Abstract
Objectives
The transposon Tn1331 possesses a region including three antibiotic resistance genes with the structure aac(6′)-Ib-attC-aadA1-attI1*-blaOXA-9-attC, which potentially includes four gene cassettes. Experimental data on the mobility of fusion cassettes as well as those on mobility of cassettes in a genetic environment such as Tn1331, which lacks an integrase gene, are limited. Therefore, experiments using pJHCMW1, a plasmid harbouring this transposon, in the presence of IntI1 supplied in trans were carried out to define which cassettes are mobile in vivo.
Methods
In vivo excision of resistance genes was investigated in Escherichia coli cells harbouring pJHCMW1 and in a recombinant clone that included the intI1 gene under the control of the Ptac promoter. Plasmid DNA was purified and subjected to PCR analysis, and DNA sequencing of PCR products was performed to determine whether excision had occurred.
Results and conclusions
In vivo recombination experiments showed that the fused aadA1-attI1*-blaOXA-9-attC gene cassette was excised in the presence of IntI1. The excision of a DNA fragment including aadA1-attI1* was also detected but at a lower frequency. The analysis of the latter recombination reaction showed that, although attI1* includes only a small fraction of the complete attI1 sequence, it is still used as a substrate by IntI1, albeit in a very inefficient manner.
Keywords: integron, integrase, site-specific recombination
Introduction
Antibiotic resistance genes have the potential to reach most, if not all, bacterial cells through a combination of mechanisms that facilitate dissemination at the cellular and molecular levels. Some of these genes are found in structures known as gene cassettes, which consist of the structural gene and a target for site-specific recombination.1,2 These gene cassettes are usually part of the integrons, which include an integrase gene that mediates excision or integration of gene cassettes.3 This results in a highly versatile mechanism for the dissemination of antibiotic resistance genes at the molecular level. The spread of these resistance genes is enhanced when integrons are included within transposons and plasmids.
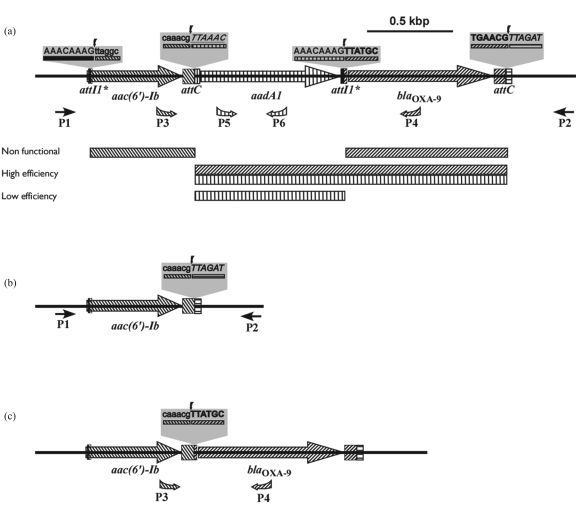
The transposon Tn1331, found in numerous multiresistance plasmids from Enterobacteriaceae including pJHCMW1,4–7 harbours a 2438 bp DNA fragment (coordinates 7302–9740, accession number AF479774) with the general structure of the variable region of class 1 integrons, but lacking the 3′- and 5′-conserved sequences.8 This region contains three resistance genes with the following genetic structure: aac(6′)-Ib-attC-aadA1-attI1*-blaOXA-9-attC (Figure 1a).8,9 In integrons, the gene located at the 5′ end of the variable region is preceded by an attI recombination site,3,10,11 which is located adjacent to the intI gene within the 5′-conserved region.12,13 In the case of Tn1331, attI is missing upstream of the aac(6′)-Ib gene (Figure 1a). Instead, an 8 bp sequence known as attI1* (AAACAAAG) is found at the beginning of the structural gene, at the location where a gene fusion between a blaTEM gene and a precursor of aac(6′)-Ib is believed to have occurred (Figure 1a).9,14 The complete attI1 site is 65 bp and includes the so-called ‘simple site’, which consists of a pair of IntI1-binding domains separated by a 7 bp spacer and two directly oriented IntI1-binding sites known as DR1 and DR2.13 The 8 bp attI1* site includes the 7 bp spacer (underlined earlier) and the nucleotide preceding it.13
Figure 1.
(a) Genetic map of the Tn1331 region including genes aac(6′)-Ib, aadA1 and blaOXA-9. The genes are shown in different patterns. Boxes with patterns represent attC loci and black boxes represent attI1* loci. The points of potential crossover reactions are indicated (downward arrows). Possible gene cassettes are indicated below the genetic map by bars of different patterns. The gene cassette including aadA1-attI1*-blaOXA-9-attC is identified by a double box with two patterns. The primers are indicated by arrows of different patterns. The genetic map is at scale, but the primers and boxes are not. (b) Genetic map of the product of excision of the aadA1-attI1*-blaOXA-9-attC gene cassette. (c) Genetic map of the product of excision of the aadA1-attI1* gene cassette.
