Abstract
The melanocortin-3 receptor (MC3R) is an important regulator of energy homeostasis, inflammation, and cardiovascular function. Inactivating mutations in MC3R gene are associated with childhood obesity. How MC3R binds to its ligands has rarely been studied. In the present study, we systematically mutated all ten acidic residues in transmembrane domains (TMs) and measured the cell surface expression levels as well as ligand binding and signaling properties of these mutants. Our results showed that of the 19 mutants stably expressed in HEK293 cells, all were expressed on the cell surface, although some mutants had decreased levels of cell surface expression. We showed that with the superpotent analog [Nle4, d-Phe7]-α-melanocyte stimulating hormone, E92, E131, D154, D158, D178, and D332 are important for ligand binding. D121 and D332 are important for binding and signaling. Further experiments using other ligands such as d-Trp8-γ-MSH, α-MSH and γ-MSH showed that different ligands induce or select different conformations. In summary, we showed that acidic residues in TMs 1 and 3 are important for ligand binding whereas the acidic residues in TMs 2 and 7 are important for both ligand binding and signaling.
Keywords: melanocortin-3 receptor, site-directed mutagenesis, ligand binding, G protein-coupled receptor, signaling, structure-function relationship
1. Introduction
The melanocortin-3 receptor (MC3R) is a member of the superfamily of G protein-coupled receptors (GPCRs). It was originally cloned independently by Cone and Gantz from rat and human, respectively [1, 2]. It is expressed in several brain regions, including the arcuate nucleus and ventromedial nuclei of the hypothalamus and limbic system [2]. It is also expressed in a variety of peripheral tissues, including the placenta, heart, and the gut [1], as well as in immune cells such as macrophages [3, 4].
During the past few years, the MC3R was increasingly recognized as an important regulator of energy homeostasis, especially fat metabolism. In gene targeting studies, MC3R and melanocortin-4 receptor (MC4R) were shown to have non-redundant roles in regulating energy homeostasis. Whereas the MC4R regulates both food intake and energy expenditure, the MC3R was shown not to affect food intake or energy expenditure [5-7]. Despite normal or decreased food intake and normal energy expenditure, MC3R knockout (KO) mice exhibited increased fat mass (approximately twice that of wild type littermates) due to increased feed efficiency [6, 7]. Mice lacking both the MC3R and MC4R showed exacerbated obesity compared to MC3R or MC4R single gene KO mice, suggesting that the two neural MCRs regulate different aspects of energy homeostasis [6]. These studies were done in C57BL/6J mice. Recently, Gettys and colleagues generated KO mice in Black Swiss;129, a mouse strain resistant to obesity. This study showed that MC3R KO produced a comparable degree of increased adiposity as the MC4R KO [8]. Recent studies showed that injections of a selective MC3R agonist D-Trp8-γ-MSH increase food intake in mice [9, 10], suggesting that MC3R is also involved in regulating food intake.
Naturally occurring mutations and polymorphisms in the MC3R gene have been identified from obese patients. So far, eight MC3R variants have been reported, including T6K, A70T, V81I, M134I, I183N, A293T, I335S, and X361S [11-15]. Functional studies revealed a number of defects, including decreased total expression, intracellular retention, and defects in receptor activation [12, 14-18]. Clinical studies showed that there is a gene-diet interaction in weight-loss program in carriers of the common polymorphic variants T6K/V81I [19], probably due to defect in substrate oxidation [20], consistent with data obtained in MC3R KO mice [21].
In addition to its role in regulating energy homeostasis, MC3R is also involved in direct regulation of the cardiovascular system, including the heart and blood pressure [22-24] and inflammation [25].
As is typical of GPCRs, the MC3R consists of the typical heptahelical structure, with an extracellular NH2 terminus and intracellular COOH terminus (schematically depicted in Fig. 1). Previous studies with the MC4R have identified several residues critical for binding to the superpotent analog of α-melanocyte stimulating hormone (MSH), [Nle4, D-Phe7]-α-MSH (NDP-MSH) [26, 27]. These include acidic residues D122 and D126 in transmembrane domain (TM) 3 as well as other residues such as hydrophobic residues in TM5 and TM6. Modeling studies (not based on rhodopsin crystal structure) suggested that the hydrophobic pocket of the ligand binding site are different between the MC3R and MC4R, whereas charge-charge interactions between the ligand and the receptor are similar in the two melanocortin receptors (MCRs) [28]. This prediction has not been verified experimentally. Since few studies have been performed to investigate the possible ligand interaction sites on the MC3R, we systematically investigated the functions of ten acidic residues in the TMs of human MC3R (hMC3R) in ligand binding and receptor activation.
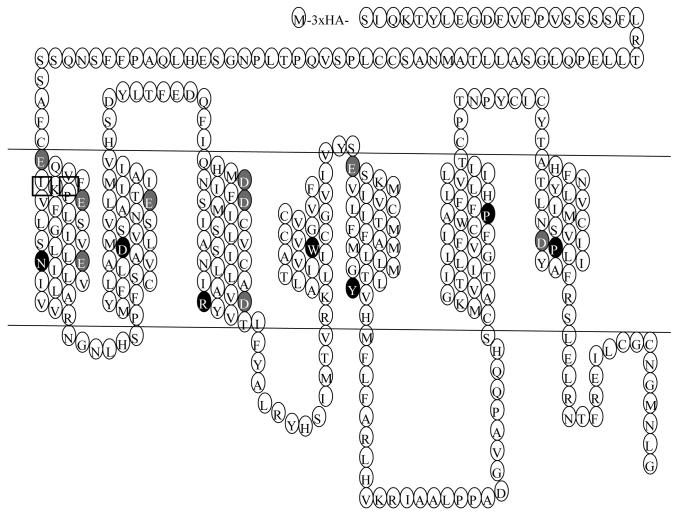
Figure 1.
Serpentine model of the human MC3R. The most highly conserved residues in each TM are indicated by the white letter on dark background. The residues mutated in this study are highlighted in white letter on gray background except D121 that is also the most highly conserved residue in TM2.
In addition to D154 (3.25) and D158 (3.29) in TM3 (corresponding to D122 and D126 in the MC4R), there are eight additional acidic residues that can potentially form a salt bridge with Arg in the pharmacaphore (HFRW) of the ligands [29]. These include: E73 (1.30), E80 (1.37), E92 (1.49), D121 (2.50), E131 (2.60), D178 (3.49), E221 (5.27), and D332 (7.49) (Fig. 1) (The numbers in the brackets are numbering based on the Ballesteros and Weinstein system, with the first number representing the helix, and the second number representing the residue's relative position to the most highly conserved residue in that TM designated as 50, decreasing towards the amino terminus, and increasing towards the carboxyl terminus). These ten acidic residues were mutated to investigate their possible roles in ligand binding and signaling.
2. Materials and methods
2.1. Hormones and supplies
NDP-MSH was purchased from Bachem (King of Prussia, PA). α- and γ-MSH were purchased from Phoenix Pharmaceuticals (Belmont, CA). D-Trp8-γ-MSH was synthesized by Peptides International (Louisville, KY). Iodinated NDP-MSH was purchased from the Peptide Radioiodination Service Center at the University of Mississippi (University, MS). Plates and flasks for tissue culture were purchased from Corning (Corning, NY). Tissue culture media, newborn calf serum, trypsin, and antibiotics were all obtained from Invitrogen (Carlsbad, CA).
