Abstract
Stimulation of the death receptor superfamily induces the activation of caspase-8 and thereby the apoptotic response. The MUC1 oncoprotein is aberrantly overexpressed by diverse human malignancies and inhibits stress-induced apoptosis. The present results demonstrate that MUC1 blocks activation of caspase-8 and apoptosis in the response of malignant cells to tumor necrosis factor α, TRAIL and FasL. The results show that MUC1 associates constitutively with caspase-8. The MUC1 cytoplasmic domain (MUC1-CD) binds directly to the caspase-8 p18 fragment upstream to the catalytic Cys-360 site. The results also demonstrate that MUC1-CD binds to FADD at the death effector domain. In nonmalignant epithelial cells, MUC1 interacts with caspase-8 and FADD as an induced response to death receptor stimulation. The functional significance of these interactions is supported by the demonstration that MUC1 competes with caspase-8 for binding to FADD and blocks recruitment of caspase-8 to the death-inducing signaling complex. These findings indicate that MUC1 is of importance to the physiologic regulation of caspase-8 activity and that overexpression of MUC1 as found in human malignancies could contribute to constitutive inhibition of death receptor signaling pathways.
Keywords: MUC1, caspase-8, death receptors, TNFα, TRAIL, FasL, apoptosis
Introduction
The apoptotic response of cells is induced by extrinsic and intrinsic pathways that confer downstream activation of the caspase cysteine proteases. The extrinsic apoptotic pathway is activated by ligand stimulation of the tumor necrosis factor α (TNFα) receptor superfamily of death receptors. These death receptors include, among others, TNF receptor 1 (TNF-R1), Fas (CD95), and TNF-related apoptosis-inducing ligand receptors 1 (TRAIL-R1/DR4) and 2 (TRAIL-R2/DR5). A death domain (DD) of 60-70 amino acids is shared by these receptors and is necessary for the induction of apoptosis (1). TNFα stimulation of TNF-R1 induces the formation of a cell membrane complex with the adaptor TRADD, TNFR-associated factor 2 (TRAF2) and receptor interacting protein 1 (RIP1) that leads to the activation of NF-κB and cell survival (2). Alternatively, TRADD and RIP1 associate with FADD and caspase-8 (2, 3). In turn, activation of caspase-8 results in the cleavage of procaspase-3 and the induction of apoptosis (4-6). Caspase-8 also cleaves Bid, a proapoptotic member of the Bcl-2 family, and thereby induces release of mitochondrial apoptogenic factors, such as cytochrome c, into the cytosol (7, 8). Stimulation of Fas with Fas ligand (FasL) or TRAIL-R1/2 receptors with TRAIL induces the recruitment of FADD and formation of the death-inducing signaling complex (DISC) (9). Recruitment of caspase-8 by FADD through their death effector domains (DEDs) is associated with dimerization of caspase-8, interdimer cleavage to the active caspase-8 p18/p10 fragments and the apoptotic response.
The human MUC1 mucin-type glycoprotein is expressed at the apical plasma membranes of normal secretory epithelial cells (10). MUC1 is translated as a single polypeptide that undergoes autoproteolysis into two subunits in the endoplasmic reticulum (11-13). The MUC1 N-terminal subunit (MUC1-N) is the mucin component with variable numbers of 20 amino acid tandem repeats that are modified by O-glycosylation (14, 15). MUC1-N forms a stable heterodimer with the MUC1 C-terminal subunit (MUC1-C) at the cell membrane and extends beyond the glycocalyx as part of a physical barrier that protects the epithelial cell surface. With transformation and loss of polarity, MUC1 is aberrantly expressed at high levels on the entire surface of carcinoma cells (10). Overexpression of MUC1 in transformed cells is also associated with accumulation of MUC1-C in the cytosol and targeting of this subunit to the nucleus (16-21) and mitochondria (22-24). MUC1-C consists of a 58 amino acid extracellular domain, a 28 amino acid transmembrane domain and a 72 amino acid cytoplasmic domain (25). The MUC1 cytoplasmic domain (MUC1-CD) functions as a substrate for c-Src (26), glycogen synthase kinase 3β (27), protein kinase Cδ(28) and c-Abl (29). MUC1-CD also interacts directly with the Wnt effector β-catenin (20, 24, 26-28, 30), the p53 tumor suppressor (18), estrogen receptor α (19), IκB kinase β (IKKβ) and IKKγ(31).
