Abstract
Matrix metalloproteinase-20 (MMP-20, enamelysin) has a highly restricted pattern of expression. In healthy tissues, MMP-20 is observed in the enamel organ and pulp organ of developing teeth and is present only as an activated enzyme. To identify other tissues that may express MMP-20, we performed a systematic mouse tissue expression screen. Among the non-tooth tissues assayed, MMP-20 transcripts were only identified in minute quantities within the large intestine. The murine Mmp20 promoter was cloned, sequenced and assessed for potential tooth-specific regulatory elements. In silico analysis identified four promoter modules that were common to Mmp20 and at least two of three co-regulated predominantly tooth-specific genes that encode ameloblastin, amelogenin, and enamelin. We asked if the highly restricted MMP-20 expression pattern was associated with a broad substrate specificity that might preclude its expression in other tissues. An iterative mixture-based random doedecamer peptide library screen with Edman sequencing of MMP-20 cleavage products revealed that, among MMPs previously screened, MMP-20 had unique substrate preferences. These preferences indicate that MMP-20 has a deep and wide catalytic pocket that can accommodate substrates with large aromatic residues in the P1′ position. Based on matrices derived from the peptide library data, we identified and then confirmed that Type V collagen is an MMP-20 substrate. Since Type V collagen is not present in dental enamel but is an otherwise widely distributed collagen, and since only active MMP-20 has been observed in teeth, our data suggests that control of MMP-20 activity is primarily regulated by transcriptional means.
Dental enamel is unique because it is the only epithelially-derived calcified tissue in vertebrates and it is the hardest substance in the body. Five predominantly enamel-specific gene products are secreted by ameloblasts of the enamel organ into the developing enamel matrix. These gene products are amelogenin, ameloblastin, enamelin, matrix metalloproteninase-20 (MMP-20), and Kallikrein 4 (KLK-4). Of these five proteins, four are secreted during early enamel development as the enamel grows to full thickness and one, KLK-4, is secreted later in development as the thick enamel layer hardens (1). Both AMBN and ENAML localize to human chromosome 4 (2, 3) whereas MMP20 localizes to human chromosome 11q22 within the MMP gene cluster (4). The amelogenin gene is present only on the X chromosome in mice (Amelx), but the Y chromosome also carries an amelogenin gene (AMELY) in humans. In males, 90% of amelogenin transcripts are expressed from AMELX with only 10% expression from AMELY (5). AMELY does not compensate for loss of AMELX, and deletion of AMELY has no effect on teeth (6). Knockout mouse studies and/or human mutations have demonstrated that a loss of function in any one of the five enamel matrix proteins will cause diseased dental enamel termed amelogenesis imperfecta (7-13). Here we focus on MMP-20 by characterizing its tissue distribution, its promoter regulatory elements shared with co-regulated genes, and its substrate specificity.
Matrix metalloproteinase-20 (MMP-20) was initially named enamelysin because it was isolated by PCR-based homology cloning from a porcine enamel organ-specific cDNA library and Northern blot tissue analysis did not detect MMP-20 expression in other tissues (14). Later, ameloblasts of the enamel organ and odontoblasts of the dental papilla were demonstrated to express MMP-20 (15-19). Even so, MMP-20 expression proved to be highly restricted, especially when compared to other members of the MMP family. In one study, 51 different cell lines were assessed by RT-PCR for MMP-20 expression and none tested positive (20). MMP-20 expression has been observed in pathologic tissues, such as in ghost cells of calcifying odontogenic cysts (21), and odontogenic tumors (22, 23). MMP-20 expression was also observed in bradykinin treated granulosa cells isolated from the follicles of porcine ovaries (24). Nevertheless, among healthy tissues, MMP-20 is predominantly expressed within enamel organ and the dental papilla. The phenotype in mice devoid of MMP-20 activity (7) and humans with homozygous MMP20 mutation (11) supports the data demonstrating tooth-specific expression. MMP-20 null mice have thin brittle enamel that tends to delaminate from the underlying dentin and individuals with the human mutation have soft, pigmented enamel that is similarly fragile. No other phenotypes were observed. Therefore, to date, MMP-20 is considered a tooth-specific MMP.
Here we take a defined approach to assess the distribution of MMP-20 expression in the mouse. Although we did find that the large intestine expressed very low levels of MMP-20 that were undetectable by Northern blotting procedures, we consider MMP-20 to be an essentially tooth-specific matrix metalloproteinase. This conclusion lead us to ask two more questions: 1) Can we identify the promoter elements directing MMP-20 tooth-specific expression, and 2) does MMP-20 possess a broad substrate specificity that might preclude its expression in other healthy tissues?
To definitively answer the first question, a cell line was required that expresses all the factors necessary for MMP-20 transcription. Although two cell lines were reported to express MMP-20 (11), we found these cells expressed so little MMP-20 that it precluded their use for promoter analyses. However we did clone, sequence, and perform in depth in silico analysis of the murine MMP-20 promoter. The MMP-20 results were then compared with 3 other co-regulated genes (Amelx, Ambn, Enam) to identify transcription factor binding sites (TFBS) and/or promoter modules that likely play a role in tooth-specific gene expression.
To answer the second question we sought to characterize the active site-mediated substrate specificity of MMP-20. An iterative mixture-based peptide library method was performed (25). The approach involves treating peptide mixtures with MMP-20 and analyzing the product pool by amino terminal Edman sequencing, which provides an indication of the relative selectivity for each amino acid at any of several positions relative to the cleavage site. The same method was previously applied to seven other MMPs, which allows direct comparison of the cleavage selectivity of MMP-20 with related proteases (25, 26). The MMP-20 peptide library results were then used to generate a weight matrix that scores each amino acid at each position relative to the scissile bond (P4-P4′). This allowed us to search the protein sequence database with the goal of identifying new MMP-20 substrates. Based on results from the peptide library screen, a new MMP-20 substrate was identified (Type V collagen) that is widely distributed throughout the body and is present in bone and dentin (27, 28), but is not present in enamel (27).
Materials and Methods
Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was extracted from day-old mouse tissues with Trizol reagent (Invitrogen) and the Access RT-PCR system (Promega) was used to reverse transcribe 1 μg of total RNA from each tissue and to amplify the resulting cDNAs. For MMP-20, mouse 5′ PCR primers annealed to exon 9 and 3′ primers annealed to exon 10. PCR primers were: mMMP20 ex9 (sense) 5′-GGC CAT ATA GAT GCT GCT GTG GA-3′, MMP20 ex10 (antisense) 5′GAG GCC AGT AGG AGA CAA AGA GGA C-3′, F-ms-β-actin (sense) 5′-TCG TGC GTG ACA TCA AGG AGA AGC-3′, R-ms-β-actin (anitsense) 5′-CAG CAC TGT GTT GGC ATA GAG GTC-3′
Southern Blot Analysis of RT-PCR Results
Thirty μl of PCR product for each tissue analyzed by RT-PCR was loaded into an agarose gel, electorphoresed, transblotted onto a nylon membrane, and fixed to the membrane by incubation at 80 °C for 1 h. The Genius system (Boehringer Mannheim) was used to perform the Southern blotting procedures. In brief, primers nested to the RT-PCR primers were used in a PCR reaction to label 339 bp of MMP-20 cDNA (20 ng/ml) with Digoxigenin-11-dUTP (DIG). The primers were: F-South-MMP20 (sense) 5′-CTG GAT TGG CTG CTG AGT CGT-3′, R-South-MMP20 (antisense) 5′-TGA GTG CAC ATA GTC CTT TT TCT-3′. The probe was incubated overnight at 50 °C in hybridization buffer consisting of 5X SSC, 0.1% N-lauroylsarcosine, 0.2% SDS, and 1.0% Genius system blocking reagent. Identification of cDNA bands resulted after incubation with anti-DIG-alkaline phosphatase followed by color development with NBT and X-phosphate solution as stated in the manufacturer's instructions.
qPCR Analysis of MMP-20 Expression in Mouse Tissues
SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) generated the cDNA for Realtime PCR analysis. Primer sequence for MMP-20 expression analyses were: fMMP20-rltm (sense) 5′-CCT CTG GCC TTG CTG TCC TT-3′, rMMP20-rltm (antisense) 5′-CCT GGG GGC CTC CTT TCT-3′. Internal control primers for mouse β-actin were: fβ-actin-rltm (sense) 5′TCA AGA TCA TTG CTC CTC CTG AGC-3′, rβ-actin-rltm (antisense) TAC TCC TGC TTG CTG ATC CAC ATC-3′. The PCR reaction mix (SYBR Green Supermix, BioRad) was composed of 100 mM KCl, 40 mM Tris HCl (pH 8.4), 0.4 mM of each dNTP, iTaq DNA polymerase 50 units/ml, 6 mM MgCl2, SYBR Green I, 20 nM fluorescein, and stabilizers. PCR temperature profile was 3 min 95 °C initial melt then; 20 s 95 °C, 30 s 65 °C for 45 cycles then 30 s 95 °C, for 1 cycle; 1 min 55 degrees C followed by stepwise temperature increases from 55 °C to 95 °C to generate the melt curve. Standard curves were generated with the primer set by use of a 10-fold dilution series ranging from 1000 ng/ml to 100 pg/ml. PCR efficiencies and relative expression levels of MMP-20 as a function of β-actin expression were calculated as previously described (29).