2.2. In vitro mutagenesis of hMC3R
The wild type (WT) hMC3R tagged at the N-terminus with 3xHA tag was obtained from Missouri S&T cDNA Resource Center (www.cDNA.org, Rolla, MO). Point mutants were generated by QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described in detail before [30]. IsoPure DNA purification kits from Denville Scientific (Metuchen, NJ) were used to prepare the plasmids for transfection. Automated DNA sequencing, performed at the DNA Sequencing Facility of University of Chicago Cancer Research Center (Chicago, IL), was used to confirm that only the intended mutations were introduced in the constructs.
2.3. Cells and transfections
HEK293T cells, obtained from American Type Culture Collection (Manassas, VA), were maintained at 5% CO2 in Dulbecco's modified Eagle's medium containing 50μg/ml gentamicin, 10 mM Hepes, and 10% newborn calf serum. Cells were plated on gelatin-coated 35 mm 6-well clusters. When the cells reached 50-70% confluency, they were transfected using the calcium precipitation method [31]. Four μg plasmid in 2 ml media was used per 35 mm well. Cells were used approximately 48h after transfection for measuring ligand binding and signaling.
2.4. Ligand binding to intact cells
Ligand binding assays were performed as described in detail before [30]. Briefly, HEK293T cells were transfected as described above. Approximately 48 hours after transfection, cells were washed two times with warm Waymouth's MB752/1 media (Sigma, St. Louis, MO) containing 1 mg/ml bovine serum albumin (BSA) and 50μg/ml gentamicin) (herein referred as Waymouth/BSA) and then incubated with 100,000 cpm of [125I]-NDP-MSH with or without different concentrations of unlabeled ligands at 37 C for 1 hour. Concentrations of the unlabeled ligands are indicated in the figures. To terminate the reaction, cells were placed directly on ice, washed twice with cold Hank's balanced salt solution containing 1 mg/ml BSA and 50μg/ml gentamicin. Cells were solubilized with 100 μl of 0.5 N NaOH. Cell lysates were then collected using cotton swabs, and counted in a gamma counter. Concentrations that result in 50% inhibition (IC50) and receptor occupancy (RO) were calculated using GraphPad Prism 4.0 software (San Diego, CA).
2.5. Signaling assay to measure ligand stimulation of intracellular cAMP production
Transient transfection of HEK293T cells was performed as described above. Two days after transfection, cells were washed twice with warm Waymouth/BSA. Then fresh Waymouth/BSA containing 0.5 mM isobutylmethylxanthine (Sigma) was added to each well. After 15 min incubation at 37 C, either buffer alone or different concentrations of ligands were added and the incubation was continued for another hour. The final concentrations of the ligands are indicated in the figures. Intracellular cAMP was extracted using 0.5 N percholoric acid containing 180μg/ml theophylline (Sigma), and measured using radioimmunoassay [32]. All determinations were performed in triplicate. Maximal responses (Emax) and concentrations that result in 50% maximal response (EC50) were calculated using Prism 4.0 software.
2.6. FACS assay of HEK293 cells stably expressing WT or mutant hMC3Rs
Cells stably expressing WT or mutant hMC3Rs were established as described before [30, 33]. The day before the experiment, cells were plated into wells of 6-well plates. On the day of the experiment, cells were washed twice with filtered phosphate-buffered saline for immunohistochemistry (PBS-IH) [in mM, consisting of 137 NaCl, 2.7 KCl, 1.4 KH2PO4, and 4.3 Na2HPO4 (pH 7.4)], fixed with 4% paraformaldehyde for 30 min, and then incubated with blocking solution (5% BSA in PBS-IH) for 1 hr. Cells were then incubated for 1 hr with Alexa Fluor® 488 Labeled HA.11 monoclonal antibody (CRP Inc., Princeton, NJ) diluted 1:200 in PBS-IH containing 1 mg/ml BSA. Cells were then washed three times with PBS-IH and assayed with a MoFlo7-color flow cytometer & high-performance sorter (Dakocytomation, Fort Collins, CO). All the steps were performed at room temperature. The expression level of the mutants was calculated as a percentage of WT expression using the formula: [mutant − pcDNA3]/[WT − pcDNA3]×100%.
2.7. Statistical analyses
Student's t test was used to determine the significance of differences in cell surface expression levels as well as the signaling and binding parameters between WT and mutant hMC3Rs. The statistical analysis was carried out using GraphPad Prism 4.0.
3. Results
3.1. Ligand binding and signaling properties of the mutant hMC3Rs using NDP-MSH as the ligand
To study the importance of acidic TM residues in ligand binding and signaling, we mutated all ten acidic TM residues by site-directed mutagenesis. For these studies, Asp was mutated to Glu (to conserve negative charge) or Gln (to eliminate negative charge). Similarly, Glu was mutated to Asp (to conserve negative charge) and Gln (to eliminate negative charge but conserve size). We reasoned that results obtained from these mutants would reveal whether the negative charge is important for ligand binding and/or receptor activation. A total of twenty mutants were generated.
We first studied the ligand binding and signaling properties of the mutants using NDP-MSH as the ligand. Since NDP-MSH is a superpotent analog, we reasoned that if a mutant cannot bind and respond to NDP-MSH, it would not be able to bind and respond to the native ligands such as α- and γ-MSH.
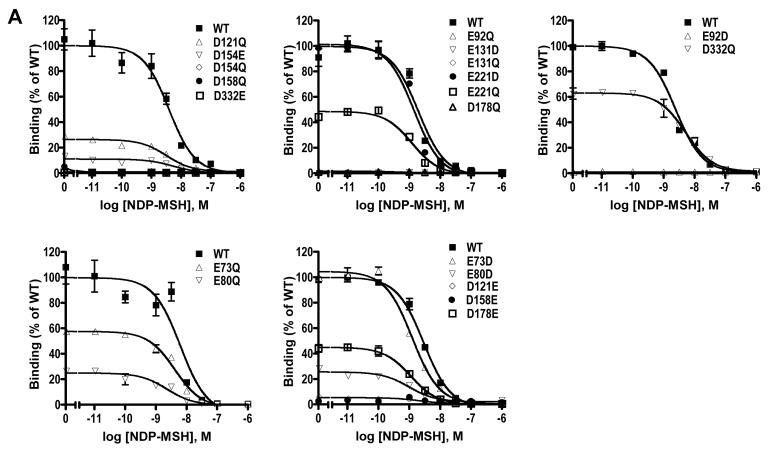
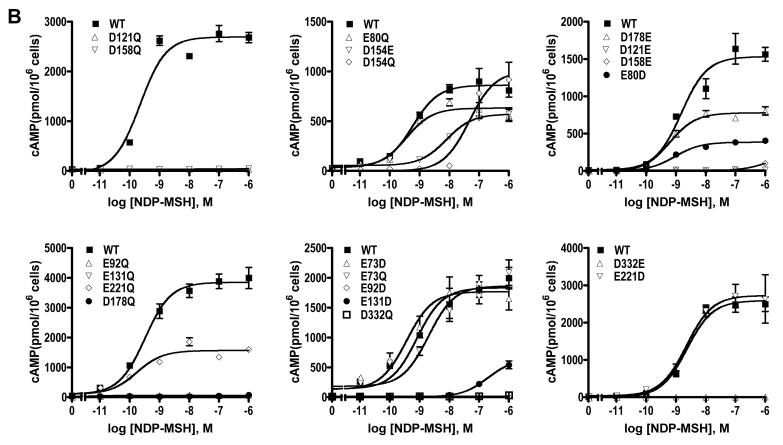
As shown in Fig. 2 and Table 1, the WT hMC3R binds to NDP-MSH with an IC50 of 2.84 nM and responds to NDP-MSH stimulation with dose-dependent increases in cAMP production. The EC50 was 0.86 nM. Of the twenty mutants, ten had no measurable ligand binding (Fig. 2A). These mutants were E92D, E92Q, D121E, E131D, E131Q, D154Q, D158E, D158Q, D178Q, and D332E. Ten mutants had no measurable signaling in response to NDP-MSH stimulation (Fig. 2B). These mutants were E92D, E92Q, D121E, D121Q, E131Q, D158E, D158Q, D178Q, D332E, and D332Q. The following five mutants had decreased ligand binding and/or signaling: E73D, E80D, E80Q, D154E, D178E, E221Q. Only three mutants, E73D, E73Q and E221D, had normal ligand binding and signaling. None of the mutants are constitutively active (data not shown).