Overexpression of MUC1 as found in human carcinomas and certain hematologic malignancies induces transformation and resistance to apoptosis induced by genotoxic agents, reactive oxygen species and hypoxia (16, 18, 20, 22, 29, 32-34). The present results demonstrate that MUC1-C binds directly to caspase-8 and FADD, blocks recruitment of caspase-8 and thereby prevents activation of the death receptor-induced extrinsic apoptotic pathway.
Material and Methods
Cell culture
Human HCT116 colon cancer cells expressing an empty vector or MUC1 (22) and human MCF-7 breast cancer cells were cultured in DMEM medium (Mediatech, Herndon, VA) with 10% heat-inactivated fetal bovine serum (FBS; Mediatech), 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine (ATCC, Manasses, VA). Human BC-1 lymphoma and U-937 leukemia cells were grown in RPMI-640 medium supplemented with 10% FBS, antibiotics and L-glutamine. Human MCF-10A breast epithelial cells were grown in mammary epithelial cell growth media (MEGM; Lonza, Walkersville, MD). Cells were treated with TRAIL (Calbiochem, San Diego, CA), Flag-tagged TRAIL (Axxora, San Diego, CA), TNFα (Sigma, St. Louis, MO) and FasL (CH11; Upstate Biotechnology Inc., Lake Placid, NY).
Silencing of MUC1 and FADD expression
The BLOCK-iT Target Screening System (Invitrogen, Carlsbad, CA) was used to generate siRNAs that target two MUC1 sequences (#1, AAGGTACCATCAATGTCCACG; or #2, AAGTTCAGTGCCCAGCTCTAC) and a control sequence (CGCTTACCGATTCAGAATGG). The RNAi cassettes were transferred to pLenti4/BLOCK-iT-DEST by LR recombination for the generation of lentiviral vectors. BC-1 cells were infected with the lentiviruses at a multiplicity of infection of 5 in the presence of 8 μg/ml polybrene. Cell clones were selected in methylcellulose semi-solid medium containing 200 μg/ml Zeocin and assayed for downregulation of MUC1 by immunoblotting. Transient transfection of the MCF-10A cells with control, MUC1 siRNA or FADD siRNA pools (Dharmacon, Lafayette, CO) was performed in the presence of Lipofectamine 2000.
Stable expression of MUC1-C in U-937 cells
PT67 cells were transfected with pLXIN or pLXIN-MUC1-C and selected in the presence of G418. Supernatants were filtered, assayed for retroviral titers using NIH3T3 cells and used for infection of U-937 cells. At 24 h after infection, the cells were seeded into methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) and single cell clones were selected in G418.
Immunoprecipitation, DISC isolation and immunoblot analysis
Cells were lysed by sonication in the presence of 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 100 μg/ml phenylmethylsulphonyl fluoride and protease inhibitor mixture. Soluble proteins were incubated with anti-caspase-8 (Axxora; BD Biosciences, San Jose, CA) for 2 h at 4°C, followed by precipitation with protein A/G beads (Pierce Biotechnologies, Rockford, IL). In certain experiments, cells were incubated with the complex of Flag-tagged TRAIL and anti-Flag (M2; Sigma), and then lysates were immunoprecipitated with protein-G-sepharose to isolate DISC complexes as described (35). Immune complexes and lysates were subjected to immunoblot analysis with anti-MUC1-C (LabVision, Fremont, CA), anti-caspase-8, anti-β-actin (Sigma, St. Louis, MO), anti-His (Invitrogen), anti-GST (EMD Biosciences, LaJolla, CA), mouse anti-FADD (Upstate Cell Signaling Solutions, Charlottesville, VA) and rabbit anti-FADD (Santa Cruz Biotechnology, Santa Cruz, CA). