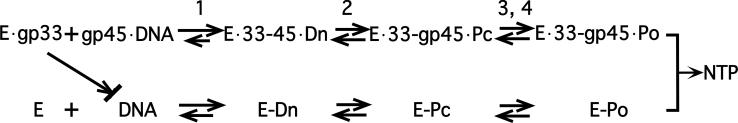Abstract
Activated transcription of the bacteriophage T4 late genes is generated by a mechanism that stands apart from the common modalities of transcriptional regulation: the activator gp45 is the viral replisome’s sliding clamp; two sliding clamp-binding proteins, gp33 and gp55, replace the host RNA polymerase (RNAP) σ subunit. We have mutagenized, reconfigured and selectively disrupted individual interactions of the sliding clamp with gp33 and gp55 and have monitored effects on transcription. The C-terminal sliding clamp-binding epitopes of gp33 and gp55 are perfectly interchangeable, but the functions of these two RNAP-sliding clamp connections differ, with only the gp33-gp45 linkage essential for activation. Formation of transcription-ready promoter complexes by the sliding clamp-activated wild-type T4 RNAP resists competition by high concentrations of the polyanion heparin. This avid formation of promoter complexes requires both linkages of the T4 late RNAP to the sliding clamp. Preopening the promoter compensates for loss of the gp55-gp45 but not the gp33-gp45 linkage. We interpret these findings in relation to the common model of transcriptional initiation in bacteria and highlight the roles of individual interactions of the promoter complex.
Keywords: RNA polymerase, promoter, gene regulation, phage T4, sliding clamp
Introduction
Interaction of RNA polymerase (RNAP) with promoters is a multistep process that includes selection of a promoter among other sequences, promoter binding and, ultimately, formation of the transcriptionally competent, open complex (Record et al, 1996). Most bacterial promoters are defined by two short sequence motifs positioned 35 and 10 base pairs (bp) upstream of the start point of transcription. Promoters are recognized by the promoter specificity subunit (s) of the RNA polymerase holoenzyme through sequence-specific interactions of s domain 4 (bound to the flap of the RNAP b subunit) with the -35 motif, and domain 2 (bound to the RNAP b’ subunit coiled coil) with the -10 motif. Additional interactions, of the two RNAP a subunit C-terminal domains (aCTDs) with DNA upstream of position ∼-40, and of other regions of s with DNA near the start point of transcription, influence promoter strength and play important regulatory roles (Haugen et al, 2006; Ross et al, 1993; Wilson and Dombroski, 1997; Zenkin et al, 2007).
Transcription of the bacteriophage T4 late genes is regulated by a mechanism that stands apart from the known modalities of transcriptional regulation. First, the late genes are served by promoters consisting solely of an 8-bp motif (consensus sequence TATAAATA) centered 10 bp upstream of the transcriptional start site. Transcription initiating at the ∼40 T4 late promoters utilizes the host cell’s RNAP core, modified (Walter et al, 1968) to inactivate interaction of its two αCTDs with DNA (by ADP-ribosylation of Arg265 in the DNA-presentation helix) and bearing two small phage-encoded subunits, the gene 33 and gene 55 products (gp33 and gp55).
Second, the RNAP β’ coiled coil and β flap of the T4 late holoenzyme are occupied by separate proteins: gp55 and gp33 bind to the RNAP core at sites that are the principal attachment points of σ70 domain 2 (Wong et al, 2003) and domain 4 (Nechaev et al, 2004), respectively. Gp55 is the promoter recognition subunit of T4 late transcription and gp55-RNAP holoenzyme suffices for accurately initiating basal late transcription in vitro (Kassavetis and Geiduschek, 1984); a limited similarity of gp55 residues ∼42-122 with σ70 domain 2 can be discerned (Gribskov and Burgess, 1986). Gp33 is the essential coactivator (Herendeen et al, 1990) of the high-level T4 late gene transcription that is required for producing the massive quantities of structural proteins that assemble into progeny virions during the final minutes of the phage multiplication cycle (Epstein et al, 1963). There is no similarity of amino acid sequence between gp33 and σ70 domain 4.
Third, T4 late transcription in vivo is coupled to concurrent DNA replication through a unique transcriptional activator, the DNA-loaded sliding clamp, gp45 (Herendeen et al, 1992; Tinker et al, 1994a; Tinker et al, 1994b). Gp45, a head-to-tail trimer with a central annulus that is lined with positively charged side chains and accommodates double-stranded DNA, interacts with the similar acidic and hydrophobic C-terminal motifs of its ligands—gp55, gp33 and the T4 DNA polymerase gp43 (Figure 1A) (Moarefi et al, 2000; Shamoo and Steitz, 1999; Trakselis et al, 2001). Gp45, the processivity factor of the T4 DNA polymerase, is loaded onto DNA at primer-template junctions by its conjugate clamp loader, the T4 gp44/gp62 complex. Because the DNA-mounted state of gp45 is transient (Fu et al, 1996), a supply of loading sites must be maintained for continual reattachment, which in turn requires continued DNA synthesis. This is the basis of the dependence of active T4 late transcription on concurrent DNA replication in vivo.
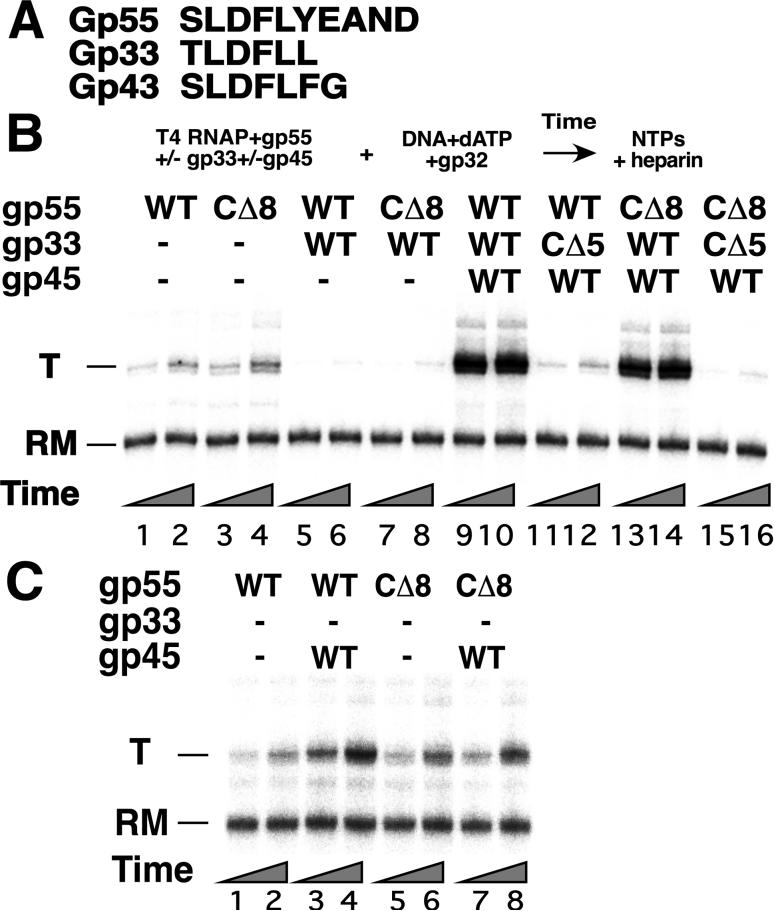
Figure 1. Interactions of the gp45 sliding clamp with gp55 and gp33 are functionally asymmetric.