Cloning and In Silico Analysis of Mmp20 5′-Flanking Region
The murine 129-strain genomic library cloned in the Lambda Fix II vector (Stratagene) was screened with a 441 bp MMP-20 cDNA probe to obtain genomic clones of MMP-20. The probe was DIG labeled (Boehringer Mannheim) as described above and plaque lifting, prehybridization, hybridization, washings of the filters, and identification of plaques with insert were performed according to the manufacture's instructions. Putative positive clones were purified by secondary and tertiary screening rounds. Genomic DNA inserts were subcloned into pZero-2 plasmid vectors (Invitrogen) for identification of insert size and sequencing.
The amelogenin, ameloblastin, enamelin, and MMP-20 promoters were assessed for common TFBS by Genomatix software applications in the region spanning 500 bp upstream of the first or only transcription start site. As explained in detail elsewhere (30), TFBS can be described by position weight matrices. The matrices are mined from the literature and the literature is cross-referenced to eliminate putatively erroneous TFBS from the matrices. This weight matrix pattern definition is superior to a fixed IUPAC consensus sequence because it represents the complete nucleotide distribution for each single position that defines the TFBS. With the nucleotide weight matrix, potential TFBS can also be quantified for their similarity to experimentally verified TFBS. The promoters were analyzed with these matrices by calculating individual conservation scores (Ci-value) for each nucleotide of a potential TFBS. The calculation is as follows
where P(i,b) is the relative frequency of nucleotide b at position i. This information is used by the MatInspector program to scan for nucleotide sequences that match patterns from a library. A matrix similarity score is then calculated which reaches 1 only if the test sequence corresponds to the most conserved nucleotide at each position of the matrix. The matrix similarity is calculated as follows
where Ci(j) is the Ci-value of position j, n is the length of the matrix, score (b,j) is the matrix value for base b at position j, and max_score is the maximum score within a matrix column at position j (for details see (31)).
Matrices for transcription factors (TFs) that bind to similar sites, as identified from different publications or research groups, are assigned to a family. Only the highest scoring, most conserved match to one of these matrices is used for further analysis. The grouping of matrices into families condenses the output because redundant matches among related TFs are eliminated. Once the individual TFBS in the promoters were found, the FrameWorker program identified modules present in at least 3 of the 4 co-regulated promoters. The modules consisted of at least two TFBS in common order, orientation, and distance from each other.
Primer Extension Analysis
Primer extension analysis to identify utilized transcription start sites was performed according to Maniatis (32). Briefly, the antisense primer (5′-CAG TAG GTT GGG GTC TGC AGT GGC AAA CTT T) hybridized to a region located 47 bp downstream of the translation start site. The primer was labeled with 32P-ATP and 105 cpm was hybridized with 10 ug of total RNA isolated from mouse unerupted first molars. Reverse transcription was performed followed by degradation of single strand RNA with RNase A. cDNA fragments were separated on a 6% polyacrylamide gel next to a sequencing reaction generated by use of the same primer.
Expression and Purification of Recombinant Human MMP-20 (rhMMP-20)
A 1466 bp cDNA fragment encompassing the entire hMMP-20 coding region was generated with PstI and was ligated in frame into the pRSET B E. coli expression vector (Invitrogen) as previously described (4). rhMMP-20 was expressed in E. coli as an N-terminal His6 fusion after induction with 0.1mM 1-thio-β-D-galactopyranoside followed by overnight incubation at 30 °C as described previously (4). rhMMP-20 was solubilized from inclusion bodies, and purified on a Ni column. The eluted protein was refolded by dialysis against 20 mM Tris/HCl (pH 7.5) 5mM CaCl2, 150 mM NaCl, 50 μM ZnCl2, and 0.01% NaN3 as describe previously (4) and the supernatant was concentrated 10 fold by use of a microcon YM-10 filter (Amicon, Inc). Enzymatically inactive control MMP-20 was prepared by point mutation of the glutamic acid at position 235 (within the HEXXH active site consensus) to an alanine (E235A).
Peptide Library Screen for MMP-20 Substrate Specificity
Analysis of MMP-20 cleavage specificity using peptide libraries was performed essentially as described previously (25) and is summarized below. To examine positions downstream of the cleavage site, we used an N-terminally acetylated random peptide dodecamer, Ac-XXXXXXXXXXXX, in which each position X is an equimolar mixture of the 19 naturally occurring amino acids excluding cysteine. The random peptide (1 mM) was incubated with rhMMP-20 (1.5 μg/ml) for 4 hr at 37°C in MMP reaction buffer (50 mM HEPES, pH 7.4, 200 mM NaCl, 10 mM CaCl2). A 10μl aliquot of this digest was loaded directly on an Applied Biosystems Procise 494 automated Edman sequencer. Because the N-termini of the substrate peptides are blocked, only the cleaved portion of the mixture is detected on the sequencer. Under these conditions, approximately 7.5% of the peptides are cleaved, meaning that 750 pmol of unblocked peptide is sequenced. On average, this corresponds to 40 pmol of each amino acid residue and is thus, as a pool, within the detection limits of standard Edman sequencing. Note that each individual peptide is present at quantities insufficient for detection; the method relies on analysis of the bulk properties of the substrate pool. The selectivity value for each residue was calculated by dividing the molar amount of the residue in a given sequencing cycle by the amount present in the starting mixture and then normalizing the data to an average value of 1. Preferences at the upstream positions were obtained using the peptide mixtures MAXXXXXLRGAARE(K-biotin) and MGXXPXXLRGGGEE(K-biotin). Peptides were digested with rhMMP-20 as above, and the reactions quenched by adding o-phenanthroline to a concentration of 20 mM. Digests (20 μl) were treated with 600 μl avidin agarose in 600 μl 50 mM NH4HCO3 with agitation for 1 hr at room temperature to remove unreacted peptides and C-terminal fragments. The slurry was transferred to a column and the unbound fraction was collected and pooled with 1.2 ml 50 mM NH4HCO3 wash. This fraction, containing the N-terminal fragments of the cleaved peptides, was evaporated to dryness under reduced pressure with centrifugation, suspended in 200 μl ddH2O and dried again. The residue was suspended in 30 μl ddH2O and applied to the Edman sequencer. Data were analyzed as described above. Identical reactions were run using the catalytically inactive E235A mutant of rhMMP-20, which did not detectably cleave any of the peptide libraries.
Collagen Digestion by MMP-20
Type III, IV, or V Collagen (250 μg/μl) from human placenta (Sigma) were each incubated with human recombinant MMP-20 (15 μg/μl) in a buffer containing 100 mM Tris/HCl pH 7.6, 200 mM NaCl, and 0.1% Brij-35 in a total volume of 200 μl. The reaction proceeded for 16 h at 37 °C and was stopped by the addition of EDTA at a final concentration of 20 mM. For SDS-PAGE analysis, sample loading buffer was added at a final concentration of 63 mM Tris/HCl, 2% SDS, 150 μl/ml glycerol, 0.05% (w/v) Bromphenol blue, and 50 μl/ml dithiothreitol. Samples (20 μl) were run on 10% SDS-PAGE gels and protein bands were stained with Coomassie Brilliant Blue.
RP-HPLC Fractionation of Type V Collagen
Human Type V collagen (Sigma) was fractionated by reversed phase-high performance liquid chromatography (RP-HPLC) using the Beckman System Gold HPLC system equipped with a Discovery C18 column (4.6 mm ID × 25 cm, Supelco, Bellefonte, PA). The column was equilibrated with 0.05% trifluoroacetic acid (buffer A) and eluted with a linear gradient of buffer B (0–100% B, over 50 min) containing 0.05% trifluoroacetic acid/80% acetonitrile at a flow rate of 1.0 ml/min at room temperature. Type V collagen eluted in the second, third and fourth peaks.
