(A) and glutamatergic synapses on CA1 interneurons of the rat foetal hippocampus. Eur J Neurosci. 2002;16:197–208. doi: 10.1046/j.1460-9568.2002.02073.x. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- Hensch TK, Stryker MP. Columnar architecture sculpted by GABA circuits in developing cat visual cortex. Science. 2004;303:1678–1681. doi: 10.1126/science.1091031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurteaux C, Lucas G, Guy N, El Yacoubi M, Thummler S, Peng XD, Noble F, Blondeau N, Widmann C, Borsotto M, et al. Deletion of the background potassium channel TREK-1 results in a depression-resistant phenotype. Nat Neurosci. 2006;9:1134–1141. doi: 10.1038/nn1749. [DOI] [PubMed] [Google Scholar]
- Hoffer MJ, Hilhorst-Hofstee Y, Knijnenburg J, Hansson KB, Engelberts AC, Laan LA, Bakker E, Rosenberg C. A 6Mb deletion in band 2q22 due to a complex chromosome rearrangement associated with severe psychomotor retardation, microcephaly and distinctive dysmorphic facial features. Eur J Med Genet. 2007;50:149–154. doi: 10.1016/j.ejmg.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Houser CR, Hendry SH, Jones EG, Vaughn JE. Morphological diversity of immunocytochemically identified GABA neurons in the monkey sensory-motor cortex. J Neurocytol. 1983;12:617–638. doi: 10.1007/BF01181527. [DOI] [PubMed] [Google Scholar]
- Inlow JK, Restifo LL. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1534/genetics.166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irintchev A, Koch M, Needham LK, Maness P, Schachner M. Impairment of sensorimotor gating in mice deficient in the cell adhesion molecule L1 or its close homologue, CHL1. Brain Res. 2004;1029:131–134. doi: 10.1016/j.brainres.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kalisman N, Silberberg G, Markram H. The neocortical microcircuit as a tabula rasa. Proc Natl Acad Sci USA. 2005;102:880–885. doi: 10.1073/pnas.0407088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri H, Fagiolini M, Hensch TK. Optimization of somatic inhibition at critical period onset in mouse visual cortex. Neuron. 2007;53:805–812. doi: 10.1016/j.neuron.2007.02.026. [DOI] [PubMed] [Google Scholar]
- Kato M, Saitoh S, Kamei A, Shiraishi H, Ueda Y, Akasaka M, Tohyama J, Akasaka N, Hayasaka K. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome) Am J Hum Genet. 2007;81:361–366. doi: 10.1086/518903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kondo S. Parvalbumin, somatostatin and cholecystokinin as chemical markers for specific GABAergic interneuron types in the rat frontal cortex. J Neurocytol. 2002;31:277–287. doi: 10.1023/a:1024126110356. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Orchestration of neuronal migration by activity of ion channels, neurotransmitter receptors, and intracellular Ca2+ fluctuations. J Neurobiol. 1998;37:110–130. [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett FS, 2nd, Mori Y, Campbell KP, Frankel WN. The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet. 1998;19:340–347. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 2004;27:400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Manent JB, Represa A. Neurotransmitters and brain maturation: early paracrine actions of GABA and glutamate modulate neuronal migration. Neuroscientist. 2007;13:268–279. doi: 10.1177/1073858406298918. [DOI] [PubMed] [Google Scholar]
- Marin O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293:872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Marshall CA, Goldman JE. Subpallial dlx2-expressing cells give rise to astrocytes and oligodendrocytes in the cerebral cortex and white matter. J Neurosci. 2002;22:9821–9830. doi: 10.1523/JNEUROSCI.22-22-09821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guijarro FJ, Brinon JG, Blasco-Ibanez JM, Okazaki K, Hidaka H, Alonso JR. Neurocalcin-immunoreactive cells in the rat hippocampus are GABAergic interneurons. Hippocampus. 1998;8:2–23. doi: 10.1002/(SICI)1098-1063(1998)8:1<2::AID-HIPO2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Marty S, Berzaghi Mda P, Berninger B. Neurotrophins and activity-dependent plasticity of cortical interneurons. Trends Neurosci. 1997;20:198–202. doi: 10.1016/s0166-2236(96)01026-0. [DOI] [PubMed] [Google Scholar]
- McConnell SK. Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J Neurosci. 1988;8:945–974. doi: 10.1523/JNEUROSCI.08-03-00945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- Metin C, Baudoin JP, Rakic S, Parnavelas JG. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur J Neurosci. 2006;23:894–900. doi: 10.1111/j.1460-9568.2006.04630.x. [DOI] [PubMed] [Google Scholar]
- Michishita M, Ikeda T, Nakashiba T, Ogawa M, Tashiro K, Honjo T, Doi K, Itohara S, Endo S. A novel gene, Btcl1, encoding CUB and LDLa domains is expressed in restricted areas of mouse brain. Biochem Biophys Res Commun. 2003;306:680–686. doi: 10.1016/s0006-291x(03)01035-0. [DOI] [PubMed] [Google Scholar]
- Mienville JM, Barker JL. Potassium current expression during prenatal corticogenesis in the rat. Neuroscience. 1997;81:163–172. doi: 10.1016/s0306-4522(97)00171-1. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M, Schachner M, Montag D. Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol Cell Biol. 2002;22:7967–7981. doi: 10.1128/MCB.22.22.7967-7981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov YM, Freund TF. Postnatal development and migration of cholecystokinin-immunoreactive interneurons in rat hippocampus. Neuroscience. 2003;120:923–939. doi: 10.1016/s0306-4522(03)00409-3. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, Takeuchi T, Itohara S, Yanagawa Y, Obata K, et al. Na(v)1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko AG, Ullrich B, Missler M, Krasnoperov V, Rosahl TW, Sudhof TC. Structure and evolution of neurexophilin. J Neurosci. 1996;16:4360–4369. doi: 10.1523/JNEUROSCI.16-14-04360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippard D, Lu L, Lee D, Saunders JC, Crenshaw EB. Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J Neurosci. 1999;19:5980–5989. doi: 10.1523/JNEUROSCI.19-14-05980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Malloy MP, Porteous DJ, Blackwood DH, Muir WJ. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet. 2005;136:26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- Pickard BS, Pieper AA, Porteous DJ, Blackwood DH, Muir WJ. The NPAS3 gene—emerging evidence for a role in psychiatric illness. Ann Med. 2006;38:439–448. doi: 10.1080/07853890600946500. [DOI] [PubMed] [Google Scholar]
- Pillai-Nair N, Panicker AK, Rodriguiz RM, Gilmore KL, Demyanenko GP, Huang JZ, Wetsel WC, Maness PF. Neural cell adhesion molecule-secreting transgenic mice display abnormalities in GABAergic interneurons and alterations in behavior. J Neurosci. 2005;25:4659–4671. doi: 10.1523/JNEUROSCI.0565-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plappert CF, Schachner M, Pilz PK. Neural cell adhesion molecule-null mice are not deficient in prepulse inhibition of the startle response. Neuroreport. 2005;16:1009–1012. doi: 10.1097/00001756-200506210-00025. [DOI] [PubMed] [Google Scholar]
- Plappert CF, Schachner M, Pilz PK. Neural cell adhesion molecule (NCAM-/-) null mice show impaired sensitization of the startle response. Genes Brain Behav. 2006;5:46–52. doi: 10.1111/j.1601-183X.2005.00132.x. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Reiner O, Coquelle FM, Peter B, Levy T, Kaplan A, Sapir T, Orr I, Barkai N, Eichele G, Bergmann S. The evolving doublecortin (DCX) superfamily. BMC Genomics. 2006;7:188. doi: 10.1186/1471-2164-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM. The early origins of autism. Sci Am. 2000;282:56–63. doi: 10.1038/scientificamerican0200-56. [DOI] [PubMed] [Google Scholar]
- Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Sutrala SR, Goossens D, Williams NM, Heyrman L, Adolfsson R, Norton N, Buckland PR, Del-Favero J. Gene copy number variation in schizophrenia. Schizophr Res. 2007 doi: 10.1016/j.schres.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Szatmari P, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Filosa A, Armentano M, Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Vieland VJ, Jenkins L, Frantz R, Bartlett CW, Goedken R, Childress D, Spence MA, Smith M, et al. Evaluation of the chromosome 2q37.3 gene CENTG2 as an autism susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2005;136:36–44. doi: 10.1002/ajmg.b.30180. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Woo NH, Lu B. Regulation of cortical interneurons by neurotrophins: from development to cognitive disorders. Neuroscientist. 2006;12:43–56. doi: 10.1177/1073858405284360. [DOI] [PubMed] [Google Scholar]
- Ylisaukko-oja T, Rehnstrom K, Auranen M, Vanhala R, Alen R, Kempas E, Ellonen P, Turunen JA, Makkonen I, Riikonen R, et al. Analysis of four neuroligin genes as candidates for autism. Eur J Hum Genet. 2005;13:1285–1292. doi: 10.1038/sj.ejhg.5201474. [DOI] [PubMed] [Google Scholar]
- Zhang B, Yuan Y, Jia Y, Yu X, Xu Q, Shen Y, Shen Y. An association study between polymorphisms in five genes in glutamate and GABA pathway and paranoid schizophrenia. Eur Psychiatry. 2005;20:45–49. doi: 10.1016/j.eurpsy.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.