A. Sequence alignment of the gp55, gp33 and gp43 C-terminal motifs. B. Basal, repressed and activated T4 late transcription of a 3′-end recessed template containing the T4 late promoter P23 with T4-modified RNAP core and gp55, gp33 and gp45, as indicated. Top: The reaction scheme: grouped components were premixed before combining, as indicated. The time allowed for promoter opening, indicated by the triangles below the lanes, was 5 and 15 min (odd- and even-numbered lanes, respectively). T: the principal transcript initiating at P23; RM: recovery marker. C. Gp45 is virtually ineffective in the absence of gp33. Transcription with the indicated components as specified in B.
In this work, we have selectively disrupted individual interactions of the gp45 sliding clamp with gp33 and gp55, and monitored the effects on transcription. Our findings allow us to interpret the mechanism of T4 late transcription in terms of the common model of transcriptional initiation in bacteria and to highlight the roles of individual interactions in the promoter complex.
Results
The functional asymmetry of gp55 and gp33 interactions with the sliding clamp
The gp45 sliding clamp binds to the T4 late promoter complex through interactions with the highly similar C-ends of gp55 and gp33 (Figure 1A). Similar acidic and hydrophobic motifs are located at the C-ends of gp55 homologues of other T4-family phages (with the possible exception of nt-1 and 65) (Comeau et al, 2007; Filée et al, 2006; Nolan et al, 2006; Petrov et al, 2006). Amino acid sequence is generally more divergent among putative gp33 homologues, especially at their N-ends but, with the exception of phages nt-1 and 65, they present similar hydrophobic and acidic 4-to 6-residue C-terminal elements (Supplemental Figure 1).
The first experiment examines the effect on transcription of eliminating either or both of these interactions with gp45 by removing the 5 and 8 C-terminal amino acids of gp33 and gp55, respectively (Figure 1B). Formation of open promoter complexes by T4-modified RNAP core (RNAPT4) and other specified components was quantified by single-round transcription assay (effectively, but see Figure 6 below) with simultaneously added heparin (to 100 μg/ml) and all 4 ribonucleoside triphosphates (NTPs). Gp45, gp55, and gp33 or deletion variants were present as noted at the top of panel B. All other components required for fully activated transcription (the T4 clamp loader; DNA with its primer-template junction clamp-loading site; and T4 single-stranded DNA-binding protein, gp32) were present throughout. In the absence of gp45, wild-type (WT) gp55 supported low-level transcription (Figure 1B, lanes 1 and 2) and removal of its C-terminal epitope did not affect this transcription (CD8, lanes 3 and 4). WT gp33 (lanes 5-8) and gp33CD5 (lanes 15, 16) inhibited this transcription, as expected (Nechaev et al, 2004; Nechaev and Geiduschek, 2006). In the presence of gp45, high-level transcription was observed with the wild-type components (lanes 9, 10) (Kolesky et al, 2002). Eliminating the gp55-gp45 interaction allowed transcription that was significantly above the basal level (compare lanes 13, 14 with lanes 1, 2), only moderately lower than fully activated transcription (lanes 9, 10) under these reaction conditions (see also Supplemental Figure 2). In contrast, eliminating the gp33-gp45 interaction essentially abolished activation (compare lanes 11, 12 with lanes 1, 2).
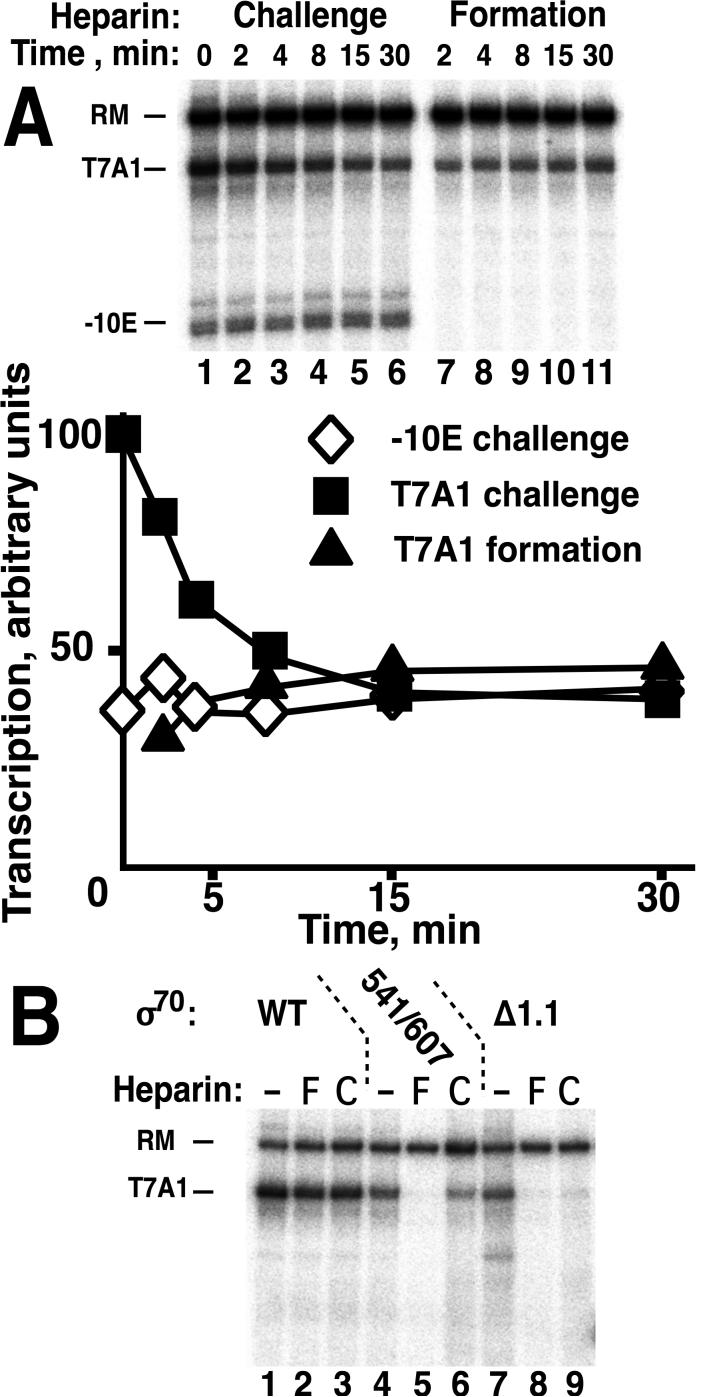
Figure 6. Formation of σ70 open promoter complexes in the presence of heparin.
A. Transcription with unmodified E. coli RNAP core and WT σ70 was carried out in reaction medium without PEG (as indicated in Figure 5 for formation and challenge of open complexes with heparin). DNA fragments containing T7A1 and -10Econ promoters were combined in equimolar amounts. Top: autoradiograph of a transcription gel. Bottom: quantification. B. Transcription with WT or mutant σ70 holoenzymes at the T7A1 promoter under conditions of promoter complex formation (F) in the presence of heparin or challenge (C), as above. R541C/L607P is the mutant in σ domain 4 that is defective in attachment to the RNAP β flap; Δ1.1 is σ (100-613), deleted for domain 1.1 only. Promoter opening time: 30 min, challenge: 15 min.
To determine the significance of the gp45-33 linkage in activation, we probed the ability of gp45 to activate transcription in reactions from which gp33 (and not just the gp45-33 connection) was absent altogether. In the absence of gp33, the gp55-gp45 interaction generated at most only a modest increase of transcription above the basal level (Figure 1C, compare lanes 3, 4 with lanes 1, 2; and compare lanes 7, 8 relative to lanes 5, 6 with lanes 3, 4 relative to lanes 1, 2), as also seen previously (Kolesky et al, 2002). In particular, promoter opening remained slow relative to fully activated transcription (Figures 1B, 1C, and data not shown). Evidently, activation of transcription by gp45 sliding clamp requires its intact connection with gp33 and does not take place in the absence of gp33.