SDS-PAGE
SDS-PAGE was performed using. Novex 4-20% Tris-Glycine Gels (Invitrogen). Samples were dissolved in Laemmli sample buffer (Bio-Rad) and electrophoresis was carried out with a current of 20 mA for 1.5 h. The gels were stained with Bio-Safe Coomassie (Bio-Rad). The apparent molecular masses of protein bands were estimated by comparison with SeeBlue® Plus2 Pre-Stained standards (Invitrogen).
Western Immunoblots
The proteins on electrophoresed gels were electrotransferred onto Hybond-P membrane (Amersham Biosciences). Polyclonal antibody for human Type V collagen was used for 30 min at dilution of 1:40,000. Secondary antibody was HRP conjugated anti-rabbit IgG (Bio-Rad). The membranes were immunostained by the chemiluminescent detection with ECL Advance™ Western blotting detection kit (Amersham Biosciences).
Results
Analysis of Newborn Mouse Tissues for MMP-20 Expression
Day old mice were sacrificed and processed for extraction of total RNA. RNA from bladder, eye, large intestine, spleen, stomach, thymus, and whole mandibles containing developing tooth buds were assessed for MMP-20 expression by RT-PCR procedures. In addition, complete day-old mice that either did or did not have their mandible and maxillas removed were processed for RT-PCR analysis of MMP-20 expression. After 30 PCR cycles, agarose gel electrophoresis revealed that only the jaw containing the developing tooth buds had appreciable levels of MMP-20 transcripts (Figure 1, middle panel). Nested primers were designed to amplify and clone the PCR product into the pPCR-Script™ AMP SK(+) vector for use in Southern blot procedures. Southern blotting of the PCR products revealed additional bands. The whole mouse, but not the whole mouse without jaws, had a distinct and well-defined band the same size as that present in the jaw-only lane (Figure 1, top panel). Interestingly, the presence of a band in the large intestine lane indicated that MMP-20 was also expressed in that tissue. The procedures were repeated with different mice at a later date with similar results. In an attempt to verify MMP-20 expression in the large intestine, 30 μg of total RNA extracted from the large intestine was electrophoresed for Northern blot procedures. However, although transcripts were clearly present in the lane containing RNA from the jaw, we were unable to detect MMP-20 expression by this approach in the large intestine (not shown).
Figure 1.

Identification of newborn mouse tissues expressing MMP-20. Tissues were dissected from newborn mice and processed for extraction of total RNA. (A) Southern blot, using a 32P-labeled nested probe to confirm the tissue-specific MMP-20 RT-PCR results; (B) RT-PCR for identification of tissue-specific MMP-20 expression. This gel was processed for the Southern blot of panel A. (C) positive RT-PCR control demonstrating that each tissue expresses β-actin. Lane 1, negative control (no cDNA); lane 2, complete mouse pup with jaws removed (no teeth); lane 3, complete mouse pup; lane 4, jaw (with teeth); lane 5, stomach; lane 6, eye; lane 7, intestine; lane 8, bladder; lane 9, spleen; lane 10, thymus; lane 11, 100 bp DNA ladder. Note that 30 cycles of PCR yielded an MMP-20 band only in the jaw containing the tooth buds (lane 4), while Southern blotting of the PCR results demonstrated MMP-20 expression in RNA extracted from the complete mouse pup (lane 3) and intestine (lane 7).
To confirm that MMP-20 was expressed in the large intestine and to quantify any differences in expression level between tooth buds and large intestine, we performed quantitative real-time PCR (qPCR) according to the method of Pfaffl (29) using β-actin as the internal reference gene. The tissues assessed for MMP-20 expression were 4-day old first molar tooth buds plus adult brain, heart, kidney, liver, lung, pancreas, spleen, stomach, large intestine, and small intestine. Of the tissues assayed, only the tooth bud and the large intestine were positive for MMP-20 transcripts. However, the MMP-20 expression level observed in the large intestine was approximately 5000 times less than that observed in the 4-day old first molar tooth buds (Table 1). These results demonstrate that MMP-20 expression is primarily confined to tooth tissues and that MMP-20 is essentially a tooth-specific matrix metalloproteinase. To identify possible regulatory elements that direct tooth-specific expression, we cloned and characterized the murine MMP-20 promoter.
Table 1.
Quantitative real-time PCR analysis of MMP-20 expression levels in mouse tissues. Cycle threshold represents the number of PCR cycles necessary for the start of logarithmic cDNA expansion. Relative expression was calculated as a function of the expression level observed in the large intestine.
| Tissue Assayed | Cycle Threshold | Relative Expression |
|---|---|---|
| Large intestine | 34.67 | 1.00 |
| 3-day EO | 22.99 | 4.71 × 103 |
| Small Intestine | nd | - |
| Brain | nd | - |
| Heart | nd | - |
| Kidney | nd | - |
| Liver | nd | - |
| Lung | nd | - |
| Pancreas | nd | - |
| Spleen | nd | - |
| Stomach | nd | - |
Cloning of the Mouse MMP-20 Promoter
A 441 bp probe spanning from 376 bp 5′ to the translation start site and extending into exon 1 was DIG labeled and used to screen a mouse (129/SvJ) genomic DNA lambda fixII phage library. After three successive screening rounds, 6 clones were isolated, plaque purified, and assessed for the presence of probe sequence by PCR analysis. Inserts from 4 of the 6 clones were excised with Not-I and subcloned into pZero-2. The insert orientation and length estimation of 5′ noncoding region were assessed by PCR analysis. The 3′ primer originally used to make the probe for phage screening was paired with a primer specific for each vector arm. Agarose gel eletrophoresis of the PCR reactions revealed insert orientation and demonstrated that each 5′ non-coding region was between approximately 8-10 Kb in length. The clone with the longest length of 5′ non-coding region was sequenced. The entire genomic clone was 16,972 bp in length and was 99-100% identical to portions of the Mus musculus BAC clone RP23-338J1. The Mmp20 ATG translation initiation codon was located at position 10,007 from our 5′ clone end (GenBank accession DQ190458).
Identification of the Mmp20 Transcription Initiation Site
Primer extension was performed using a 32P-labeled primer (5′-CAG TAG GTT GGG GTC TGC AGT GGC AAA CTT T) that hybridized 47 bp 3′ of the ATG translation initiation codon. The labeled primer annealed to the genomic clone to generate the sequencing ladder and was also used on total RNA isolated from mouse enamel organ (Figure 2) to identify the transcription start site (TSS). To independently confirm the enamelysin TSS, the entire 16,972 bp genomic fragment was assessed for consensus start sites by in silico analysis http://www.fruitfly.org/seq_tools/promoter.html. As shown in Figure 2, good agreement exists between the experimental data and the computer generated data. Thus, the entire 16,972 bp genomic fragment contained 9,956 bp of enamelysin promoter upstream of the TSS, a 173 bp first exon which includes 50 bp of 3′ untranslated region and the translation start, the first intron of 6,762 bp, and the remaining sequence represented the first 80 bp of the second exon. Next, we asked if the enamelysin promoter contains consensus transcription factor binding sites (TSBS) that direct its tooth-specific expression.
Figure 2.
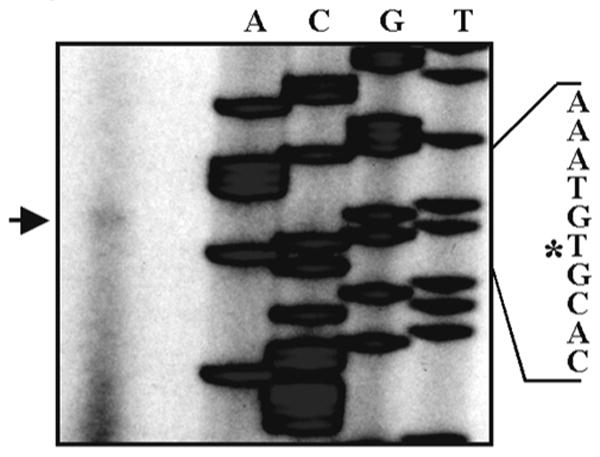
Location of the transcription start site in the murine MMP-20 gene. The transcription start was mapped by primer extension and in silico analysis. Each lane of the sequencing reaction was labeled with A, C, G, or T to identify the nucleotide sequenced within the genomic clone. The arrow designates the primer extension product. The asterisk designates the nucleotide position of the transcription start site that was identified by both primer extension and in silico analysis.