At the 3′ end of the aadA1 gene, instead of the usual attC site (also known as 59 base element10), there is a copy of attI1* (Figure 1a), which may have been formed by an illegitimate recombination event between attC located 3′ of aadA1 of an integron and attI1 located 5′ of blaOXA-9 of another integron in which the blaOXA-9 gene cassette is adjacent to the 5′-conserved sequence.8,15,16
The Tn1331 2438 bp DNA region described in the preceding paragraphs has the potential to include four gene cassettes, assuming that the excision of a gene cassette requires at least one intact attC site at one boundary. Three of the potential gene cassettes include a single resistance gene, whereas the fourth includes aadA1 and blaOXA-9 (Figure 1a). However, the characteristics discussed earlier suggest that not all of them may be excised in the presence of the IntI1 integrase. To define which ones are mobile in vivo, we performed excision experiments in E. coli cells carrying pJHCMW1 and in a recombinant plasmid that included the intI1 gene under the control of the Ptac promoter.
Materials and methods
Bacterial strains and plasmids
E. coli TOP10 (Invitrogen) cells were used for all assays. Plasmid pJHCMW1 was originally isolated from Klebsiella pneumoniae JHCK1.4 Plasmid pLQ369 is a recombinant clone that includes the intI1 gene under the control of the Ptac promoter.17
In vivo recombination assays
Recombination assays to detect excision of resistance genes were performed using E. coli TOP10 cells harbouring pJHCMW1 and pLQ369, following the protocol described by Gravel et al.,17 with slight modifications. The assays were carried out by inducing the expression of the intI1 gene in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside followed by incubation of the cell suspension at 37°C for 3 h in Luria broth (0.5% yeast extract, 1.5% tryptone and 0.5% NaCl) with shaking. To determine whether recombination had occurred, plasmid DNA was extracted from the cells using the QIAspin miniprep kit (Qiagen) and used as template in the PCR analysis. PCR reactions were carried out using the QIAGEN Taq master mix, and the products were detected by agarose gel electrophoresis. PCR reactions were carried out using the primers shown in Table 1. Control assays were performed using E. coli TOP10 that carried only pJHCMW1.
Table 1.
Nucleotide sequences of the primers used for PCR
| Oligonucleotide | Sequence | Coordinates (accession number AF479774) |
|---|---|---|
| P1 | 5′-GCCTCGTGATACGCCTATTTT | 7111-7131 |
| P2 | 5′-GCCCTTCTGATGAAGCGTC | 10080-10061 |
| P3 | 5′-GCAAGGTACCGTAACCACCC | 7820-7839 |
| P4 | 5′-GCTGCGAGAACCAGACAACAG | 9168-9148 |
| P5 | 5′-TGCTGGCCGTACATTTG | 8057-8073 |
| P6 | 5′-TCATTGCGCTGCCATTC | 8321-8305 |
DNA sequencing
Prior to sequencing, the PCR products were cloned into the pCR2.1 vector as recommended by the supplier (Invitrogen). The recombinant clones were purified using the QIAspin miniprep kit (Qiagen) and used as templates for sequencing reactions. Sequencing was performed on both DNA strands using an ABIPrism 3100 BioAnalyzer and Taq FS Terminator Chemistry (Taq FS, Perkin-Elmer) at the Utah State University sequencing facility. Sequences were examined and assembled with Sequencher 4.7 software (Gene Codes Corp.).
Results and discussion
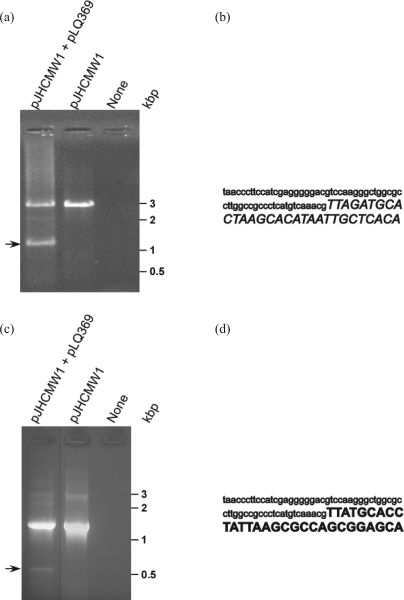
Plasmid DNA extracted from E. coli cells harbouring pJHCMW1 and pLQ369, after inducing expression of intI1, was used as a template in an amplification reaction with primers P1 and P2. Figure 2(a) shows the agarose gel electrophoresis analysis of the products of this reaction. There were two amplicons: one with the size expected of amplification of the region encompassing aac(6′)-Ib-attC-aadA1-attI1*-blaOXA-9-attC (Figure 1a), and the other had the size expected of a region lacking the aadA1 and blaOXA-9 genes (schematically shown in Figure 1b). The nature of the latter amplicon was confirmed by nucleotide sequencing. Figure 2(b) shows that the region encompassed by the IntI1 recombination site within the attC site located downstream of aac(6′)-Ib and that by the recombination site within the attC site located downstream of blaOXA-9 were missing from the template. Furthermore, the nucleotide sequence of this amplicon shows that the excision occurred at the expected sites for an IntI1-mediated site-specific recombination event between two attC sites (Figure 2a), a result consistent with the IntI1-mediated excision of a gene cassette consisting of aadA1-attI1*-blaOXA-9-attC.
Figure 2.
(a and c) Agarose gel electrophoresis of amplicons obtained using plasmid DNA extracted from cells cultured in the conditions indicated in the text as template and the primers P1 and P2 (a) or P3 and P4 (c). The 1191 bp fragment (arrow) in the gel of (a) corresponds to the amplicon obtained when excision of the fragment aadA1-attI1*-blaOXA-9-attC has occurred. The 568 bp fragment (arrow) in the gel in (c) corresponds to the amplicon obtained when excision of the fragment aadA1-attI1* has occurred. The line between the first and second lanes shows that they were not contiguous in the original gel. (b) Nucleotide sequence of the region surrounding the recombination site in the 1191 bp amplicon. (d) Nucleotide sequence of the region surrounding the recombination site in the 568 bp amplicon. The format of the fonts in the sequences shown in (b) and (d) corresponds to that in the genetic map shown in Figure 1(a).
Amplification reactions using primers P1 and P6 or P5 and P2 (Figure 1a) each showed a unique band consistent with intact pJHCMW1 DNA (data not shown), indicating that the excision of the putative gene cassettes aac(6′)-Ib-attC and blaOXA-9-attC was not detected in these experiments.
To detect the product of excision of the aadA1-attI1* gene cassette, an amplification reaction was carried out using the P3 and P4 primers (Figure 1a). Figure 2(c) shows that a faint band corresponding in size to a fragment amplified from a template missing the aadA1-attI1* gene cassette was detected (see a map detailing this excision in Figure 1c). The band intensity indicates that this template is present in a small fraction of the plasmid molecules. This experiment was repeated several times, and in all cases, the same size fragment was detected. This suggests that the excision between the attC locus located upstream of aadA1 and attI1* occurred infrequently. The fact that the deletion product is the result of IntI1-mediated site-specific recombination between attC and attI1* was confirmed by nucleotide sequencing (Figure 2d). As a consequence of the overloading to detect this band, we also observed an unidentified band of 3 kb that was also present in the control that lacked pLQ369. It was somewhat surprising to observe an IntI1-mediated recombination between attC and attI1*, which is a markedly truncated IntI1 recombination site. This site has been the subject of previous studies. There is a structure in Tn1404 that consists of the prototype aadB gene cassette,18 followed by an aadA10 gene cassette that contains attI1* instead of a complete attC.19 Recombination studies failed to detect IntI1-mediated deletion of the aadA10 cassette, suggesting that, in this environment, attI1* is either non-functional or so inefficient that PCR analysis was unable to detect the products.19 In the case of Tn1331, low but detectable recombination between attC and attI1* was consistently observed, demonstrating that attI* is functional.
In conclusion, our in vivo recombination experiments show that the fused gene cassette including aadA1-attI1*-blaOXA-9-attC and the imperfect gene cassette aadA1-attI1* can be excised. However, the excision efficiency of the latter is much lower. Conversely, no excision of the gene cassettes containing only aac(6′)-Ib or blaOXA-9 was observed. Although there are prototype attC sites downstream of both genes, the usual IntI1 target sequences located at the 5′ ends are missing. blaOXA-9 is preceded by attI1*, whereas in the case of aac(6′)-Ib, attI1* is located towards the beginning, but within the gene (Figure 1a). These results suggest that the attI1* locus does not serve as a substrate for IntI1-mediated recombination with attC when it is located 5′ of a gene. In contrast, it can serve as a substrate for the integrase when located 3′ of a gene, albeit inefficiently. As these experiments were designed to detect the excision of the gene cassette, the question of whether IntI1 also mediates the integration of a gene cassette with these characteristics remains unanswered.
The combination of gene exchange at the cellular and molecular levels results in the virtual elimination of barriers between bacteria, allowing antibiotic resistance genes to reach virtually all bacterial cells.20 IntI1-mediated excision and integration of gene cassettes play a crucial role in the mobilization of antibiotic resistance genes.3 Studies on structural21 and mechanistic17,22 aspects of the IntI1-mediated reaction will not only increase our understanding of the capture and spread of resistance traits, but also other genetic traits that may have an impact in genomic evolution.
Funding
This study was supported by the Public Health Service grant 2R15AI047115 (to M. E. T.) from the National Institutes of Health and BID 1728 OC/AR PICT 13431 (to D. C.). D. C. is a career member of CONICET. T. R. P. was supported by the Cal State Fullerton MARC U*STAR Program grant 2T34GM008612-12 from the National Institutes of Health. M. S. R. was supported in part by fellowships from the International Union of Microbiological Societies and CONICET (postdoctoral).
Transparency declarations
None to declare.
References
- 1.Hall RM, Brookes DE, Stokes HW. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–59. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 2.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 3.Bonomo R, Hujer A, Hujer K. Integrons and superintegrons. In: Bonomo R, Tolmasky ME, editors. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. Washington, DC: ASM Press; 2007. pp. 331–8. [Google Scholar]
- 4.Woloj M, Tolmasky ME, Roberts MC, et al. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 1986;29:315–9. doi: 10.1128/aac.29.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soler Bistue AJ, Birshan D, Tomaras AP, et al. Klebsiella pneumoniae multiresistance plasmid pMET1: similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS ONE. 2008;3:e1800. doi: 10.1371/journal.pone.0001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamorro RM, Actis LA, Crosa JH, et al. Dissemination of plasmid-mediated amikacin resistance among pathogenic Klebsiella pneumoniae. Medicina (B Aires) 1990;50:543–7. [PubMed] [Google Scholar]
- 7.Garcia DC, Woloj GM, Pineiro S, et al. An 8-year study of resistance to amikacin in Gram-negative bacilli isolates from patients with nosocomial infection at one hospital in Argentina. J Med Microbiol. 1995;42:283–90. doi: 10.1099/00222615-42-4-283. [DOI] [PubMed] [Google Scholar]
- 8.Sarno R, McGillivary G, Sherratt DJ, et al. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother. 2002;46:3422–7. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolmasky ME. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid. 1990;24:218–26. doi: 10.1016/0147-619x(90)90005-w. [DOI] [PubMed] [Google Scholar]
- 10.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–83. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 11.Rowe-Magnus DA, Mazel D. Integrons: natural tools for bacterial genome evolution. Curr Opin Microbiol. 2001;4:565–9. doi: 10.1016/s1369-5274(00)00252-6. [DOI] [PubMed] [Google Scholar]
- 12.Ouellette M, Roy PH. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 1987;15:10055. doi: 10.1093/nar/15.23.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partridge SR, Recchia GD, Scaramuzzi C, et al. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146:2855–64. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 14.Dery KJ, Soballe B, Witherspoon MS, et al. The aminoglycoside 6′-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell's cytoplasm. Antimicrob Agents Chemother. 2003;47:2897–902. doi: 10.1128/AAC.47.9.2897-2902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolmasky ME, Crosa JH. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 16.Tolmasky ME. Aminoglycoside-modifying enzymes: characteristics, localization, and dissemination. In: Bonomo R, Tolmasky ME, editors. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. Washington, DC: ASM Press; 2007. pp. 265–8. [Google Scholar]
- 17.Gravel A, Messier N, Roy PH. Point mutations in the integron integrase IntI1 that affect recombination and/or substrate recognition. J Bacteriol. 1998;180:5437–42. doi: 10.1128/jb.180.20.5437-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron FH, Groot Obbink DJ, Ackerman VP, et al. Nucleotide sequence of the AAD(2″) aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986;14:8625–35. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge SR, Collis CM, Hall RM. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob Agents Chemother. 2002;46:2400–8. doi: 10.1128/AAC.46.8.2400-2408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolmasky ME. Overview of dissemination mechanisms of gene coding for resistance to antibiotics. In: Bonomo R, Tolmasky ME, editors. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. Washington, DC: ASM Press; 2007. pp. 267–70. [Google Scholar]
- 21.MacDonald D, Demarre G, Bouvier M, et al. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–62. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 22.Gravel A, Fournier B, Roy PH. DNA complexes obtained with the integron integrase IntI1 at the attI1 site. Nucleic Acids Res. 1998;26:4347–55. doi: 10.1093/nar/26.19.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]