Thus, the principal finding of this experiment is that eliminating the interactions of gp55 and gp33 with the sliding clamp has entirely different consequences, suggesting that these two linkages play different roles in transcriptional activation.
The C-terminal interaction motifs of gp33, gp55 and gp43 are interchangeable
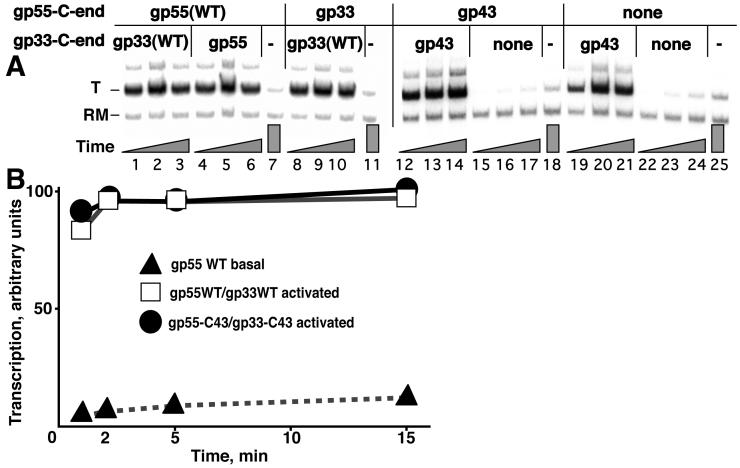
The similar C-terminal motifs of gp33, gp55 and gp43 are necessary for binding to gp45 in solution; the C-termini of gp55 as well as gp43 are also sufficient for gp45 binding (Alley et al, 1999; Wong and Geiduschek, 1998). To test the functional equivalence of these motifs, they were exchanged in gp55 and gp33 in all combinations and tested for compatibility with transcriptional activation. The results of one such experiment are shown in Figure 2A. Gp55 with its 8 C-terminal residues replaced by the 5-residue C-terminal motif of gp33 or gp43 was as competent for transcriptional activation as the wild-type protein (compare lanes 8-10 with lanes 1-3, and data not shown). Placing the identical C-terminal motif into gp33 and gp55 had no effect on transcriptional activation, and it did not matter whether the motif was taken from gp33, gp43 or gp55 (compare lanes 12-14 and 8-10 with lanes 1-3, and data not shown). Regardless of which C-terminal motif was present on gp55, the gp33-gp45 interaction was essential for activity (e.g., lanes 15-17) and placement of each C-terminal motif in gp33 allowed significant activation in the absence of the gp55-gp45 interaction (compare lanes 19-21 with lanes 1-3 and lane 25; Supplemental Figure 2).
Figure 2. The C-terminal motifs of gp55, gp33 and gp43 are interchangeable.
A. Activated transcription with unmodified E. coli RNAP core, gp45 and with gp55 and gp33 bearing the indicated C-terminal motifs; (-): no gp33 added. Time allowed for promoter opening (gray triangles): 2, 5 and 15 min. B. Time course of transcription with gp55 and gp33 wild-type (WT) or bearing the C-terminal motif of gp43. All components were added for activated transcription; gp33 was omitted for (effectively) basal transcription.
The replacement of both of the gp55 and gp33 C-terminal motifs by the gp43 motif is of special interest because the gp43-gp45 interaction has been extensively characterized (Trakselis et al, 2001). An experiment to compare promoter opening of this combination with the wild type is shown in Figure 2B. Substitution with the gp43 motif had no effect on transcriptional activation. The same result was observed under a variety of reaction conditions: with different proportions of polymerase to DNA, at the even lower temperature of 15°C, in reaction medium without polyethylene glycol (PEG) and with RNAPT4 as well as unmodified E. coli RNAP core (RNAPU). These results eliminate the possibility that differences between their C-termini direct gp33 and gp55 to different binding sites on the sliding clamp and also argue against the possibility that the different roles of gp55 and gp33 in transcriptional activation are specified by differing gp45 attachment sites.
Activation sustained solely by the gp33-gp45 interaction
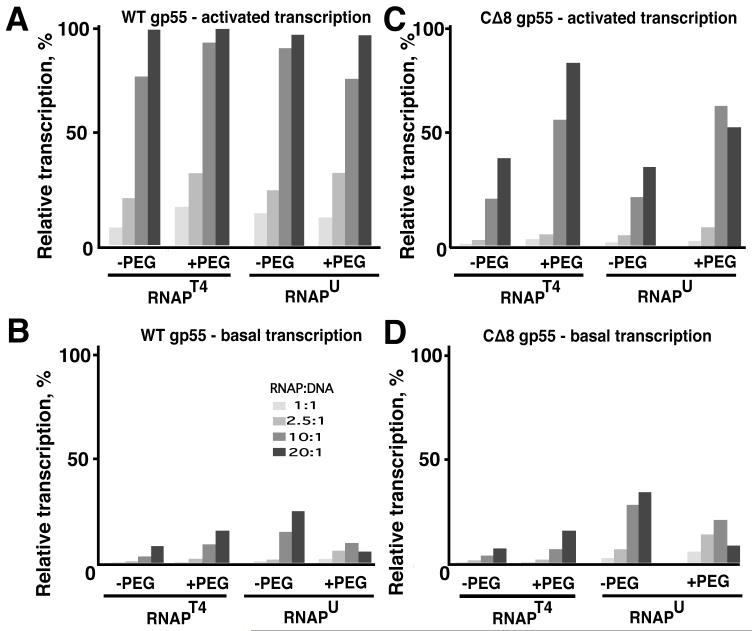
The characteristics of this “one-armed” activation were further examined in an experiment that exploited the following properties of T4 late transcription. First, fully activated transcription with wild-type gp55 and gp33 is robust, in the sense that it is unaffected by PEG and manifested with T4-modified as well as unmodified RNAP (RNAPT4 and RNAPU, respectively) (Figure 3A). Second, gp55-driven basal transcription (that is, in the absence of gp45) is markedly more active with RNAPU than with RNAPT4 (Figure 3B and data not shown). This is consistent with a significant contribution by the unmodified αCTD to basal transcriptional activity at this late promoter, which has very AT-rich segments at bp -51 to -37 and bp -82 to -74 (only 2 GC bp out of 15 and 1 out of 9, respectively), and elimination of that contribution when Arg265 in the αCTD is ADP-ribosylated, as it is in RNAPT4. Third, PEG specifically inhibits basal late transcription with RNAPU but stimulates basal transcription with RNAPT4 (Figure 3B). PEG also generally stimulates transcription by σ70-RNAPU at promoters commonly used by us (T7A1, λPR, galP1, lacUV5) under similar reaction conditions. While the exact mechanisms underlying these effects of PEG are not understood, its role as a macromolecular crowding agent suggests that gp55- and RNAPU-specific inhibition by PEG is due to fixing unmodified αCTDs to DNA or to the body of the gp55-RNAP in a conformation that prevents transcription. We note that nonspecific DNA binding by gp55-RNAPU is not impaired by PEG (Nechaev and Geiduschek, 2006), and data not shown).
Figure 3. Transcription in the absence of the gp45-gp55 interaction.
A.-D. Transcription with T4-modified (RNAPT4) or unmodified (RNAPU) core enzyme and the indicated gp55. All components were added for activated transcription; gp33 was omitted for basal transcription. Different molar proportions of RNAP to DNA were compared (1:1, 2.5:1, 10:1 and 20:1, in increasing order). Promoter opening time: 5 min. The reaction medium lacked PEG or contained 5% PEG 3350, as indicated.
With RNAPU core and in the absence of the gp55-gp45 interaction (CΔ8gp55 holoenzyme), the gp33-gp45 interaction increased transcription in PEG-containing reaction medium but had no effect on transcription in the absence of PEG (Figure 3 C, D and Supplemental Figure 3). Stated another way, when the αCTDs were functional for DNA binding (and correspondingly elevated promoter activity), the gp33-gp45 interaction was either nonfunctional or unable to further increase transcription. It is anticipated that the two transcription-elevating interactions, AT-rich upstream DNA with the αCTD and the sliding clamp with the promoter complex, are mutually exclusive because they compete for space on the same DNA segment. This finding suggests that both linkages of the sliding clamp to the RNAPU-promoter complex through the C-ends of gp55 and gp33 are required to exclude the αCTDs from the DNA segment that is occupied by gp45 (Tinker et al, 1994b) or, possibly, to prevent a gp45-αCTD clash that results in misaligning gp45 with the promoter complex. Of course, this clash does not materialize in the normal phage multiplication cycle because of the attendant αCTD modification.
Notably, sliding clamp activation of transcription sustained solely by the gp33-gp45 interaction was significantly lower than fully activated transcription (compare panels A and C), indicating the importance of both the gp45-gp55 and the gp45-gp33 interactions. We indicate below that the two interactions with the sliding clamp play nonoverlapping roles, but the data to this point make one distinction clear: barring interference by the αCTD, the gp33-sliding clamp linkage partially activates transcription by gp55-RNAP, while the gp55-sliding clamp linkage does not exert a significant effect. When there is interference by the αCTD, as with RNAPU in the absence of PEG, one-armed activation is ineffective and transcription does not rise above the basal level (Supplemental Figure 3).
Mutational analysis of the gp33 and gp55 linkages to the sliding clamp
The gp33 and gp55 segments that link the C-terminal epitopes with the rest of the protein are divergent, even among the T-even family phages that are capable of infecting E. coli (Supplemental Figure 1). Accordingly, they might be thought of as nonspecific linkers for attaching the sliding clamp to the late promoter complex. We have subjected these connector segments to mutagenesis. The original objective was to examine constraints on their lengths, but the analysis has yielded additional insights into their functional and structural characteristics. Effects of mutations on transcriptional activation were assessed, with RNAPU in PEG-containing reaction medium, by measuring the rate of promoter opening for single rounds of transcription (Figure 4 and Supplemental Figures 4 and 5). For certain gp33 mutants (Supplemental Figure 4), changes in the affinity for RNAP core were also assessed by measuring the dependence of repression of basal transcription (by RNAPU in reaction medium without PEG) on the concentration of gp33 (Nechaev et al, 2004). Gp55 linker mutants were assessed for basal transcription and found to be unaffected.
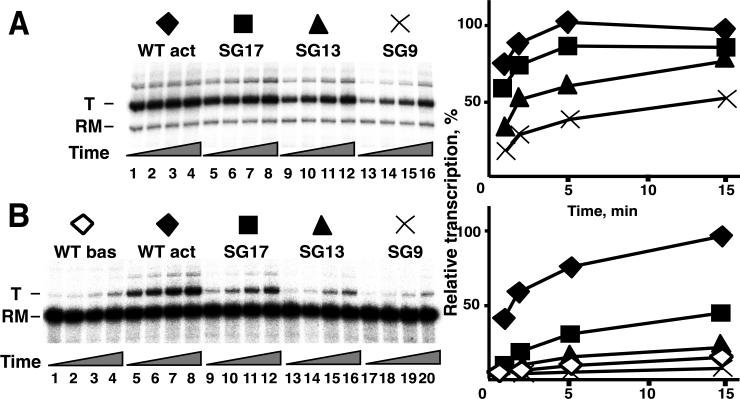
Figure 4. Activated transcription with variant gp55 in which the entire connector (amino acids 159-175) has been replaced by a flexible linker, and effects of shortening this SG linker.
Transcription in reaction medium containing 5% PEG 3350 at 25°C was analyzed at a 10-fold molar excess of unmodified E. coli RNAP core relative to DNA (panel A) or with equimolar proportions (panel B). Components were assembled as shown at the top of Figure 1B and open complexes were allowed to form for the time indicated before addition of NTPs and heparin.
For gp55, small deletions or insertions in the linker, for example removing residues 162-165 (TPGA) or adding SGGGC after residue 165 (165ins5), did not significantly affect transcriptional activation (Supplemental Figure 5A). More extensive substitutions of sequence within this segment were also without substantial effect on activated transcription under the assayed conditions. For example, removing residues 159-165 (TYRTPGA) reduced transcription to the same level as removing the C-terminal epitope, consistent with the connector to the sliding clamp having been excessively shortened. Replacing these 7 residues with the flexible 7-amino acid linker SGGGCSG (Δ159-165-ins7) restored essentially full activity (Supplemental Figure 4B). The gp55 double mutant P163A/P175A was also fully competent for transcriptional activation (data not shown). Even replacement of the entire residue 159-175 segment with the simple 17-residue linker (SG)8C allowed retention of a very substantial level of activated transcription when assayed at polymerase excess (Figure 4A, lanes 4-8). Incrementally shortening the SG linker by 4 and 8 residues impaired transcription (Figure 4A, lanes 9-16), although the ability to bind gp45 (tested by pull-down assay) was retained. A defect of the SG17-linker mutant was noted at a 10-fold lower enzyme concentration, that is, at DNA excess over active RNAP (Figure 4B). More detailed analysis of this defect (and more generally of gp55 linkers that retain the sliding clamp-binding motif but are conditionally impaired for gp45-dependent activation) might provide new insights into the role of the gp55 linker in specifying the orientation of gp45 in the promoter complex. However, the retention of very substantial activation by the sliding clamp in spite of the drastic SG17 substitution under any reaction condition is striking.
Comparable changes of the gp33 connector severely impaired transcriptional activation. For example, inserting the 5-residue flexible linker SGGGC between residues 101 and 102 severely diminished activation (Supplemental Figure 5C). Deleting residues 98-102 (SVVRC) was also strongly deleterious, and replacement with a flexible linker (SGGGC) was not tolerated (data not shown). A double-alanine substitution scan also showed the sensitivity of gp33 function to changes in the residue 92-107 segment (Supplemental Figure 4), with the largest effect of AA substitution at residues 94-97 (L94A/L95A and R96A/P97A), minimal effects of substitution at residues 100 and 101 (V100A/R101A), and substantial sensitivity to substitution at residues 102-105 (C102A/E103A and K104A/T105A). These double-alanine substitutions did also affect polymerase core binding but, at the high molar excess (12X) of gp33 over core enzyme that was used for the activation assay, deficiencies of core occupancy would not account entirely for the observed effects on transcriptional activation (Supplemental Figure 4).
The principal conclusion to be drawn from these experiments is that the gp55 connector is at least partly unstructured and flexible and that it tolerates lengthening. Requiring the gp55 linker to span the space between the sliding clamp and the body of gp55-RNAP in the activated promoter complex imposes a minimum length requirement. Extensive amino acid substitution of the gp55 linker is compatible with transcriptional activation and flexibly extending the linker is also tolerated. In contrast, the gp33 connector is highly constrained and mutations therein affect transcriptional activation as well as core binding functions.
Avid formation of the activated T4 late promoter complex
Heparin (Walter et al, 1967) and other polyanions bind RNAPs and are commonly used as competitive inhibitors of transcription. In the extensive analysis of the phage λPR and lacUV5 promoters that establishes the general kinetic scheme of the reaction pathway to initiation of transcription (Record et al, 1996), heparin has been used to trap free RNAP while leaving stably bound (isomerized), closed as well as open promoter complexes untouched, at least over limited periods of time. That heparin also directly attacks certain preformed, open σ70 promoter complexes was originally shown for the very strong phage T7A1 early promoter (Pfeffer et al, 1977) and has also been noted for the rrnB promoter (Barker, 2001).
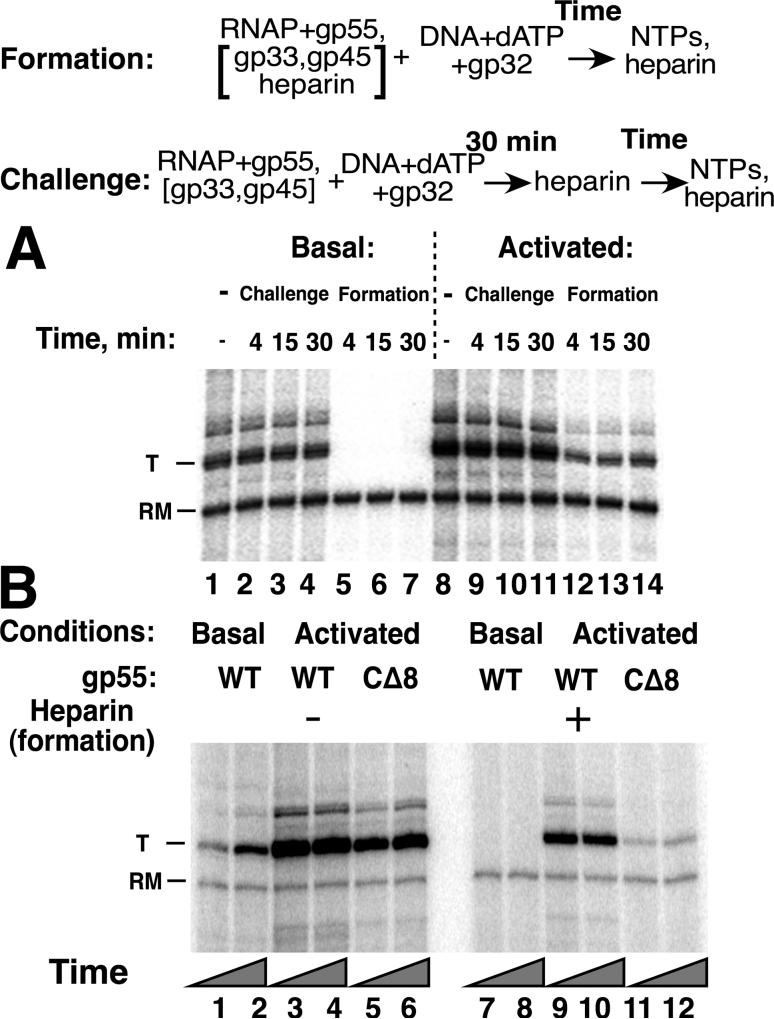
It was therefore unexpected to find the outcome of the experiment summarized in Figure 5A, which showed formation of open, transcription-competent activated T4 late promoter complexes in competition with a high concentration of heparin. In contrast, basal transcription was completely inhibited by pre-added heparin (Figure 5A, compare lanes 12-14 with lanes 5-7). Preformed activated and basal open promoter complexes were comparably stable to challenge by subsequently added heparin (compare lanes 9-11 with lane 8 and lanes 2-4 with lane 1). Formation of promoter complexes in the continuous presence of heparin was significantly diminished when the gp55-gp45 connector was disrupted (Figure 5B, compare lanes 11, 12 with lanes 9, 10).
Figure 5. Formation of T4 late open promoter complexes in the presence of heparin.
Top: Reaction schemes for formation of promoter complexes in the presence of heparin and for challenge of preformed promoter complexes by heparin. A. Transcription with unmodified E. coli RNAP core, WT gp55 under basal and activated conditions (as for Figure 2B) in reaction medium without PEG. For formation of complexes in the presence of heparin, the latter was added to RNAP to a final concentration of 100 ng/μl prior to DNA (lanes 5-7 and 12-14). For challenge, heparin was added (to the same concentration) to promoter complexes preformed for 30 min in the absence of heparin (lanes 2- 4 and 9-11). Transcription not challenged by heparin is shown in lanes 1 and 8. B. Formation of open promoter complexes in the presence of heparin with WT or CΔ8gp55; 100 ng/μl heparin was added to RNAP prior to DNA for reactions shown in lanes 7-12. Promoter opening time 5 or 15 min (odd- and even-numbered lanes, respectively).
In view of the newly identified avid formation of activated T4 late promoter complexes, 2 σ70 promoters were also tested: T7A1 (with UP, -35 and -10 sites) and E-10con, a galP1-based promoter with only a consensus extended -10 site. Under the same reaction conditions as above, preformed E-10con promoter complexes were stable (Figure 6A, lanes 1-6) but did not form in the presence of heparin (Figure 6A, lanes 7-11). Preformed T7A1 promoter complexes were less stable but did form in the presence of heparin. An additional surprising result was finding that T7A1 DNA, heparin and RNAP came to equilibrium and reached the same final state relatively quickly, regardless of order of mixing, under the conditions of this experiment. The ability to form transcription complexes in the face of competition by heparin was lost when the σ70 domain 4-β flap interaction was disrupted by mutations R541C/L607P (Gregory et al, 2004), and diminished when σ70 lacked its N-terminal domain 1.1 (Figure 6B). Thus, a parallel can be drawn between these σ70 and gp55 promoters: sufficiently avid formation of promoter complexes to overcome the heparin competitor characterized those promoter complexes with the more extensive set of protein-protein as well as protein-DNA interactions and was sensitive to loss of these individual interactions.
Interactions with the sliding clamp and promoter opening
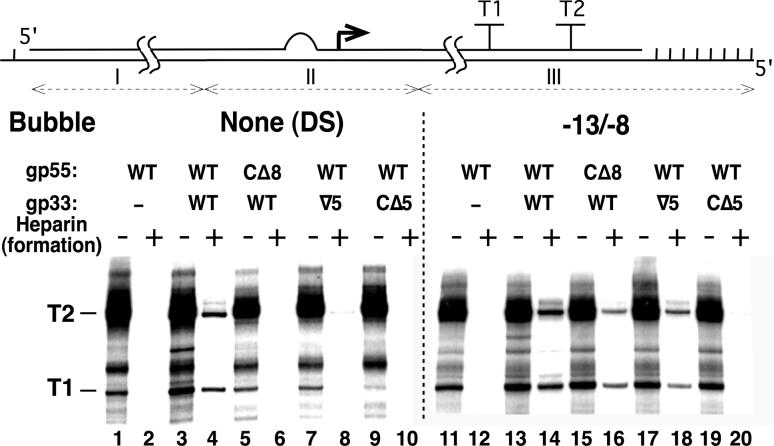
The next experiment explores whether losses of transcriptional activation (Figure 1) and avidity (Figure 5) that are generated by eliminating the sliding clamp interaction of gp33 or gp55 can be compensated by partially preopening the T4 late promoter. A DNA template with a clamp-loading site and an unpaired DNA segment between bp -13 and -8 (retaining the sequence of the nontranscribed strand) but otherwise closely resembling the standard DNA template was assembled as described in Methods and validated for specific transcription (Supplemental Figure 6). The control DNA with its fully double-stranded promoter was prepared and tested in the same way. Transcription with these templates generated a high extraneous background (Figure 7, odd-numbered lanes) that was eliminated in the presence of heparin, allowing the late transcripts to be clearly distinguished (even-numbered lanes). On the double-stranded promoter template, only wild-type activated late promoter complexes were able to form in the presence of heparin (lane 4). Mutating gp33 by inserting a 5-residue flexible linker (lane 8) and disconnecting gp55 or gp33 from the sliding clamp (lanes 6 and 10, respectively) eliminated avid formation of promoter complexes.