In Silico Analysis of the MMP-20 Promoter
No known cell line exists that expresses even moderate levels of MMP-20, so traditional promoter analyses were not possible. However, as a necessary first step, we performed in silico analysis to identify candidate TFBS that may direct predominantly tooth-specific gene expression. Enamelysin, amelogenin, ameloblastin, and enamelin are primarily expressed in the tooth and their promoters are co-regulated (15, 16, 19, 29, 33). The murine promoter sequences were analyzed for TFBS common to at least three of the four promoters. Twenty-nine common elements were identified, eleven of which were common to all four promoters. However, based on the pattern of binding sites detected, the sequences displayed no overall similarity. That is, the binding sites among the promoters were not conserved in number, orientation (location on plus or minus strand) and/or relative distance from the TSS. Overall, no conserved patterns of individual TFBS were detectable among the four promoters. However, sequence subsets containing conserved modules were present. A module contains at least two TFBS in conserved order and conserved distance from each other and may enhance or antagonize specific activation or repression of a gene (30). Interestingly, although Amelx localizes to the X chromosome and the Ambn is on mouse chromosome 5, they appeared to share several common patterns in their enhancer sequences. Both enhancers were enriched in binding sites for transcription factors related to developmental processes such as GATA, HOXF and CLOX. The MMP-20 enhancer by contrast contained very few of these binding sites. Pair-wise sequence analysis using FrameWorker software demonstrated that the amelogenin, ameloblastin, and enamelin promoters had more enhancer modules in common with each other when compared to the MMP-20 promoter (not shown). These data lend support to the suggestion that AMBN, AMELX, and ENAM have arisen from an ancestral enamel matrix protein gene and that AMELX was subsequently translocated (34). Nine promoter modules were identified that were common to MMP-20 and to at least two of the three remaining tooth-specific genes. Four of these modules were highly conserved with respect to their distance relative to the TSS and with respect to the distance between the two TFBS present in the module. The modules identified were HOMF/BRNF, BRNF/TBPF, HOXF/BRNF, and HOXF/HOXC (Figure 3, Table 2). The ameloblastin and MMP-20 BRNF binding site in the HOMF/BRNF module was the same BRNF site found in the BRNF/TBPF module (Figure 3, horizontal brackets). The two modules were unrelated in the enamelin promoter. The HOXF/BRNF module present in the amelogenin, enamelin, and MMP-20 promoters was independent of the two other modules containing a BRNF binding site. Interestingly, different forms of a HOXF/HOXC module were conserved either between amelogenin and MMP-20 or between enamelin and MMP-20 (Figure 3). Conformation that each of these modules plays a role in tooth-specific gene expression awaits the development of a suitable cell line containing the appropriate transcription factors for natural expression of ameloblastin, amelogenin, enamelin, and MMP-20. However, the conserved orientation and distance between TFBS in each module and the relatively conserved distance of each module from their respective transcription start site are unique promoter characteristics that suggest involvement in tooth-specific gene expression.
Figure 3.
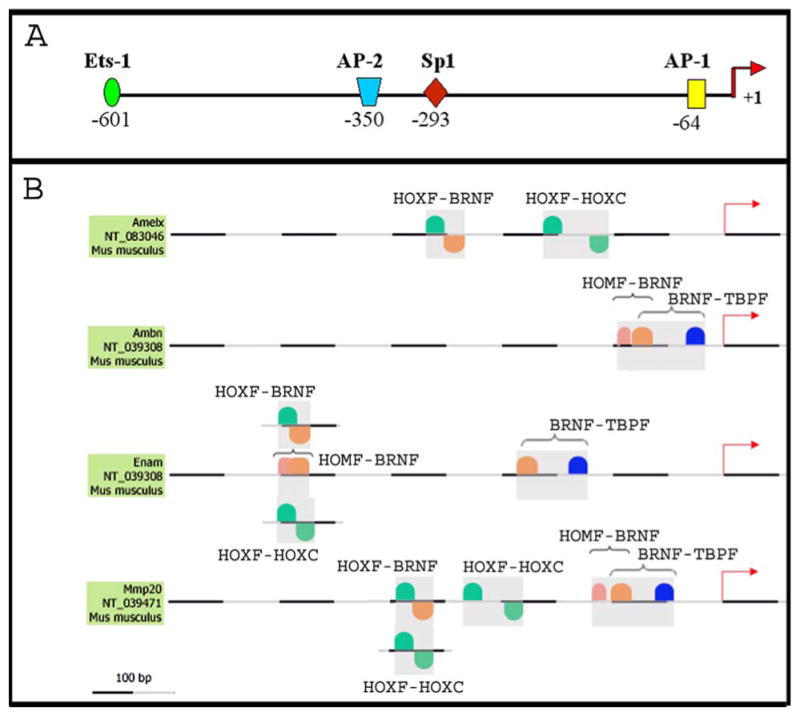
In silico analysis of the MMP-20 promoter and the co-regulated amelogenin, ameloblastin, and enamelin promoters. (A) MMP-20 promoter schematic displaying the identity and distance from the transcription start site of four transcription factor binding sites (TFBS) commonly observed in the MMP family of enzymes; (B) promoter analysis using FrameWorker software to identify shared promoter modules in co-regulated tooth-specific genes. Modules contain two or more TFBS and were identified based on their probability of binding individual transcription factors and based on each module's relative lack of abundance within the genome. Nine modules each with two TFBS were identified among the co-regulated genes and four of these were deemed likely to direct tooth-specific expression. Three of the four promoters contained each of the four identified modules (see Table 2). GenBank accession numbers are listed beneath the name of each promoter.
Table 2.
MatInspector promoter analysis of modules present in three of the four co-regulated tooth-specific genes. Modules denote the family of transcription factors with members that bind to the core binding site. TFBS nucleotides in capitol letters denote the core sequence. Lower case nucleotides in bold and underlined denote base pairs that are observed in the TFBS at an occurrence of greater than 60%. TFBS, transcription factor binding site; TSS, transcription start site.
| Ambn | Amelx | Enam | Mmp20 | |
|---|---|---|---|---|
| Module 1 | HOMF/BRNF | HOMF/BRNF | HOMF/BRNF | |
| TFBS | TAATt/gCACAttttt | TAATt/atagtTAAT | TAATt/acatgaACTAt | |
| Strand Specificity | +/+ | +/+ | +/+ | |
| bp Separating TFBS | 16 | 12 | 20 | |
| bp from TSS | 64 | 375 | 83 | |
| Module 2 | BRNF/TPBF | BRNF/TPBF | BRNF/TPBF | |
| TFBS | gCACAttttt/ttTAAA | atagaTAAT/aTAAA | acatgaACTAt/aTAAAa | |
| Strand Specificity | +/+ | +/+ | +/+ | |
| bp Separating TFBS | 48 | 46 | 39 | |
| bp from TSS | 18 | 124 | 45 | |
| Module 3 | HOXF/BRNF | HOXF/BRNF | HOXF/HOXF/BRNF | |
| TFBS | TAAA/tatATTT | TAAT/atagtTAAT | TAAA/cTAAT/tttATTT | |
| Strand Specificity | +/- | +/- | +/+/- | |
| bp Separating TFBS | 17 | 11 | 42, 16 | |
| bp from TSS | 236 | 375 | 262 | |
| Module 4 | HOXF/HOXC | HOXF/HOXC | HOXF/HOXF/HOXC & HOXF/HOXC | |
| TFBS | TTAATc/aTGATtttt | TAAT/TGATatat | TAAA/cTAAT/tTGATttat & TAAT/gtGATTgac | |
| Strand Specificity | +/- | +/- | +/+/- & +/- | |
| bp Separating TFBS | 42 | 17 | 44, 37 & 18 | |
| bp from TSS | 106 | 370 | 261 & 181 |
Substrate Specificity of MMP-20
Mixture-based oriented peptide libraries (25) were used to assess MMP-20 cleavage site specificity. We started with a random dodecamer library, which was used to obtain the substrate preferences for the positions downstream of the cleavage site. The relative amounts of amino acid found at the four positions immediately C-terminal to the cleavage site are shown in Figure 4. As is typical for MMPs, MMP-20 is highly selective for hydrophobic residues at the P1′ position. The preferred amino acid at P1′ was leucine, but MMP-20 also strongly selected methionine and tyrosine to a comparable extent. While the so-called deep pocket MMPs like MMP-3 can accommodate aromatic residues such as tyrosine at the P1′ position, aliphatic residues are consistently selected over aromatic ones (25, 35). Thus, MMP-20 is unique in the extent to which it selects aromatic residues in the P1′ position. MMP-20 was relatively non-selective at the P2′ position and had a slight preference for smaller residues at P3′, which has been observed with some but not all MMPs (25, 26, 35-37).