Figure 2.
Ligand binding and signaling properties of the WT and mutant hMC3Rs with NDP-MSH as the ligand. HEK293T cells were transiently transfected with the indicated hMC3R constructs and binding and signaling properties of the hMC3Rs were measured as described in Materials and Methods. In A, different concentrations of unlabeled NDP-MSH were used to displace the binding of 125I-NDP-MSH to hMC3Rs on intact cells. Results shown are expressed as % of WT binding ± range from duplicate determinations within one experiment. In B, HEK293T cells transiently transfected with the indicated hMC3R constructs were stimulated with different concentrations of NDP-MSH. Intracellular cAMP levels were measured using radioimmunoassay. Results are expressed as the mean ± SEM of triplicate determinations within one experiment. All experiments were performed at least three times (see Table 1 for the number of experiments done for each hMC3R).
Table 1.
NDP-MSH-Stimulated Signaling and Ligand Binding of WT and Mutant hMC3Rs
| hMC3R | n | NDP-MSH-stimulated cAMP |
NDP-MSH binding |
||
|---|---|---|---|---|---|
| EC50 (nM) | Emax (% WT) | IC50 (nM) | RO (% WT) | ||
| WT | 19 | 0.86 ± 0.13 | 100 | 2.84 ± 0.48 | 100 |
| E73D | 4 | 0.52 ± 0.16 | 73 ± 17 | 1.35 ± 0.13a | 95 ± 13 |
| E73Q | 3 | 1.23 ± 0.37 | 139± 52 | 1.61 ± 0.15 | 107 ± 14 |
| E80D | 3 | 1.42 ± 0.69 | 22 ± 5b | 1.03 ± 0.32a | 39 ± 7c |
| E80Q | 3 | 0.30 ± 0.08 | 56 ± 13a | 1.22 ± 0.29 | 58 ± 7c |
| E92D | 3 | ND* | ND* | ND* | ND* |
| E92Q | 3 | ND* | ND* | ND* | ND* |
| D121E | 3 | ND* | ND* | ND* | ND* |
| D121Q | 3 | ND* | ND* | 2.26 ± 0.56 | 16 ± 5 b |
| E131D | 3 | 139.16 ± 29.59 b | 22 ± 7 | ND* | ND* |
| E131Q | 4 | ND* | ND* | ND* | ND* |
| D154E | 3 | 4.24 ± 1.38a | 87 ± 13 | 3.07 ± 1.12 | 17 ± 3c |
| D154Q | 3 | 117.25 ± 23.86b | 103 ± 13 | ND* | ND* |
| D158E | 3 | ND* | ND* | ND* | ND* |
| D158Q | 3 | ND* | ND* | ND* | ND* |
| D178E | 3 | 0.56 ± 0.11 | 56 ± 15 | 1.53 ± 0.19 | 53 ± 5c |
| D178Q | 3 | ND* | ND* | ND* | ND* |
| E221D | 3 | 2.22 ± 0.92 | 201 ± 54 | 2.17 ± 0.80 | 112 ± 10 |
| E221Q | 3 | 1.52 ± 0.24 | 101 ± 45 | 1.39 ± 0.24 | 93 ± 12 |
| D332E | 4 | ND* | ND* | ND* | ND* |
| D332Q | 3 | ND* | ND* | 1.77 ± 0.27 | 57 ± 7c |
ND, could not be detected.
significantly different from WT hMC3R, p<0.05
significantly different from WT hMC3R, p<0.01
significantly different from WT hMC3R, p<0.001
The data are expressed as the mean ± SEM of three or four independent experiments. The maximal response under NDP-MSH stimulation was 2308 ± 362 pmol cAMP/106 cells for WT hMC3R. Binding is shown as receptor occupancy (RO) that is a crude estimate of the relative maximal binding level.
3.2. Cell surface expression of the hMC3R mutants
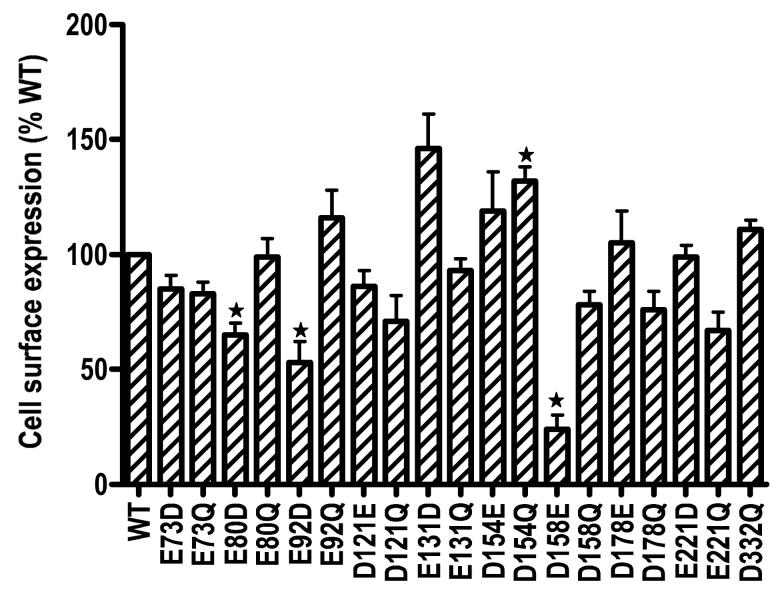
Flow cytometry was used to determine the cell surface expression of the mutant receptors. Cells stably transfected with the empty vector was used to correct for background staining. We were unable to establish cells stably expressing D332E hMC3R after three separate transfections. Of the 19 mutants that were stably expressed in HEK293 cells, our data showed that they were all expressed on the cell surface (Fig. 3). Three mutants, including E80D, E92D and D158E had decreased cell surface expression. E131D and D154Q hMC3R had increased cell surface expression (Fig. 3).
Figure 3.
Cell surface expression of the WT and mutant hMC3Rs. Cells stably expressing WT or mutant hMC3Rs were stained with HA monoclonal antibody for the 3xHA epitope at the N terminus of the hMC3R. Results are expressed as % of WT expression levels. Star (✩) indicates significantly different from WT hMC3R.
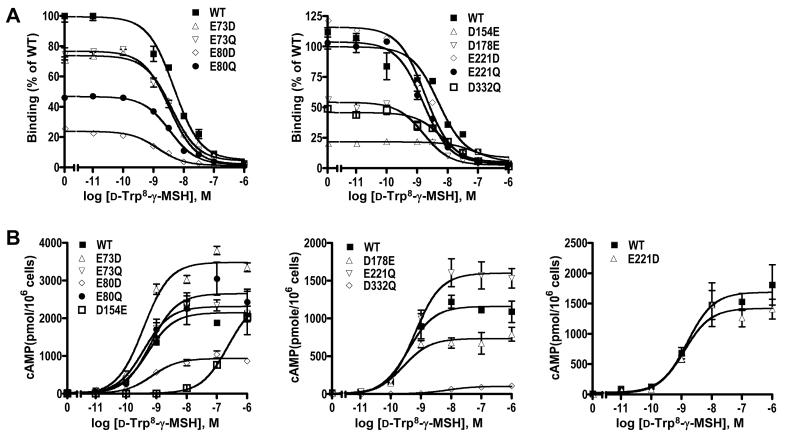
3.3. Ligand binding and signaling properties of the mutant hMC3Rs using d-Trp8-γ-MSH as the ligand
D-Trp8-γ-MSH is a superpotent analogue of γ-MSH discovered by Hruby and colleagues [34]. They found that D-Trp8-γ-MSH is even more selective than the natural γ-MSH towards MC3R [34]. With this analog, the IC50 is 6.7 nM for hMC3R, and 600 nM for hMC4R; the EC50 is 0.33 nM for hMC3R, and 100 nM for hMC4R [34]. Therefore, the analog is about 2 orders of magnitude more selective at hMC3R in terms of binding, and 300 fold more selective towards hMC3R in terms of signaling [34].
We studied the ligand binding and signaling properties of the nine mutants that had detectable ligand binding and signaling when NDP-MSH was used as the ligand. As shown in Fig. 4 and Table 2, the WT hMC3R bound to d-Trp8-γ-MSH with an IC50 of 6.23 nM, and responded to d-Trp8-γ-MSH stimulation with increased cAMP production. The EC50 was 0.69 nM. These results are in good agreement with the results obtained by Hruby and colleagues [34]. All the other mutants except D154E bound to d-Trp8-γ-MSH with similar IC50 as the WT hMC3R (Table 2). All other mutants (except D154E and D332Q) responded to d-Trp8-γ-MSH stimulation with similar EC50 as the WT hMC3R (Table 2). The maximal responses were decreased with E80D and D332Q (Table 2).
Figure 4.
Ligand binding and signaling properties of the WT and mutant hMC3Rs with D-Trp8-γ-MSH as the ligand. HEK293T cells were transiently transfected with the indicated hMC3R constructs and binding and signaling properties of the hMC3Rs were measured as described in Materials and Methods. In A, different concentrations of unlabeled D-Trp8-γ-MSH were used to displace the binding of 125I-NDP-MSH to hMC3Rs on intact cells. Results shown are expressed as % of WT binding ± range from duplicate determinations within one experiment. In B, HEK293T cells transiently transfected with the indicated hMC3R constructs were stimulated with different concentrations of D-Trp8-γ-MSH. Intracellular cAMP levels were measured using radioimmunoassay. Results are expressed as the mean ± SEM of triplicate determinations within one experiment. All experiments were performed at least three times (see Table 2 for the number of experiments done for each hMC3R).
Table 2.
Ligand Binding and D-Trp8-γ-Stimulated cAMP Production of WT and Mutant hMC3Rs
| hMC3R | n |
d-Trp8-γ-MSH-stimulated cAMP |
d-Trp8-γ-MSH binding |
|
|---|---|---|---|---|
| EC50 (nM) | Emax (% WT) | IC50 (nM) | ||
| WT | 8 | 0.69 ± 0.15 | 100 | 6.23 ± 1.80 |
| E73D | 4 | 0.42 ± 0.06 | 130 ± 45 | 5.17 ± 2.41 |
| E73Q | 4 | 0.70 ± 0.26 | 87 ± 21 | 5.24 ± 1.78 |
| E80D | 4 | 0.71 ± 0.27 | 32 ± 5 c | 2.57 ± 1.02 |
| E80Q | 3 | 0.84 ± 0.31 | 99 ± 18 | 2.61 ± 0.46 |
| D154E | 3 | 231.80 ± 75.56a | 88 ± 31 | 106.24 ± 30.66c |
| D178E | 3 | 0.50 ± 0.13 | 64 ± 14 | 3.84 ± 1.79 |
| E221D | 3 | 0.65 ± 0.28 | 135 ± 53 | 5.30 ± 1.79 |
| E221Q | 3 | 0.58 ± 0.02 | 215 ± 58 | 5.25 ± 2.45 |
| D332Q | 3 | 16.99 ± 4.83a | 9 ± 1 c | 8.24 ± 1.85 |
significantly different from WT hMC3R, p<0.05
significantly different from WT hMC3R, p<0.01
significantly different from WT hMC3R, p<0.001
The data are expressed as the mean ± SEM of three or four independent experiments. The maximal response under d-Trp8-γ-MSH stimulation was 2162 ± 338 pmol cAMP/106 cells for WT hMC3R.
3.4. Ligand binding and signaling properties of the mutant hMC3Rs using α- or γ-MSH as the ligand
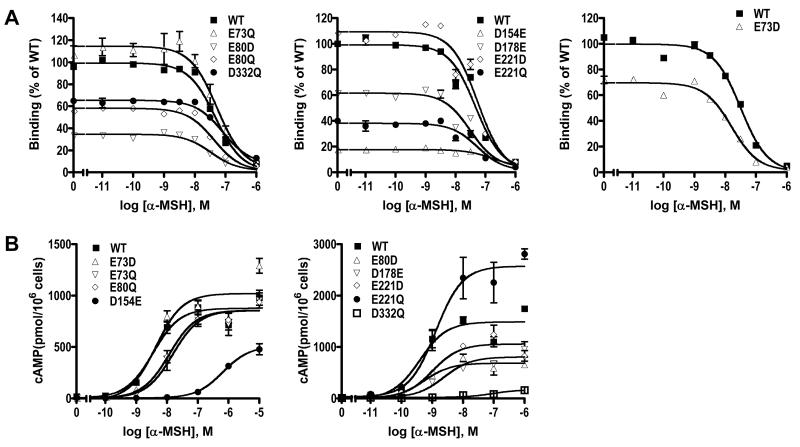
We also studied the binding and signaling properties of the nine mutants with either α- or γ- MSH used as the ligand. With α-MSH as the ligand, we showed that except for D154E, which showed minimal binding, and D332Q, the other seven mutants bind to α-MSH with similar IC50 as the WT hMC3R (Fig. 5 and Table 3). In fact, the IC50 for D178E was significantly decreased compared with WT hMC3R. In signaling assays, D154E and D332Q showed increased EC50 as compared to the WT hMC3R. The other seven mutants had similar EC50 as the WT hMC3R, although the Emax for E80D, D154E, D178E, and D332Q were significantly decreased (Fig. 5 and Table 3).
Figure 5.
Ligand binding and signaling properties of the WT and mutant hMC3Rs with α-MSH as the ligand. HEK293T cells were transiently transfected with the indicated hMC3R constructs and binding and signaling properties of the hMC3Rs were measured as described in Materials and Methods. In A, different concentrations of unlabeled α-MSH were used to displace the binding of 125I-NDP-MSH to hMC3Rs on intact cells. Results shown are expressed as % of WT binding ± range from duplicate determinations within one experiment. In B, HEK293T cells transiently transfected with the indicated hMC3R constructs were stimulated with different concentrations of α-MSH. Intracellular cAMP levels were measured using radioimmunoassay. Results are expressed as the mean ± SEM of triplicate determinations within one experiment. All experiments were performed at least three times (see Table 3 for the number of experiments done for each hMC3R).
Table 3.
Ligand Binding and α-MSH-Stimulated cAMP Production of WT and Mutant hMC3Rs
| hMC3R | n | 〈-MSH-stimulated cAMP |
〈-MSH binding |
|
|---|---|---|---|---|
| EC50 (nM) | Emax (% WT) | IC50 (nM) | ||
| WT | 9 | 5.39 ± 1.85 | 100 | 61.18 ± 6.90 |
| E73D | 4 | 3.64 ± 0.33 | 121 ± 24 | 58.62 ± 8.59 |
| E73Q | 4 | 8.35 ± 3.59 | 110 ± 12 | 78.25 ± 13.27 |
| E80D | 4 | 4.21 ± 0.73 | 38 ± 4b | 31.87 ± 6.75 |
| E80Q | 4 | 11.13 ± 4.69 | 89 ± 5 | 43.56 ± 4.34 |
| D154E | 3 | 107.60 ± 53.77 | 37 ± 17 | 592.53 ± 249.75 |
| D178E | 4 | 16.42 ± 5.35 | 44 ± 9a | 39.06 ± 3.04a |
| E221D | 4 | 5.03 ± 2.58 | 80 ± 25 | 48.95 ± 4.67 |
| E221Q | 5 | 13.16 ± 5.34 | 109 ± 38 | 66.49 ± 9.79 |
| D332Q | 5 | 49.09 ± 19.65 | 14 ± 3c | 178.34 ± 39.62a |
significantly different from WT hMC3R, p<0.05
significantly different from WT hMC3R, p<0.01
significantly different from WT hMC3R, p<0.001
The data are expressed as the mean ± SEM of three to five independent experiments. The maximal response under 〈-MSH stimulation was 1356 ± 180 pmol cAMP/106 cells for WT hMC3R.
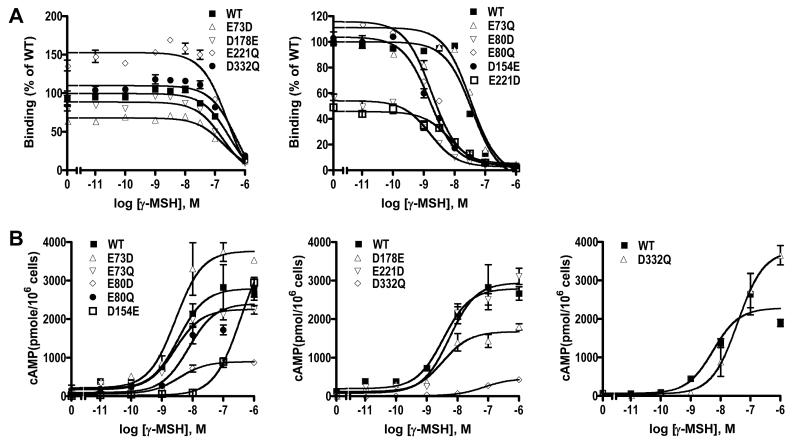
We also measured the ligand binding and signaling responses to γ-MSH stimulation. Our results showed that no binding and signaling could be detected for D154E. The other eight mutants had similar IC50 as the WT hMC3R (Fig. 6 and Table 4). The signaling of E73D, E73Q, E80D, E80Q, and E221Q, were normal. The EC50 for D178E and E221Q were increased. Although D332Q had binding to γ-MSH, no signaling could be measured (Fig. 6 and Table 4).
Figure 6.
Ligand binding and signaling properties of the WT and mutant hMC3Rs with γ-MSH as the ligand. HEK293T cells were transiently transfected with the indicated hMC3R constructs and binding and signaling properties of the hMC3Rs were measured as described in Materials and Methods. In A, different concentrations of unlabeled γ-MSH were used to displace the binding of 125I-NDP-MSH to hMC3Rs on intact cells. Results shown are expressed as % of WT binding ± range from duplicate determinations within one experiment. In B, HEK293T cells transiently transfected with the indicated hMC3R constructs were stimulated with different concentrations of γ-MSH. Intracellular cAMP levels were measured using radioimmunoassay. Results are expressed as the mean ± SEM of triplicate determinations within one experiment. All experiments were performed at least three times (see Table 4 for the number of experiments done for each hMC3R).
Table 4.
Ligand Binding and γ-MSH-Stimulated cAMP Production of WT and Mutant hMC3Rs
| hMC3R | n | γ-MSH-stimulated cAMP |
γ-MSH binding |
|
|---|---|---|---|---|
| EC50 (nM) | Emax (% WT) | IC50 (nM) | ||
| WT | 5 | 37.35 ± 9.55 | 100 | 218.90 ± 46.59 |
| E73D | 4 | 40.08 ± 10.48 | 164 ± 1 c | 197.48 ± 17.71 |
| E73Q | 3 | 43.38 ± 12.39 | 116 ± 18 | 212.77 ± 100.42 |
| E80D | 4 | 71.63 ± 13.02 | 133 ± 56 | 128.13 ± 35.67 |
| E80Q | 4 | 64.00 ± 23.44 | 123 ± 24 | 254.88 ± 51.84 |
| D154E | 3 | ND* | ND* | ND* |
| D178E | 4 | 171.56 ± 44.45 | 85 ± 20 | 169.57 ± 30.49 |
| E221D | 4 | 141.90 ± 86.46 | 119 ± 33 | 190.94 ± 46.32 |
| E221Q | 3 | 260.67 ± 118.50 | 222 ± 71 | 217.90 ± 35.11 |
| D332Q | 3 | ND* | ND* | 397.10 ± 156.45 |
ND, could not be detected.
significantly different from WT hMC3R, p<0.05
significantly different from WT hMC3R, p<0.01
significantly different from WT hMC3R, p<0.001
The data are expressed as the mean ± SEM of 3-5 independent experiments. The maximal response under γ-MSH stimulation was 1324 ± 309 pmol cAMP/106 cells for WT hMC3R.
4. Discussion
Extensive studies of the structure-activity relationships of the melanocortins have revealed the pharmacophore His-Phe-Arg-Trp (reviewed in [35]). Site-directed mutagenesis and molecular modeling of the MC4R revealed two types of interactions between the agonist and the receptor: ionic interaction between the positively charged residues in the ligand and the acidic residues in TMs 2 and 3 of the receptor, and the hydrophobic interaction between the aromatic residues in the ligand and aromatic and hydrophobic residues in TMs 4, 6 and 7 [26, 27, 36].
We hypothesized that the acidic residues in the TMs of the MC3R also form ionic interactions with the basic residues in the agonists. Therefore, in this study, we investigated the functions of all ten acidic residues in the TMs of the MC3R. Some of these residues are predicted to be important in ligand binding based on previous studies in other MCRs, whereas some of these residues are also predicted to be important in signaling based on studies in GPCRs in general. For example, D121 (2.50) is the most conserved residue in TM2. Extensive studies in other GPCRs have shown that this Asp is in close proximity with and may form hydrogen bonds with N7.49 in TM7 bridged by water molecules [37-40]. Mutations of this Asp frequently result in a defect in receptor-G protein coupling [40-43], including the MC4R ([44] and our unpublished observations). Recent pioneering structural studies in β2-adrenergic receptor showed that water-mediated hydrogen bonds link D2.50 with N7.49, N1.50, as well as other residues [45]. D178 (3.49) belongs to the highly conserved DRY motif in Family A GPCRs. From the studies on rhodopsin and adrenergic receptors, it was suggested that the Asp (3.49) in the DRY motif (Glu in rhodopsin) becomes protonated upon G protein binding and receptor activation (see [46] and references therein) therefore acting as a molecular switch that activates the G protein. Similarly, D332 (7.49) belongs to the highly conserved N/DPxxY motif. Mutations in this motif affect receptor expression, trafficking, ligand affinity, G protein coupling, and association with small G proteins [47-50]. Therefore, mutagenesis studies with these residues should also reveal their roles in signaling.
Flow cytometry showed that all mutants (except one we could not test; we could not establish the stable cell line expressing D332E MC3R) were expressed on the cell surface. Cell surface expression of E80D, E92D, and D158E were significantly decreased compared with WT hMC3R. Binding and signaling defects of these mutants could be due, at least partially, to the decreased cell surface expression. D154Q had significant increase in cell surface expression. The expressions of the other mutants were not significantly different from WT receptor expression (Fig. 3).
Since all mutants examined were expressed on the cell surface, we measured their ligand binding and signaling properties. We first used the superpotent analog NDP-MSH. These data are shown in Fig. 2 and Table 1. A careful inspection of the ligand binding and signaling of the mutants with the cell surface expression revealed that E92, D121, E131, D158, and D332 are important for ligand binding and signaling. Both mutations at these loci resulted in defective ligand binding and signaling. Mutations at E80 caused decreased binding and signaling. Mutations at D154 primarily affect ligand binding: the maximal binding of D154E was estimated to be about 20% of WT hMC3R with similar IC50 whereas binding of D154Q could not be detected. Both D154E and D154Q had similar maximal responses as WT hMC3R, although the EC50 for D154Q was increased 136-fold. Similar phenomenon has been observed before [30, 51-53]. We suggest that this might be due to the presence of spare receptor in this transient expression system.
Comparison of the two mutations at the same locus revealed a few interesting observations. For example, at D154, no binding could be measured with D154Q whereas some binding could be measured with D154E, suggesting that the negative charge is important for binding to NDP- MSH. Similar observations were made at D178. However, at D332, the size rather than the charge might be more important for binding to NDP-MSH. D332Q could bind to NDP-MSH whereas there was no binding for D332E, which could be due to the lack of expression at the cell surface. We were unable to establish cells stably expressing D332E mutant for flow cytometry.
Comparison of ligand binding and signaling showed that D121 and D332 are important for both ligand binding and signaling. Mutants D121Q and D332Q could bind to NDP-MSH but no signaling could be measured. These data are consistent with previous observations in other GPCRs (see above).
For the mutants that showed binding and signaling to NDP-MSH, we further studied their ligand binding and signaling properties to other ligands, including d-Trp8-γ-MSH, α-MSH, and γ-MSH. A total of nine mutants were chosen for these experiments. d-Trp8-γ-MSH is a superpotent ligand discovered by Hruby and colleagues [34]. In ligand binding experiments with d-Trp8-γ-MSH as the competitor, the IC50 of D154E was increased 17-fold. The other mutants had similar IC50 as the WT MC3R. The EC50 of D154E was also dramatically increased (335-fold) (Fig. 4 and Table 2). When the natural ligand γ-MSH was used as the ligand, no measurable ligand binding and signaling could be measured. No signaling could be measured with D332Q after γ-MSH stimulation (Fig. 6 and Table 4). When α-MSH was used to stimulate the nine mutants, D154E and D332Q had increased EC50 (Fig. 5 and Table 3).
The data obtained with D332Q showed that the MC3R could exist in different conformations after binding to different ligands. The mutant showed no signaling activity to NDP- and γ-MSH stimulations whereas some signaling could be measured with d-Trp8-γ-MSH and α-MSH stimulations. In studies with M1 muscarinic receptor, distinct modes of receptor activation were also observed when several ligands were used to study the mutant receptors [54]. Similarly, in gonadotropin-releasing hormone (GnRH) receptor, differential binding of GnRH I and II to the human GnRH receptor was shown to either selectively stabilize or induce different active conformations [55].
Previously, Yang and colleagues performed alanine mutagenesis with some of the residues reported in this study [56]. Although we made more conservative mutations, similar conclusions were reached in the two studies. For example, both studies showed that D121 and D332 are important for signaling. Therefore our data with more conservative changes further support the importance of negative charge in these residues for ligand binding. By using multiple ligands, we showed that D332Q could respond to d-Trp8-γ-MSH and α-MSH to a small degree (Figs. 3 and 5, Tables 2 and 3).
The mutations that affect ligand binding can reside deep in the transmembrane domain. For example, E92 is close to the intracellular surface. Both E92D and E92Q are expressed on the cell surface but no ligand binding could be determined (Fig. 2 and Table 1). Similar observations were obtained before. In the GnRH receptor, mutations in the cytoplasmic end of TMs were found to affect ligand binding [55]. These mutations likely affect the conformation of the receptor rather than directly disrupt ligand-receptor interaction. Further studies using other techniques such as affinity cross-linking are needed to address whether the mutants shown herein that affect binding disrupt the binding pocket(s) per se or cause conformational change that indirectly affect ligand binding.
In summary, the results presented showed that acidic residues in TMs 1 and 3 are important for ligand binding whereas those in TMs 2 and 7 are important for both binding and signaling. These are important constraints for modeling ligand binding and G protein coupling/activation in the MC3R.
Acknowledgements
This study was supported by NIH grant R15DK077213 and intramural grants from Auburn University (Biogrant and Animal Health and Disease Research Program at College of Veterinary Medicine at Auburn University). We thank the Flow Cytometry Core Facility at Auburn University College of Veterinary Medicine (Dr. R. C. Bird, director) for their assistance in the flow cytometry experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
None.
References
- 1.Gantz I, Konda Y, Tashiro T, Shimoto Y, Miwa H, Munzert G, et al. Molecular cloning of a novel melanocortin receptor. J Biol Chem. 1993;268:8246–50. [PubMed] [Google Scholar]
- 2.Roselli-Rehfuss L, Mountjoy KG, Robbins LS, Mortrud MT, Low MJ, Tatro JB, et al. Identification of a receptor for © melanotropin and other proopiomelanocortin peptides in the hypothalamus and limbic system. Proc Natl Acad Sci U S A. 1993;90:8856–60. doi: 10.1073/pnas.90.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Getting SJ, Christian HC, Lam CW, Gavins FN, Flower RJ, Schioth HB, et al. Redundancy of a functional melanocortin 1 receptor in the anti-inflammatory actions of melanocortin peptides: studies in the recessive yellow (e/e) mouse suggest an important role for melanocortin 3 receptor. J Immunol. 2003;170:3323–30. doi: 10.4049/jimmunol.170.6.3323. [DOI] [PubMed] [Google Scholar]
- 4.Getting SJ, Di Filippo C, Christian HC, Lam CW, Rossi F, D'Amico M, et al. MC-3 receptor and the inflammatory mechanisms activated in acute myocardial infarct. J Leukoc Biol. 2004;76:845–53. doi: 10.1189/jlb.0306175. [DOI] [PubMed] [Google Scholar]
- 5.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan XM, Yu H, et al. Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet. 2000;26:97–102. doi: 10.1038/79254. [DOI] [PubMed] [Google Scholar]
- 7.Butler AA, Kesterson RA, Khong K, Cullen MJ, Pelleymounter MA, Dekoning J, et al. A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology. 2000;141:3518–21. doi: 10.1210/endo.141.9.7791. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Kilroy GE, Henagan TM, Prpic-Uhing V, Richards WG, Bannon AW, et al. Targeted deletion of melanocortin receptor subtypes 3 and 4, but not CART, alters nutrient partitioning and compromises behavioral and metabolic responses to leptin. FASEB J. 2005;19:1482–91. doi: 10.1096/fj.05-3851com. [DOI] [PubMed] [Google Scholar]
- 9.Marks DL, Hruby V, Brookhart G, Cone RD. The regulation of food intake by selective stimulation of the type 3 melanocortin receptor (MC3R) Peptides. 2006;27:259–64. doi: 10.1016/j.peptides.2005.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M, Kim A, Conwell IM, Hruby V, Mayorov A, Cai M, et al. Effects of selective modulation of the central melanocortin-3-receptor on food intake and hypothalamic POMC expression. Peptides. 2008 doi: 10.1016/j.peptides.2007.11.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hani EH, Dupont S, Durand E, Dina C, Gallina S, Gantz I, et al. Naturally occurring mutations in the melanocortin receptor 3 gene are not associated with type 2 diabetes mellitus in French Caucasians. J Clin Endocrinol Metab. 2001;86:2895–8. doi: 10.1210/jcem.86.6.7589. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Poh LK, Kek BL, Loke KY. The role of melanocortin 3 receptor gene in childhood obesity. Diabetes. 2007;56:2622–30. doi: 10.2337/db07-0225. [DOI] [PubMed] [Google Scholar]
- 13.Lee YS, Poh LK, Loke KY. A novel melanocortin 3 receptor gene (MC3R) mutation associated with severe obesity. J Clin Endocrinol Metab. 2002;87:1423–6. doi: 10.1210/jcem.87.3.8461. [DOI] [PubMed] [Google Scholar]
- 14.Tao YX. Functional characterization of novel melanocortin-3 receptor mutations identified from obese subjects. Biochim Biophys Acta. 2007;1772:1167–74. doi: 10.1016/j.bbadis.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Mencarelli M, Walker GE, Maestrini S, Alberti L, Verti B, Brunani A, et al. Sporadic mutations in melanocortin receptor 3 in morbid obese individuals. Eur J Hum Genet. 2008;16:581–6. doi: 10.1038/sj.ejhg.5202005. [DOI] [PubMed] [Google Scholar]
- 16.Tao YX, Segaloff DL. Functional characterization of melanocortin-3 receptor variants identify a loss-of-function mutation involving an amino acid critical for G protein-coupled receptor activation. J Clin Endocrinol Metab. 2004;89:3936–42. doi: 10.1210/jc.2004-0367. [DOI] [PubMed] [Google Scholar]
- 17.Rached M, Buronfosse A, Begeot M, Penhoat A. Inactivation and intracellular retention of the human I183N mutated melanocortin 3 receptor associated with obesity. Biochim Biophys Acta. 2004;1689:229–34. doi: 10.1016/j.bbadis.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Feng N, Young SF, Aguilera G, Puricelli E, Adler-Wailes DC, Sebring NG, et al. Co-occurrence of two partially inactivating polymorphisms of MC3R is associated with pediatric-onset obesity. Diabetes. 2005;54:2663–7. doi: 10.2337/diabetes.54.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoro N, Perrone L, Cirillo G, Raimondo P, Amato A, Brienza C, et al. Effect of the melanocortin-3 receptor C17A and G241A variants on weight loss in childhood obesity. Am J Clin Nutr. 2007;85:950–3. doi: 10.1093/ajcn/85.4.950. [DOI] [PubMed] [Google Scholar]
- 20.Rutanen J, Pihlajamaki J, Vanttinen M, Salmenniemi U, Ruotsalainen E, Kuulasmaa T, et al. Single nucleotide polymorphisms of the melanocortin-3 receptor gene are associated with substrate oxidation and first-phase insulin secretion in offspring of type 2 diabetic subjects. J Clin Endocrinol Metab. 2007;92:1112–7. doi: 10.1210/jc.2006-1201. [DOI] [PubMed] [Google Scholar]
- 21.Sutton GM, Trevaskis JL, Hulver MW, McMillan RP, Markward NJ, Babin MJ, et al. Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or -4 receptors. Endocrinology. 2006;147:2183–96. doi: 10.1210/en.2005-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Versteeg DH, Van Bergen P, Adan RA, De Wildt DJ. Melanocortins and cardiovascular regulation. Eur J Pharmacol. 1998;360:1–14. doi: 10.1016/s0014-2999(98)00615-3. [DOI] [PubMed] [Google Scholar]
- 23.Ni XP, Pearce D, Butler AA, Cone RD, Humphreys MH. Genetic disruption of ©-melanocyte-stimulating hormone signaling leads to salt-sensitive hypertension in the mouse. J Clin Invest. 2003;111:1251–8. doi: 10.1172/JCI16993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphreys MH. ©-MSH, sodium metabolism, and salt-sensitive hypertension. Am J Physiol. 2004;286:R417–R30. doi: 10.1152/ajpregu.00365.2003. [DOI] [PubMed] [Google Scholar]
- 25.Getting SJ, Lam CW, Leoni G, Gavins FN, Grieco P, Perretti M. [D-Trp8]-gamma-melanocyte-stimulating hormone exhibits anti-inflammatory efficacy in mice bearing a nonfunctional MC1R (recessive yellow e/e mouse) Mol Pharmacol. 2006;70:1850–5. doi: 10.1124/mol.106.028878. [DOI] [PubMed] [Google Scholar]
- 26.Yang YK, Fong TM, Dickinson CJ, Mao C, Li JY, Tota MR, et al. Molecular determinants of ligand binding to the human melanocortin-4 receptor. Biochemistry. 2000;39:14900–11. doi: 10.1021/bi001684q. [DOI] [PubMed] [Google Scholar]
- 27.Haskell-Luevano C, Cone RD, Monck EK, Wan YP. Structure activity studies of the melanocortin-4 receptor by in vitro mutagenesis: identification of agouti-related protein (AGRP), melanocortin agonist and synthetic peptide antagonist interaction determinants. Biochemistry. 2001;40:6164–79. doi: 10.1021/bi010025q. [DOI] [PubMed] [Google Scholar]
- 28.Lee EJ, Lee SH, Jung JW, Lee W, Kim BJ, Park KW, et al. Differential regulation of cAMP-mediated gene transcription and ligand selectivity by MC3R and MC4R melanocortin receptors. Eur J Biochem. 2001;268:582–91. doi: 10.1046/j.1432-1327.2001.01900.x. [DOI] [PubMed] [Google Scholar]
- 29.Hruby VJ, Wilkes BC, Hadley ME, Al-Obeidi F, Sawyer TK, Staples DJ, et al. 〈-Melanotropin: the minimal active sequence in the frog skin bioassay. J Med Chem. 1987;30:2126–30. doi: 10.1021/jm00394a033. [DOI] [PubMed] [Google Scholar]
- 30.Tao YX, Segaloff DL. Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology. 2003;144:4544–51. doi: 10.1210/en.2003-0524. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–52. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan ZC, Sartin JL, Tao YX. Molecular cloning and pharmacological characterization of porcine melanocortin-3 receptor. J Endocrinol. 2008;196:139–48. doi: 10.1677/JOE-07-0403. [DOI] [PubMed] [Google Scholar]
- 33.Tao YX, Johnson NB, Segaloff DL. Constitutive and agonist-dependent self-association of the cell surface human lutropin receptor. J Biol Chem. 2004;279:5904–14. doi: 10.1074/jbc.M311162200. [DOI] [PubMed] [Google Scholar]
- 34.Grieco P, Balse PM, Weinberg D, MacNeil T, Hruby VJ. D-Amino acid scan of ©-melanocyte-stimulating hormone: importance of Trp8 on human MC3 receptor selectivity. J Med Chem. 2000;43:4998–5002. doi: 10.1021/jm000211e. [DOI] [PubMed] [Google Scholar]
- 35.Holder JR, Haskell-Luevano C. Melanocortin ligands: 30 years of structure-activity relationship (SAR) studies. Med Res Rev. 2004;24:325–56. doi: 10.1002/med.10064. [DOI] [PubMed] [Google Scholar]
- 36.Nickolls SA, Cismowski MI, Wang X, Wolff M, Conlon PJ, Maki RA. Molecular determinants of melanocortin 4 receptor ligand binding and MC4/MC3 receptor selectivity. J Pharmacol Exp Ther. 2003;304:1217–27. doi: 10.1124/jpet.102.044974. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Flanagan C, Ballesteros JA, Konvicka K, Davidson JS, Weinstein H, et al. A reciprocal mutation supports helix 2 and helix 7 proximity in the gonadotropin-releasing hormone receptor. Mol Pharmacol. 1994;45:165–70. [PubMed] [Google Scholar]
- 38.Wilson MH, Highfield HA, Limbird LE. The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol Pharmacol. 2001;59:929–38. doi: 10.1124/mol.59.4.929. [DOI] [PubMed] [Google Scholar]
- 39.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 40.Bee MS, Hulme EC. Functional analysis of transmembrane domain 2 of the M1 muscarinic acetylcholine receptor. J Biol Chem. 2007;282:32471–9. doi: 10.1074/jbc.M703909200. [DOI] [PubMed] [Google Scholar]
- 41.Ceresa BP, Limbird LE. Mutation of an aspartate residue highly conserved among G-protein- coupled receptors results in nonreciprocal disruption of α2-adrenergic receptor-G-protein interactions. J Biol Chem. 1994;269:29557–64. [PubMed] [Google Scholar]
- 42.Tao Q, Abood ME. Mutation of a highly conserved aspartate residue in the second transmembrane domain of the cannabinoid receptors, CB1 and CB2, disrupts G-protein coupling. J Pharmacol Exp Ther. 1998;285:651–8. [PubMed] [Google Scholar]
- 43.Roche JP, Bounds S, Brown S, Mackie K. A mutation in the second transmembrane region of the CB1 receptor selectively disrupts G protein signaling and prevents receptor internalization. Mol Pharmacol. 1999;56:611–8. doi: 10.1124/mol.56.3.611. [DOI] [PubMed] [Google Scholar]
- 44.Biebermann H, Krude H, Elsner A, Chubanov V, Gudermann T, Gruters A. Autosomal-dominant mode of inheritance of a melanocortin-4 receptor mutation in a patient with severe early-onset obesity is due to a dominant-negative effect caused by receptor dimerization. Diabetes. 2003;52:2984–8. doi: 10.2337/diabetes.52.12.2984. [DOI] [PubMed] [Google Scholar]
- 45.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–673. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 46.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 47.Barak LS, Menard L, Ferguson SS, Colapietro AM, Caron MG. The conserved seven-transmembrane sequence NP(X)2,3Y of the G-protein-coupled receptor superfamily regulates multiple properties of the β2-adrenergic receptor. Biochemistry. 1995;34:15407–14. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 48.Hunyady L, Bor M, Baukal AJ, Balla T, Catt KJ. A conserved NPLFY sequence contributes to agonist binding and signal transduction but is not an internalization signal for the type 1 angiotensin II receptor. J Biol Chem. 1995;270:16602–9. doi: 10.1074/jbc.270.28.16602. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell R, McCulloch D, Lutz E, Johnson M, MacKenzie C, Fennell M, et al. Rhodopsin-family receptors associate with small G proteins to activate phospholipase D. Nature. 1998;392:411–4. doi: 10.1038/32937. [DOI] [PubMed] [Google Scholar]
- 50.Prioleau C, Visiers I, Ebersole BJ, Weinstein H, Sealfon SC. Conserved helix 7 tyrosine acts as a multistate conformational switch in the 5HT2C receptor. Identification of a novel “locked-on” phenotype and double revertant mutations. J Biol Chem. 2002;277:36577–84. doi: 10.1074/jbc.M206223200. [DOI] [PubMed] [Google Scholar]
- 51.Tao YX. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol Cell Endocrinol. 2005;239:1–14. doi: 10.1016/j.mce.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Tao YX, Segaloff DL. Functional analyses of melanocortin-4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J Clin Endocrinol Metab. 2005;90:5632–8. doi: 10.1210/jc.2005-0519. [DOI] [PubMed] [Google Scholar]
- 53.Tao YX. Inactivating mutations of G protein-coupled receptors and diseases: Structure-function insights and therapeutic implications. Pharmacol Ther. 2006;111:949–73. doi: 10.1016/j.pharmthera.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Spalding TA, Ma JN, Ott TR, Friberg M, Bajpai A, Bradley SR, et al. Structural requirements of transmembrane domain 3 for activation by the M1 muscarinic receptor agonists AC-42, AC-260584, clozapine, and N-desmethylclozapine: evidence for three distinct modes of receptor activation. Mol Pharmacol. 2006;70:1974–83. doi: 10.1124/mol.106.024901. [DOI] [PubMed] [Google Scholar]
- 55.Lu ZL, Gallagher R, Sellar R, Coetsee M, Millar RP. Mutations remote from the human gonadotropin-releasing hormone (GnRH) receptor-binding sites specifically increase binding affinity for GnRH II but not GnRH I: evidence for ligand-selective, receptor-active conformations. J Biol Chem. 2005;280:29796–803. doi: 10.1074/jbc.M413520200. [DOI] [PubMed] [Google Scholar]
- 56.Chen M, Aprahamian CJ, Celik A, Georgeson KE, Garvey WT, Harmon CM, et al. Molecular characterization of human melanocortin-3 receptor ligand-receptor interaction. Biochemistry. 2006;45:1128–37. doi: 10.1021/bi0521792. [DOI] [PubMed] [Google Scholar]