Figure 7. Formation of open T4 late promoter complexes on a partially preopened (bubble) template in the presence of heparin.
Top: The 3′-end recessed template containing a partially preopened T4 late P23 promoter (not drawn to scale). Roman numerals indicate DNA fragments that were ligated to obtain the template. Horizontal lines indicate its DNA strands and vertical marks indicate its single-stranded downstream end of the template. The unpaired region (bubble), P23 transcription start and terminators T1 and T2 are indicated. Bottom: Promoter complexes were formed with unmodified E. coli RNAP core and the indicated proteins in the absence of heparin or in the presence of 100 μg/mL heparin (odd- and even-numbered lanes, respectively) during promoter complex assembly, in reaction medium without PEG. Time of promoter complex formation before addition of NTPs: 15 min.
Preopening the promoter rescued the transcription defect generated by disconnecting gp33 from the sliding clamp (compare transcript T1 in lanes 19 and 9) and, less drastically, the deficits of the other two mutants (compare lanes 17, 15 and 13 with lanes 7, 5 and 3, respectively). Promoter preopening restored avid formation to transcription complexes lacking the gp55-sliding clamp linkage (compare lanes 16 and 6) but not complexes lacking the gp33-sliding clamp linkage (compare lanes 20 and 19), or basal transcription complexes (lane 12).
The ability to rescue the deleterious effect of the gp55CΔ8 mutation by preopening the promoter implies that loss of the gp55-gp45 connection affects a late step along the reaction pathway leading to initiation of transcription (i.e., following recruitment of RNAP to the promoter). The two gp33 mutations affecting the linkage to the sliding clamp respond differently to promoter preopening. Restoration of activated transcription to both mutants by preopening the promoter would be consistent with the gp33-gp45 connection also affecting a late step in transcriptional initiation. However, avid formation of open complexes was only restored to the gp33 5-residue insertion mutant (Figure 7, lane 18), which was also quantitatively less severely impaired than gp33CΔ5 (Figure 7, lanes 7 and 9; Figure 6B; Supplemental Figure 5C; and data not shown).
Discussion
In this work, we have disrupted connections of the gp45 sliding clamp to individual T4 late transcription components, gp55 and gp33, which occupy the sites on RNAP core that bind σ70 domains 2 and 4. We show that the C-terminal motifs of gp33 and gp55, which allow attachment to the sliding clamp, are fully interchangeable (and can also be exchanged for the corresponding epitope of the gp43 DNA polymerase). Nevertheless, the loss of the gp45-gp33 and gp45-gp55 connections has entirely different consequences. Loss of the gp45-gp55 linkage still allows appreciable activation of transcription (although subject to interference by the unmodified αCTD of E. coli RNAP, and only seen cleanly when the interfering DNA interaction is eliminated, as it is with T4-modified RNAP). Loss of the gp45-gp33 connection effectively abolishes transcription. Mutagenesis of the gp33 and gp55 linkers to the sliding clamp yields a complementary picture: the gp33 linker is highly constrained and mutations in it affect transcriptional activation as well as RNAP core-binding functions. We show that partially preopening the promoter compensates for defects generated by severing the gp33 and gp55 connections with the sliding clamp, implying roles for both of these connections in promoter opening. We also show that, surprisingly, fully activated T4 late open promoter complexes form in the presence of a high concentration of heparin. Severing either the gp33 or the gp55 connection to the sliding clamp respectively abolishes or severely diminishes this avid promoter complex formation, and partly preopening the promoter compensates for loss of the gp45-gp55 connection but not the gp45-gp33 connection.
The gp45-gp33 connection and a unified model of initiation at the T4 late promoter
Gp55, the homologue of □70 domain 2, supports a basal level of specific transcription from T4 late promoters in the absence of gp45. Basal transcription is sustained by a sequence-nonspecific DNA binding site of RNAP core that remains available in gp55 holoenzyme (Nechaev et al, 2006) but is blocked by proteins, such as □70 domain 4 and gp33, that bind to the RNAP □ flap and thereby alter the DNA-binding specificity of RNAP. □ domains 4 enable RNAP holoenzymes to recognize the -35 promoter motif; gp33 prevents RNAP from interacting with unmodified double-stranded DNA (Nechaev et al, 2006). Given that the gp33-gp45 connection is required for transcriptional activation by the gp45 sliding clamp (Figure 1B and 1C), gp45 can be viewed as a DNA modification that is specifically recognized by gp33. In that sense, gp33, the T4 late counterpart of □70 domain 4, can be viewed as a specificity switch that enables RNAP to interact with DNA that contains gp45, but excludes RNAP from double-stranded DNA that lacks gp45. Transcription initiation on a T4 late promoter in the presence and absence of the gp45 sliding clamp might therefore be viewed in terms of two different promoters (one of which includes gp45) engaged by two different holoenzymes (one containing gp55 only and the other gp55 and gp33) and be represented by two pathways (Figure 8). Gp33-containing holoenzyme is barely able to recognize a gp45-less (naked DNA) promoter; conversely, gp55 holoenzyme is not stimulated (or only barely affected) by gp45 (Figures 1 and 8) (Kolesky et al, 2002). T4 late transcription is a transcription system that (noncovalentely) modifies its DNA and carries a □-like component (gp33) that specifically recognizes this modification.
Figure 8. A simplified kinetic model of T4 late transcription.
Intermediate states in the reaction sequence leading to the formation of the first internucleotide bond are indicated. E: gp55-RNAP; E·gp33: the gp33- and gp55-bearing RNAP; gp45·DNA: transcription template loaded with sliding clamps; Dn: the initial nonspecific RNAP-DNA encounter complex; Pi and Po: closed and open promoter complexes. The conversion of the initial closed Pc complex to the initiation-ready Po complex is a multi-step process. At the λPR promoter, a competitor-resistant closed complex is a significantly accumulating kinetic intermediate (Davis et al, 2007; Record et al, 1996). Whether that is the case for basal or activated transcription at the late promoter remains to be determined, so steps 3 and 4 have been lumped together.
Special kinetic properties of T4 late transcription
It is useful to consider the distinctive properties of T4 late transcription in terms of the common steps of transcriptional initiation (Record et al, 1996). Initial recognition of the closed T4 late promoter (Figure 8, step 2) is not highly selective in either the basal or activated reaction pathway, certainly much less so than recognition of the λPR promoter by σ70-RNAP, for example (Cook and deHaseth, 2007). In the case of basal transcription, this relative promiscuity of binding is contributed by a nonspecific DNA-binding site of the RNAP core that remains exposed in the gp55 holoenzyme (E in Figure 8). This nonspecific site is blocked in the gp33-gp55 holoenzyme (E·33) by gp33 (Nechaev et al, 2006), but tethering the activated holoenzyme to DNA-mounted gp45 must contribute to general, nonspecific DNA binding. As a consequence, closed complexes on the T4 gene 23 late promoter (in its AT-rich surround) are not distinguished from their nonspecific background in standard DNase footprinting experiments (Nechaev et al, 2006). In contrast, the open complex and the site for initiating transcription are precisely selected. How restoration of selectivity at steps 3 and 4 might be secured (whether by highly sequence-selective binding of gp55 to strand-separated DNA, for example) is an interesting question that merits further analysis.
Tethering RNAP to the sliding clamp must also change the processivity of DNA scanning to locate the promoter (step 2). However, it is unlikely that more rapid, processive promoter searching contributes to transcriptional activation in vitro, because steps 1 and 2 are not likely to be rate-limiting for transcriptional initiation, especially when relatively short (<1 kbp) templates are used, as is the case in these experiments.
Connections of gp45 to gp55 and gp33 in the activated promoter complex together enforce rapid promoter opening (Kolesky et al, 2002), possibly through formation of a compact structure that draws the sliding clamp to the upstream end of the promoter complex (comparable with a model of the gp45-gp43 DNA polymerase holoenzyme; (Trakselis et al, 2001). It is clear that the T4 late transcription system affords ready manipulation of individual interactions within the promoter complex and is therefore a significant addition to the repertoire of experimental systems for analyzing individual steps of transcription initiation such as promoter recognition and promoter opening.
Avid promoter complex formation
Heparin is a potent polyanionic competitor of DNA for binding RNAP. It is remarkable that activated T4 late promoter complexes form in competition with a high concentration of heparin. This process required all the components and interactions of the T4 late promoter complex, including both linkages to the sliding clamp (Figure 5). Such avid formation of transcription-ready promoter complexes has not been described but, contrary to expectation, was found not to be unique to the activated T4 late promoter and was also seen with σ70 RNAP at the T7A1 promoter. The T7A1 promoter also offers multiple sites of RNAP interaction and disruption of individual protein-protein interactions (by σ70 domain 4 with the β flap and domain 1.1 with the major cleft) as well as protein-DNA interactions eliminated avid formation of promoter complexes (Figure 6B and data not shown). T4 late promoters and the T7A1 early promoter are among the strongest promoters known. We suggest that avidity reflects the ability of these promoters to efficiently compete for RNAP in vivo.
Gp33 and late transcription in other T4-group phages
Gp33 is a functional chameleon. As the essential mediator of transcriptional activation by the sliding clamp, it provides the simplest and most direct example of coactivator function (Herendeen et al, 1990). Since it also represses basal transcription (Nechaev et al, 2006), it is appropriate to think of it as operating the switch between the two transcriptional initiation pathways (Figure 8) or as the enforcer of the dependence of late transcription on concurrent DNA replication. It can also be presented as a surrogate of σ domain 4, in that it also binds to the RNAP β flap and also interacts with a transcriptional activator (as σ domain 4 does with λcI, for example), although, unlike σ domain 4, it does not contribute sequence-specific promoter recognition. Remarkably, gp33 bears no recognizable resemblance to σ domain 4, at least at the level of amino acid sequence. It is not difficult to imagine the evolution or synthetic construction of a σ-family domain 4 derivative with the functionalities that appear to be essential for gp33: binding to the β flap (more tightly than does σ70 domain 4); obstructing the RNAP core’s nonspecific DNA-binding site (which σ70 domain 4 does also); and binding to the DNA-mounted sliding clamp. Instead, the provenance of T4 gp33 and of its divergent homologues in other T4-family phages remains a conundrum.
In closing, we briefly consider how transcription of the late genes might be regulated by other T4-family phages. In view of the presence of similar short acidic and hydrophobic motifs at the C-termini of the gp55 and gp33 homologues (Supplemental Figure 1) and of gp43 homologues (JM Nolan, http://phage.bioc.tulane.edu), it is likely that the interactions between T4 gp55-RNAP, gp33 and the sliding clamp that have been dissected in this work play similar or identical roles in the multiplication cycles of the other E. coli phages. The additional implication is that these other multiplication cycles also couple late transcription to concurrent DNA replication. (Only the two-residue shorter C-terminal motifs of RB49 and phi-1 gp33 stand out as potentially different.) The amino acid sequences of the gp55 and gp33 attachment sites on gp45—the monomer-monomer interfaces and the lateral surface patches—vary considerably among these seven phages (Nolan, 2005). We expect that networks of specific amino acid side chain interactions probably are not precisely conserved but that the general mechanism is. Needless to say, these conjectures indicate avenues of further analysis relating to this remarkable system of transcriptional regulation.
Materials and Methods
Plasmids
Plasmids for overproduction of untagged WT E. coli σ70 and its derivative Δ1.1 (amino acids 100-613) have been described (Nechaev et al, 2006). A plasmid for overproduction of σ70 mutant R541C/L607P was a gift from A. Hochschild. Gp33 and gp55 mutants were constructed as derivatives of plasmid pET21-His6-gp33 (Kolesky et al, 2002) and its gp55 counterpart. Plasmids containing the P23 T4 late promoter and an extended -10 promoter were described by Kolesky et al (1999) and Nechaev and Geiduschek (2006), respectively.
Proteins
The preparation of wild-type gp55 and gp33 (and their mutant derivatives), E. coli RNAP core C-terminally His6-tagged in the β’ subunit, and the T4-modified form of this RNAP core lacking RpbA has been described or referenced (Kolesky et al, 1999). Untagged σ70 and derivatives were overproduced in E. coli BL21(DE3) and purified from inclusion bodies, dissolved in 7M urea-containing denaturing buffer and dialyzed against storage buffer (50% (v/v) glycerol, 40mM Tris-HCl, pH 8.0, 200mM NaCl, 1mM DTT, 1mM EDTA) to final protein concentrations of ∼50 μM, as described or referenced for preparation of untagged gp55 (Kolesky et al, 1999).
Transcription templates
Duplex DNA templates for transcription were prepared by PCR amplification using Vent DNA polymerase. A 126-bp extended -10 promoter fragment (bp -79 to +47) was amplified out of plasmid pE-10 as described (Nechaev et al, 2006). The 166-bp T7A1 promoter fragment yields a 73-nt run-off transcript. Double-stranded, 3′-end recessed T4 late templates were prepared by Exo III treatment as described (Kolesky et al, 2002). Briefly, an ∼580-bp PCR fragment containing the P23 T4 late promoter and downstream rrnB terminators amplified from placO-SK110-rrnB (T1+T2) was cleaved with Hind III and Kpn I to generate ExoIII-susceptible downstream and ExoIII-resistant upstream ends and reacted with ExoIII to generate ∼100 nt of single-stranded downstream DNA. Transcripts initiating at P23 and terminating at T1 and T2 are ∼110 and ∼270 nt in length, respectively.
Partially heteroduplex and 3′-endrecessed transcription templates were prepared by ligating 3 DNA fragments. The upstream fragment was prepared by PCR-amplifying a gene 33 segment from pET21gp33 and treatment with XhoI. The middle fragment was prepared by annealing synthetic DNA oligonucleotides (5′-unphosphorylated) corresponding to bp -40 to +35 of the P23 promoter with single-stranded ends allowing ligation to XhoI and KpnI sites at its upstream and downstream ends, respectively. The bp -13/-8 promoter mismatch was created by changing the template (transcribed) strand to create AA and TT non-complementarities. The downstream 721-bp fragment was prepared by PCR-amplifying the pE-10 plasmid with the same primers that were used for preparation of 3′-end recessed templates (Kolesky et al, 2002), treating with KpnI and ExoIII to create an ∼100-bp single-stranded 3′-end recessed downstream end. The upstream, middle and downstream fragments were combined in molar ratios of 2:1:2, respectively, and joined with T4 DNA ligase at a final concentration of 50 fmol/□l middle fragment. After heat-inactivation at 65°C for 20 min, 1 □l of the ligation mix was used for transcription in each reaction without further purification.
In vitro transcription
Single-round transcription was carried out at 25°C (except as noted) in Standard Reaction Buffer (Nechaev et al, 2006) containing 200 mM K acetate, 33 mM Tris acetate, pH 7.8, 10 mM Mg acetate, 1 mM DTT, 0.12% (w/v) Tween 20 augmented with 5% (w/v) PEG 3350, except as noted. Standard concentrations of components in the assembled transcription mix were 4 nM DNA, 40 nM RNAP core, a 12-fold molar excess of gp33 and gp55 (each) relative to core, 400 nM (trimer) gp45, 160 nM gp44/62 clamp loader complex, 750 nM gp32 (single-stranded DNA-binding protein), 0.4 mM dATP, 1 mM ATP, GTP, 100 μMPCTP and 100 μM [α-32P]UTP. DNA and gp32 in 10 μL volume were added to all other proteins in 10 μL volume for the times noted for individual experiments before combining with NTPs and heparin in 5 μL volume for 5 min of RNA synthesis. For experiments shown in Figure 3 and Figure 4, RNAP concentration varied as indicated and the molar excess of gp33 and gp55 was maintained at 12:1. For experiments shown in Figure 7 and Supplemental Figure 5, the DNA concentration was 2 nM and RNAP core was at 40 nM. Sample processing and analysis followed standard procedure (Kolesky et al, 1999).
Supplementary Material
Acknowledgements
We are grateful to G.A. Kassavetis for many discussions and advice during the course of this work, and to G.A. Kassavetis, M. Ouhammouch and K. Severinov for a critical reading of the manuscript. We also thank R. Gourse and R.M. Saecker for information and L.E. Parker for editorial help and acknowledge support of this research by the National Institute of General Medical Sciences of the NIH.
References
- Alley SC, Jones AD, Soumillion P, Benkovic SJ. The carboxyl terminus of the bacteriophage T4 DNA polymerase contacts its sliding clamp at the subunit interface. J Biol Chem. 1999;274:24485–24489. doi: 10.1074/jbc.274.35.24485. [DOI] [PubMed] [Google Scholar]
- Barker M. Regulation without protein transcription factors: Intrinsic properties of Escherichia coli promoters that lead to their regulation. University of Wisconsin, Vol. PhD.; 2001. [Google Scholar]
- Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. Modular architecture of the T4 phage superfamily: A conserved core genome and a plastic periphery. Virology. 2007;362:384–396. doi: 10.1016/j.virol.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Cook VM, deHaseth PL. Strand opening-deficient Escherichia coli RNA polymerase facilitates investigation of closed complexes with promoter DNA: effects of DNA sequence and temperature. J Biol Chem. 2007;282:21319–21326. doi: 10.1074/jbc.M702232200. [DOI] [PubMed] [Google Scholar]
- Davis CA, Bingman CA, Landick R, Record MT, Jr., Saecker RM. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein R, Bolle A, Steinberg C, Kellenberger E, Boy de la Tour E, Chevalley R, Degar R, Susman M, Genhardt G, Lielausis A. Physiological studies of conditional lethal mutants of bacteriophage T4D. In Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press. 1963;28:375–394. [Google Scholar]
- Filée J, Bapteste E, Susko E, Krisch HM. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol Biol Evol. 2006;23:1688–1696. doi: 10.1093/molbev/msl036. [DOI] [PubMed] [Google Scholar]
- Fu TJ, Sanders GM, O’Donnell M, Geiduschek EP. Dynamics of DNA-tracking by two sliding-clamp proteins. EMBO J. 1996;15:4414–4422. [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci U S A. 2004;101:4554–4559. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M, Burgess RR. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkman MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Herendeen DR, Kassavetis GA, Geiduschek EP. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992;256:1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Herendeen DR, Williams KP, Kassavetis GA, Geiduschek EP. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990;248:573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Geiduschek EP. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984;81:5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky S, Ouhammouch M, Brody EN, Geiduschek EP. Sigma competition: the contest between bacteriophage T4 middle and late transcription. J Mol Biol. 1999;291:267–281. doi: 10.1006/jmbi.1999.2953. [DOI] [PubMed] [Google Scholar]
- Kolesky SE, Ouhammouch M, Geiduschek EP. The mechanism of transcriptional activation by the topologically DNA-linked sliding clamp of bacteriophage T4. J Mol Biol. 2002;321:767–784. doi: 10.1016/s0022-2836(02)00732-5. [DOI] [PubMed] [Google Scholar]
- Moarefi I, Jeruzalmi D, Turner J, O’Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Geiduschek EP. The role of an upstream promoter interaction in initiation of bacterial transcription. EMBO J. 2006;25:1700–1709. doi: 10.1038/sj.emboj.7601069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Kamali-Moghaddam M, Andre E, Léonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan J. Genomes of the T4-like phages. Tulane University; 2005. [Google Scholar]
- Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. Genetic diversity among five T4-like bacteriophages. Virol J. 2006;3:30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR, Stahl SJ, Chamberlin MJ. Binding of Escherichia coli RNA polymerase to T7 DNA: Displacement of holoenzyme from promoter complexes by heparin. J Biol Chem. 1977;252:5403–5407. [PubMed] [Google Scholar]
- Record M, Reznikoff W, Craig M, McQuade K, Schlax P. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F, editor. Escherichia coli and Salmonella. ASM Press; Washington, DC: 1996. pp. 792–820. [Google Scholar]
- Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Tinker RL, Kassavetis GA, Geiduschek EP. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J. 1994a;13:5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker RL, Williams KP, Kassavetis GA, Geiduschek EP. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994b;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Trakselis MA, Mayer MU, Ishmael FT, Roccasecca RM, Benkovic SJ. Dynamic protein interactions in the bacteriophage T4 replisome. Trends Biochem Sci. 2001;26:566–572. doi: 10.1016/s0968-0004(01)01929-6. [DOI] [PubMed] [Google Scholar]
- Walter G, Seifert W, Zillig W. Modified DNA-dependent RNA polymerase from E. coli infected with bacteriophage T4. Biochem Biophys Res Commun. 1968;30:240–247. doi: 10.1016/0006-291x(68)90441-5. [DOI] [PubMed] [Google Scholar]
- Walter G, Zillig W, Palm P, Fuchs E. Initiation of DNA-dependent RNA synthesis and the effect of heparin on RNA polymerase. Eur J Biochem. 1967;3:194–201. doi: 10.1111/j.1432-1033.1967.tb19515.x. [DOI] [PubMed] [Google Scholar]
- Wilson C, Dombroski AJ. Region 1 of simga70 is required for efficent isomerization and initiation of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- Wong K, Geiduschek EP. Activator-sigma interaction: A hydrophobic segment mediates the interaction of a sigma family promoter recognition protein with a sliding clamp transcription activator. J Mol Biol. 1998;284:195–203. doi: 10.1006/jmbi.1998.2166. [DOI] [PubMed] [Google Scholar]
- Wong K, Kassavetis GA, Leonetti JP, Geiduschek EP. Mutational and functional analysis of a segment of the sigma family bacteriophage T4 late promoter recognition protein gp55. J Biol Chem. 2003;278:7073–7080. doi: 10.1074/jbc.M211447200. [DOI] [PubMed] [Google Scholar]
- Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recogntion of the -10 promoter element. EMBO J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.