Figure 4.

Cleavage site specificity of MMP-20. The graphs on the right show the relative distribution of amino acid residues at positions C-terminal (P1′-P4′) to the MMP-20 cleavage site. These sites were identified by sequencing an N-terminally acetylated random peptide dodecamer (Ac-XXXXXXXXXXXX). Data were normalized so that a value of 1 corresponded to the average quantity per amino acid in a given sequencing cycle and would indicate no selectivity. Tryptophan was not included in the analysis because of poor yield during sequencing. The figures on the left represent the positions amino terminal to the MMP-20 cleavage site. For the P3 position data shown were generated using the library MAXXXXXLRGAARE(K-biotin). For all other positions the P3 proline library MGXXPXXLRGGGEE(K-biotin) was used. Glutamine and threonine were omitted in some cycles because of high background on the sequencer. Data were normalized as for the primed sites.
In order to determine the cleavage specificity at sites N-terminal to the cleavage site, we used two different peptide mixtures of reduced complexity. Initially the library MAXXXXXLRGAARE(K-biotin) was used, which includes several fixed residues intended to direct MMP cleavage to the X-L bond. As demonstrated for other MMPs, data from this library indicated a preference for proline at the P3 position, but also revealed an unusually strong preference for alanine (Figure 4). Specificity at the remaining upstream positions was determined using the library MGXXPXXLRGGGEE(K-biotin) in a similar manner. Fixing a proline residue at P3 directed a greater proportion of the cleavage to the intended site and thus increased the signal on the Edman sequencer relative to background for cycles corresponding to the less selective upstream positions. In contrast to most MMPs that prefer hydrophobic residues (25, 35, 36, 38), MMP-20 displayed an unusual selection for histidine at the P2 position.
To confirm that the peptide library results accurately reflect the true MMP-20 substrate specificity, we compared these results to previously identified MMP-20 cleavage sites present in the most abundant enamel matrix protein amelogenin. Previously, recombinant porcine MMP-20 was incubated with recombinant porcine amelogenin and the resulting cleavage products were identified by mass spectrometry (39). Presented in Table 3 are the preferred amino acids for positions P3-P3′ that direct hydrolysis at the scissile bond as identified by the peptide library screen (upper table). Also presented in Table 3 are the MMP-20 generated amelogenin cleavage sites that were previously identified by mass spectrometry (lower table). Five of the seven sites have a hydrophobic residue at P1′ (most have leucine), which supports this position as the most selective for MMP-20. The two sites that lack a larger hydrophobic residue at this position have a highly selected proline at the P3 position. This presumably compensates for the lack of a large hydrophobic residue at P1′. Though none of the cleavage sites within amelogenin closely match the consensus as defined using peptide libraries at all positions, inspection of the full amelogenin primary sequence revealed no potential sites that appeared more optimal than the authentic cleavage sites.
Table 3.
Comparison of MMP-20 peptide library cleavage site selectivity with cleavage sites previously demonstrated to occur in amelogenin. (Upper table) data generated by incubation of enamelysin with mixture-based oriented peptide libraries (see Figure 4) followed by amino acid sequencing. Numbers in the top row are preceded by a “P” to indicate the position of an amino acid relative to the scissile bond between P1 and P1′. Numbers in parenthesizes represent the selectivity value as defined in Figure 4. (Lower table) previously generated data identifying MMP-20 cleavage sites in porcine amelogenin of 173 amino acids in length (38). Numbers in the first column identify the new amelogenin C-terminus after cleavage by MMP-20. Amino acids in bold and underlined were selected by the mixture-based oriented library as promoting hydrolysis of the scissile bond.
| Amino Acid Position | P3 | P2 | P1 | P1′ | P2′ | P3′ |
|---|---|---|---|---|---|---|
| A (2.4) | H (2.5) | S (3.2) | L (4.1) | R (1.6) | A (2.1) | |
| P (2.0) | Y (1.8) | Q (1.8) | M (3.1) | T (1.6) | S (2.0) | |
| V (1.4) | Q (1.5) | N (1.7) | Y (3.3) | V (1.4) | T (1.7) | |
| S (1.4) | I (2.1) | |||||
| Demonstrated Enamelysin Cleavage Sites for Porcine Amelogenin | ||||||
| 148 | M | F | S | M | Q | S |
| 147 | P | M | F | S | M | Q |
| 136 | I | Q | P | L | L | P |
| 107 | P | L | P | A | Q | Q |
| 105 | N | L | P | L | P | A |
| 63 | S | H | A | L | Q | P |
| 45 | G | G | W | L | H | H |
Prediction and Identification of a Novel MMP-20 Substrate
We identified several novel potential substrates by searching the SWIS-PROT database using the Scansite program (40) with matrices derived from the MMP-20 library data. Among the substrates identified were collagens III and V. MMP-20 was predicted to cleave the collagen V alpha 1 chain precursor (GPRG-LLGPK), the collagen V alpha 3 chain precursor (PRLG-LQGP), and the collagen III alpha 1 chain precursor (GPQG-LQGL). Recombinant human MMP-20 was assessed for its ability to cleave Type III, IV, or V collagens. Type IV collagen was previously demonstrated as an MMP-20 substrate (41) so it served as the positive control. SDS-PAGE analysis demonstrated that Type V, but not Type III collagen, was cleaved by MMP-20 (Figure 5, top panel). The Type V collagen results were repeated with three more MMP-20 digestions to verify the original result (Figure 5, bottom panel).
Figure 5.
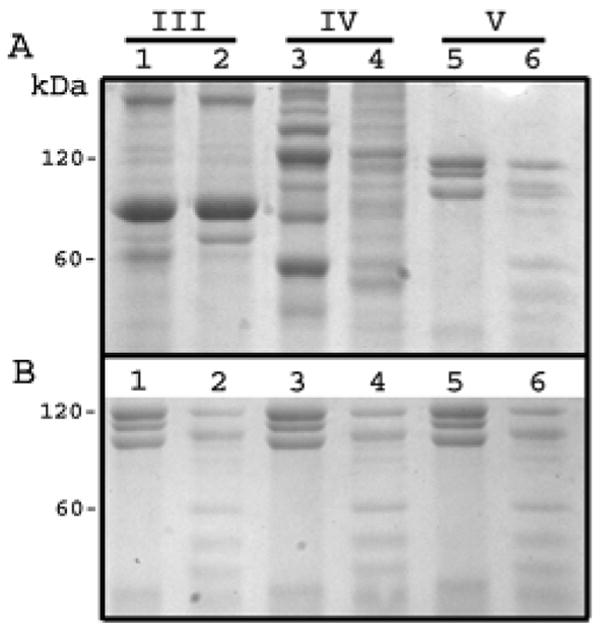
Hydrolysis of Type V but not Type III collagen by MMP-20. SDS PAGE analysis of potential MMP-20 collagen substrates. Odd numbered lanes, no enzyme added. Even numbered lanes, collagens (250 μg/ml) were incubated with MMP-20 (15 μg/ml) at 37°C for 16 h followed by Coomassie Brilliant Blue staining; (A) Type III, IV, and V collagens were electrophoresised in lanes 1-2, 3-4, and 5-6 respectively. Type IV collagen served as the positive control. Although type III collagen was not cleaved by MMP-20, it appeared that Type V was cleaved. To confirm that the Type V collagen cleavage results in panel A were not a result of protein under-loading or artifact; (B) the same experiment was repeated with the same procedure on three more Type V controls and corresponding enzyme treated samples. All three sample sets confirmed the results shown in panel A.
To confirm that MMP-20 cleaves Type V collagen and not a contaminate present within the collagen preparation, the Type V collagen preparation was divided into four fractions (Figure 6, top panel) by reversed-phase high-pressure liquid chromatography (RP-HPLC). Each fraction was electrophoresed on a 4-20% SDS-PAGE gel where the protein was either stained or underwent Western blotting procedures with a collagen Type V-specific antibody. The first protein fraction was identified as contaminate(s) because it did not stain with Coomassie Brilliant Blue and was not recognized by the Type V collagen antibody (Figure 6, bottom panel). The remaining three fractions, however, did contain Type V collagen. Fraction 3 appeared to be an intermediate comprised of proteins from both fractions 2 and 4 (Figure 6, bottom panel). Therefore only fractions 2 and 4 were selected for further analysis. Initially, a blank run and fractionation of just rhMMP-20 was performed to demonstrate the absence of contaminates and to demonstrate that MMP-20 was not a significant component of the Type V collagen fractions (Figure 7). Next, fractions 2 and 4 of the original Type V collagen preparation were individually incubated with rhMMP-20 and were assessed by SDS-PAGE and Western blotting for proteolysis (Figures 8-9). MMP-20 cleaved Type V collagen from both fractions. These results demonstrate conclusively that Type V collagen is an MMP-20 substrate and they further validate the peptide library as a useful tool for identifying novel enzyme substrates (25).
Figure 6.
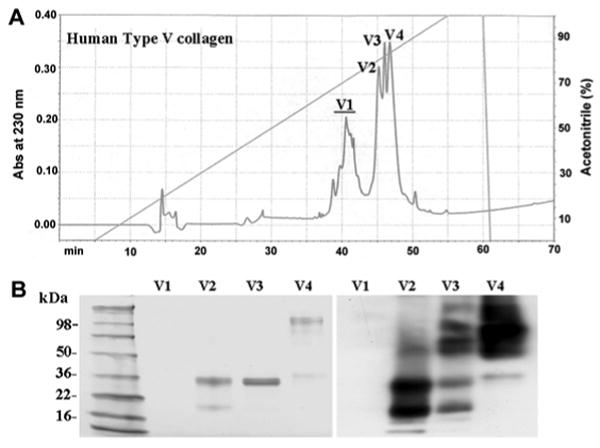
Separation and Identification of Type V collagen fractions. (A) chromatogram of Type V collagen fractionated using a Discovery C18 RP-HPLC column, showing four chromatographic peaks; (B) 4-20% SDS-PAGE stained with CBB (left) and a Western blot identifying the Type V collagen bands (right). Note that fraction V3 contains a mixture of bands that were also present in either V2 or V4.
Figure 7.

Blank and rhMMP-20 chromatograph controls. (A) blank chromatograph containing just 0.05% trifluoroacetic acid demonstrating the absence of contaminates; (B) chromatograph of rhMMP-20 in the absence of Type V collagen. No peaks occurred in the 45-50 min range were the Type V collagen fractions were previously identified.
Figure 8.
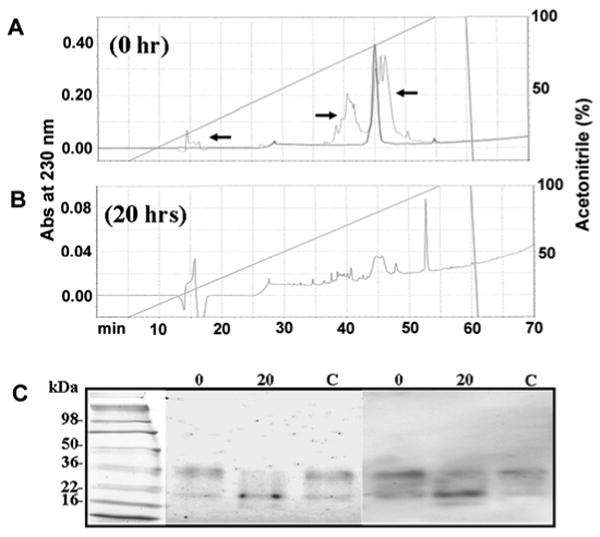
Type V collagen from fraction V2 is an MMP-20 substrate. (A) chromatogram of purified fraction V2 demonstrating a high level of V2 purity. Note that arrows point to a background trace from the chromatogram in Figure 6 demonstrating that the peak shown was from the original V2 fraction. (B) chromatogram of fraction V2 after 20 h digestion with rhMMP-20. Note that the peak at approximately 45 min was substantially diminished after the digestion. (C) SDS-PAGE gel (left) and Western blot (right) demonstrating that fraction V2 Type V collagen was digested after 20 h exposure to rhMMP-20 (lane 20) as compared to the 0 h digestion (lane 0) and the Type V collagen without rhMMP-20 (lane C).
Figure 9.

Type V collagen from fraction V4 is an MMP-20 substrate. (A) chromatogram of purified fraction V4 demonstrating a high level of V4 purity. Note that arrows point to a background trace from the chromatogram in Figure 6 demonstrating that the peak shown was from the original V4 fraction. (B) chromatogram of fraction V4 after 20 h digestion with rhMMP-20. Note that the peak at approximately 47 min was diminished after the digestion. (C) SDS-PAGE gel (left) and Western blot (right) demonstrating that fraction V4 Type V collagen was digested after 20 h exposure to rhMMP-20 (lane 20) as compared to the 0 h digestion (lane 0) and the Type V collagen without rhMMP-20 (lane C).
Discussion
Since MMP-20 has a highly restricted tooth-specific pattern of expression, we sought to take a more defined approach to identify other tissues that might express MMP-20. RT-PCR followed by Southern blotting did detect MMP-20 transcripts in the large intestine (Figure 1). However, the expression levels in the intestine were so low that they were undetectable by Northern blot analysis and were approximately 5000 times lower than the levels observed in 4 day-old tooth buds (Table 1). Recently, a dot blot MMP array was used to demonstrate that MMP-20 is expressed in human fetal and adult lung (42). However, these results were not confirmed by any other means and we have previously demonstrated by Northern blot analysis of porcine lung (14), and in this study by qPCR of mouse lung, that no detectible MMP-20 transcripts were observed. So, although the possibility exists that human lung expresses MMP-20, we remain confident that MMP-20 is essentially a tooth-specific MMP. This conclusion lead us to ask if we could identify putative promoter elements that direct MMP-20 tooth-specific expression and to ask if MMP-20 possesses a broad substrate specificity that might necessitate its limited expression in other tissues. Interestingly, the MMP-20 zymogen has not been observed on zymograms of extracted porcine enamel proteins. Zymography did reveal two bands of approximately 46 and 41 kDa that resulted from MMP-20 activity (18). However, when highly purified porcine MMP-20 was resolved into the 46 and 41 kDa bands by SDS-PAGE and casein zymography followed by transblotting for Edman sequencing and mass spectrometry, both bands were demonstrated to represent active MMP-20 (no propeptide) with the correct active form N-terminus (43). These data demonstrated that the porcine MMP-20 zymogen, if present, is in very low quantities within developing enamel. Furthermore, the results suggest that MMP-20 is either very easily activated and/or autoactivates soon after its secretion into the enamel matrix. If these activation properties were generally applicable to other tissues, then restricted expression would be the primary means to control MMP-20 enzymatic activity without the associated inhibition of other MMPs as would occur if tissue inhibitors of MMPs were induced. Since this suggests that MMP-20 activity is regulated primarily at the transcriptional level, we were interested in characterizing the MMP-20 promoter to identify potential tooth-specific regulatory elements.
Since amelogenin, ameloblastin, enamelin, and MMP-20 are co-regulated predominantly tooth-specific gene products, we assessed their promoters for shared regulatory elements. Conserved patterns of individual TFBS were identified using a large library of quality checked nucleotide weight matrices that are compiled by Genomatix. This approach was used previously to link diabetes-associated genes to insulin/glucose signaling pathways. Without a priori knowledge, legitimate co-regulated genes were identified (44). Although no pattern could be identified that was conserved among all four of the tooth gene promoters, four modules containing two TFBS in conserved orientation and distance from each other were identified in three of the four sequences. The modules were HOMF/BRNF, BRNF/TBPF, HOXF/BRNF, and HOXF/HOXC.
The HOMF family is composed of homeodomain transcription factors including Dlx and Msx both of which are important factors necessary for tooth development. Deletion of Dlx1 and Dlx2 from the mouse genome results in the loss of maxillary molars (45) whereas the expression of Dlx5 and Dlx6 compensates for this loss and allows molar development in the mandibles of these animals (reviewed in (46)). A Dlx3 mutation was demonstrated to cause defective dental enamel (amelogenesis imperfecta) and enlarged pulp chambers (taurodontism) in humans (47). Dlx and Msx will heterodimerize (48) and both Msx1 and Msx2 are also expressed in developing teeth. Mutations in the human and mouse MSX1 gene cause orofacial clefting and tooth agenesis while Msx2-deficient mice have enamel organs that fail to develop properly (reviewed in (49)). Notably, Msx2 was demonstrated to play a role in the spatiotemporal expression of the amelogenin gene (50). Therefore, members of the HOXF family of transcription factors are essential for proper tooth development and have even been demonstrated to regulate amelogenin gene expression.
The HOXF family is also composed of homeodomain transcription factors including Barx2 and goosecoid. Although Barx2 expression has not yet been demonstrated in tooth tissues, it is expressed in several mesenchymal and epithelial tissues during development and was demonstrated to serve as a regulator of hair follicle remodeling (51). In goosecoid null mice the teeth developed normally, but the underlying skeletal elements that support the teeth were absent (52). The HOXC family includes the HOX-PBX complexes that play a role in specifying regional identity along the anteroposterior axis of developing embryos (53). The BRNF family contains Brn POU domain factors that are widely distributed and are predominately associated with the differentiation, regulation, and proliferation of neural stem cells and neurons (reviewed in (54)). The TBPF family contains the TATA-binding protein factors that are commonly found in regulated genes. Thus, the in silico analysis did identify potential regulatory modules that were shared among the amelogenin, ameloblastin, enamelin and MMP-20 promoters indicating that a complex network of regulatory elements may be responsible for each gene's coordinated expression during enamel development. The identified modules are ideal candidates for in vitro mutation analysis to characterize tooth-specific expression. However, it will be necessary to identify a suitable cell line for these experiments as none is now known to exist.
The peptide library screen indicated that while MMP-20 was typical of MMPs by its primary preference for hydrophobic residues at the P1′ position, the high selectivity at this position for the aromatic tryrosine residue was unique, indicating that MMP-20 has a deep catalytic pocket (Figure 4). Crystal structures and mutagenesis of various MMPs have largely implicated a single residue, corresponding to Arg214 of human MMP-1, in controlling depth of the S1′ pocket (55;56). Shallow pocket MMPs have a relatively large residue at this position (arginine or tyrosine) that restricts access of larger residues to the pocket, whereas the deep and intermediate pocket MMPs have leucine at this position. MMP-20 by contrast has a threonine residue at the analogous position, suggesting it may have a wider S1′ pocket more accommodating to aromatic residues. These observations support recent work indicating that a continuum of shapes and sizes exists for the S1′ pocket among various MMPs (57). MMP-20 secondary selections were also distinctive and included the unusual preference for histidine at the P2 position. To confirm that the peptide library results were representative cleavages that occur in vivo, these results were compared to the known MMP-20 cleavage sites present in amelogenin. Although some features of the library-derived MMP-20 cleavage motif were represented in amelogenin cleavage sites, no individual site was a particularly close match to the consensus (Table 3). This may indicate that amelogenin cleavage sites are suboptimal, and that relatively high levels of MMP-20 activity are required to degrade its physiological substrate. This notion supports the observation of the “laddering effect” of amelogenin in the enamel matrix where proteolytic cleavage gradually reduces the size of amelogenins as new uncleaved amelogenins are secreted into the matrix (58). Thus, SDS-PAGE of enamel matrix proteins reveals a laddering of amelogenin bands that persists until MMP-20 is no longer secreted into the matrix. Alternatively, efficient cleavage of amelogenin may require docking interactions distal from the active site that are not revealed by our peptide library studies. It is also possible that fixing the P′ positions in the library prior to identifying the P positions biases the selection of preferred amino acids for the unknown P positions. This would occur if cooperativity between subsites existed, which was demonstrated for several other MMPs (26, 37, 59, 60). It should also be noted that the library results were generated with recombinant human MMP-20 while the amelogenin cleavage sites were previously identified by use of recombinant porcine MMP-20. Exceptions aside, the peptide library screen and the previously published amelogenin data (39) did predominantly coincide, indicating that MMP-20 has a unique substrate specificity among the MMP family of enzymes.
Based on the peptide library results we searched for a novel MMP-20 substrate. Both Type III and Type V collagens were identified as potential MMP-20 substrates. Type III collagen is essential for collagen I fibrillogenesis and for normal cardiovascular development (61) and Type V collagen controls the initiation of collagen fibril assembly. Murine deficiency of the major Type V collagen chain, pro-α1(V), causes death in early embryogenesis and is associated with a virtual absence of collagen fibrils (62). Therefore, the presence of Type I collagen is associated with the presence of Type V collagen. Dentin and bone contain both of these collagens (27, 28). However, enamel contains neither (27). Although, odontoblasts do express low levels of MMP-20 (15), it is unclear what role MMP-20 plays in processing Type V collagen in dentin, because dentin contains other MMPs, such as MMP-2, -9, and -10 (63) that are also capable of cleaving Type V collagen. We would argue that since MMP-20 is easily activated and has a broad substrate specificity, its restricted pattern of expression is necessitated to prevent unrestrained matrix degradation.
Taken together, the results demonstrate that MMP-20 has a highly restricted tooth-specific pattern of expression, has a large catalytic pocket with a broad substrate specificity, is capable of cleaving a widely distributed collagen that is responsible for collagen fibril assembly, and its activity is likely regulated by transcriptional means.
Acknowledgments
We thank Jun Xue for her technical expertise.
Abbreviations
- Abs
absorbance
- MMP
matrix metalloproteinase
- KLK4
kallikrein-4
- rh
recombanant human
- RP-HPLC
reversed-phase high pressure liquid chromatography
- TFA
trifluoroacetic acid
- TFBS
transcription factor binding site
- TSS
transcription start site
- HOMF
homeodomain transcription factors
- BRNF
Brn POU domain factors
- TBPF
tata-binding protein factor
- HOXF
factors with moderate activity to homeodomain consensus sequence
- HOXC
HOX-PBX complexes
Footnotes
This work was supported by National Institute of Dental and Craniofacial Research grants DE14084 (to J.D.B.) and DE 13237 (subproject 4 to J.D.B.), by NIH grant GM56203 (to L.C.C.), by a Special Fellow award from the Leukemia and Lymphoma Society (to B.E.T.), and was conducted in a laboratory renovated with NIH grant support CO6RR11244.
References
- 1.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 2.Hu CC, Hart TC, Dupont BR, Chen JJ, Sun X, Qian Q, Zhang CH, Jiang H, Mattern VL, Wright JT, Simmer JP. Cloning human enamelin cDNA, chromosomal localization, and analysis of expression during tooth development. J Dent Res. 2000;79:912–919. doi: 10.1177/00220345000790040501. [DOI] [PubMed] [Google Scholar]
- 3.MacDougall M, DuPont BR, Simmons D, Reus B, Krebsbach P, Karrman C, Holmgren G, Leach RJ, Forsman K. Ameloblastin gene (AMBN) maps within the critical region for autosomal dominant amelogenesis imperfecta at chromosome 4q21. Genomics. 1997;41:115–118. doi: 10.1006/geno.1997.4643. [DOI] [PubMed] [Google Scholar]
- 4.Llano E, Pendas AM, Knauper V, Sorsa T, Salo T, Salido E, Murphy G, Simmer JP, Bartlett JD, Lopez-Otin C. Identification and structural and functional characterization of human enamelysin (MMP-20) Biochemistry. 1997;36:15101–15108. doi: 10.1021/bi972120y. [DOI] [PubMed] [Google Scholar]
- 5.Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- 6.Lattanzi W, Di Giacomo MC, Lenato GM, Chimienti G, Voglino G, Resta N, Pepe G, Guanti G. A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum Genet. 2005;116:395–401. doi: 10.1007/s00439-004-1238-z. [DOI] [PubMed] [Google Scholar]
- 7.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 8.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 10.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 13.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett JD, Ryu OH, Xue J, Simmer JP, Margolis HC. Enamelysin mRNA displays a developmentally defined pattern of expression and encodes a protein which degrades amelogenin. Connect Tissue Res. 1998;39:101–109. doi: 10.3109/03008209809023916. [DOI] [PubMed] [Google Scholar]
- 16.Bègue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 17.Caterina J, Shi J, Sun X, Qian Q, Yamada S, Liu Y, Krakora S, Bartlett JD, Yamada Y, Engler JA, Birkedal-Hansen H, Simmer JP. Cloning, characterization, and expression analysis of mouse enamelysin. J Dent Res. 2000;79:1697–1703. doi: 10.1177/00220345000790091001. [DOI] [PubMed] [Google Scholar]
- 18.Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, Wakida K, Simmer JP, Yamada Y, Bartlett JD. Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J Dent Res. 1998;77:1580–1588. doi: 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- 19.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 20.Grant GM, Giambernardi TA, Grant AM, Klebe RJ. Overview of expression of matrix metalloproteinases (MMP-17, MMP-18, and MMP-20) in cultured human cells. Matrix Biol. 1999;18:145–148. doi: 10.1016/s0945-053x(99)00003-7. [DOI] [PubMed] [Google Scholar]
- 21.Takata T, Zhao M, Nikai H, Uchida T, Wang T. Ghost cells in calcifying odontogenic cyst express enamel-related proteins. Histochem J. 2000;32:223–229. doi: 10.1023/a:1004051017425. [DOI] [PubMed] [Google Scholar]
- 22.Takata T, Zhao M, Uchida T, Wang T, Aoki T, Bartlett JD, Nikai H. Immunohistochemical detection and distribution of enamelysin (MMP-20) in human odontogenic tumors. J Dent Res. 2000;79:1608–1613. doi: 10.1177/00220345000790081401. [DOI] [PubMed] [Google Scholar]
- 23.Väänänen A, Tjaderhane L, Eklund L, Heljasvaara R, Pihlajaniemi T, Herva R, Ding Y, Bartlett JD, Salo T. Expression of collagen XVIII and MMP-20 in developing teeth and odontogenic tumors. Matrix Biol. 2004;23:153–161. doi: 10.1016/j.matbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Kimura A, Kihara T, Ohkura R, Ogiwara K, Takahashi T. Localization of bradykinin B(2) receptor in the follicles of porcine ovary and increased expression of matrix metalloproteinase-3 and -20 in cultured granulosa cells by bradykinin treatment. Biol Reprod. 2001;65:1462–1470. doi: 10.1095/biolreprod65.5.1462. [DOI] [PubMed] [Google Scholar]
- 25.Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 26.Park HI, Turk BE, Gerkema FE, Cantley LC, Sang QX. Peptide substrate specificities and protein cleavage sites of human endometase/matrilysin-2/matrix metalloproteinase-26. J Biol Chem. 2002;277:35168–35175. doi: 10.1074/jbc.M205071200. [DOI] [PubMed] [Google Scholar]
- 27.Bronckers AL, Gay S, Lyaruu DM, Gay RE, Miller EJ. Localization of type V collagen with monoclonal antibodies in developing dental and peridental tissues of the rat and hamster. Coll Relat Res. 1986;6:1–13. doi: 10.1016/s0174-173x(86)80029-2. [DOI] [PubMed] [Google Scholar]
- 28.Lukinmaa PL, Waltimo J. Immunohistochemical localization of types I, V, and VI collagen in human permanent teeth and periodontal ligament. J Dent Res. 1992;71:391–397. doi: 10.1177/00220345920710020801. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 31.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- 33.Hu JC, Sun X, Zhang C, Simmer JP. A comparison of enamelin and amelogenin expression in developing mouse molars. Eur J Oral Sci. 2001;109:125–132. doi: 10.1034/j.1600-0722.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Deng SJ, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, Carter HL, III, Neugebauer J, Pahel G, Weiner MP, Moss ML. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J Biol Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 37.Kridel SJ, Chen E, Kotra LP, Howard EW, Mobashery S, Smith JW. Substrate hydrolysis by matrix metalloproteinase-9. J Biol Chem. 2001;276:20572–20578. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- 38.McGeehan GM, Bickett DM, Green M, Kassel D, Wiseman JS, Berman J. Characterization of the peptide substrate specificities of interstitial collagenase and 92-kDa gelatinase. Implications for substrate optimization. J Biol Chem. 1994;269:32814–32820. [PubMed] [Google Scholar]
- 39.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 40.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 41.Väänänen A, Srinivas R, Parikka M, Palosaari H, Bartlett JD, Iwata K, Grenman R, Stenman UH, Sorsa T, Salo T. Expression and regulation of MMP-20 in human tongue carcinoma cells. J Dent Res. 2001;80:1884–1889. doi: 10.1177/00220345010800100501. [DOI] [PubMed] [Google Scholar]
- 42.Ryu J, Vicencio AG, Yeager ME, Kashgarian M, Haddad GG, Eickelberg O. Differential expression of matrix metalloproteinases and their inhibitors in human and mouse lung development. Thromb Haemost. 2005;94:175–183. doi: 10.1160/TH04-10-0656. [DOI] [PubMed] [Google Scholar]
- 43.Yamada Y, Yamakoshi Y, Gerlach RF, Hu CC, Matsumoto K, Fukae M, Oida S, Bartlett JD, Simmer JP. Purification and Characterization of Enamelysin from Secretory Stage Pig Enamel. Archives of Comparative Biology of Tooth Enamel (Japan) 2003;8:21–25. [Google Scholar]
- 44.Dohr S, Klingenhoff A, Maier H, Hrabe de AM, Werner T, Schneider R. Linking disease-associated genes to regulatory networks via promoter organization. Nucleic Acids Res. 2005;33:864–872. doi: 10.1093/nar/gki230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- 46.Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 47.Dong J, Amor D, Aldred MJ, Gu T, Escamilla M, MacDougall M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am J Med Genet A. 2005;133:138–141. doi: 10.1002/ajmg.a.30521. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, bate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429–442. doi: 10.1038/sj.cr.7290185. [DOI] [PubMed] [Google Scholar]
- 50.Zhou YL, Lei Y, Snead ML. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J Biol Chem. 2000;275:29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- 51.Olson LE, Zhang J, Taylor H, Rose DW, Rosenfeld MG. Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proc Natl Acad Sci U S A. 2005;102:3708–3713. doi: 10.1073/pnas.0500519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada G, Mansouri A, Torres M, Stuart ET, Blum M, Schultz M, De Robertis EM, Gruss P. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- 53.Peterson RL, Papenbrock T, Davda MM, Awgulewitsch A. The murine Hoxc cluster contains five neighboring AbdB-related Hox genes that show unique spatially coordinated expression in posterior embryonic subregions. Mech Dev. 1994;47:253–260. doi: 10.1016/0925-4773(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 54.Andersen B, Rosenfeld MG. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev. 2001;22:2–35. doi: 10.1210/edrv.22.1.0421. [DOI] [PubMed] [Google Scholar]
- 55.Bode W, Fernandez-Catalan C, Tschesche H, Grams F, Nagase H, Maskos K. Structural properties of matrix metalloproteinases. Cell Mol Life Sci. 1999;55:639–652. doi: 10.1007/s000180050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welch AR, Holman CM, Huber M, Brenner MC, Browner MF, Van Wart HE. Understanding the P1′ specificity of the matrix metalloproteinases: effect of S1′ pocket mutations in matrilysin and stromelysin-1. Biochemistry. 1996;35:10103–10109. doi: 10.1021/bi9601969. [DOI] [PubMed] [Google Scholar]
- 57.Park HI, Jin Y, Hurst DR, Monroe CA, Lee S, Schwartz MA, Sang QX. The intermediate S1′ pocket of the endometase/matrilysin-2 active site revealed by enzyme inhibition kinetic studies, protein sequence analyses, and homology modeling. J Biol Chem. 2003;278:51646–51653. doi: 10.1074/jbc.M310109200. [DOI] [PubMed] [Google Scholar]
- 58.Smith CE, Pompura JR, Borenstein S, Fazel A, Nanci A. Degradation and loss of matrix proteins from developing enamel. Anat Rec. 1989;224:292–316. doi: 10.1002/ar.1092240219. [DOI] [PubMed] [Google Scholar]
- 59.Chen EI, Kridel SJ, Howard EW, Li W, Godzik A, Smith JW. A unique substrate recognition profile for matrix metalloproteinase-2. J Biol Chem. 2002;277:4485–4491. doi: 10.1074/jbc.M109469200. [DOI] [PubMed] [Google Scholar]
- 60.Kridel SJ, Sawai H, Ratnikov BI, Chen EI, Li W, Godzik A, Strongin AY, Smith JW. A unique substrate binding mode discriminates membrane type-1 matrix metalloproteinase from other matrix metalloproteinases. J Biol Chem. 2002;277:23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- 61.Liu X, Wu H, Byrne M, Krane S, Jaenisch R. Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc Natl Acad Sci U S A. 1997;94:1852–1856. doi: 10.1073/pnas.94.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wenstrup RJ, Florer JB, Brunskill EW, Bell SM, Chervoneva I, Birk DE. Type V collagen controls the initiation of collagen fibril assembly. J Biol Chem. 2004;279:53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- 63.Palosaari H, Pennington CJ, Larmas M, Edwards DR, Tjaderhane L, Salo T. Expression profile of matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs in mature human odontoblasts and pulp tissue. Eur J Oral Sci. 2003;111:117–127. doi: 10.1034/j.1600-0722.2003.00026.x. [DOI] [PubMed] [Google Scholar]


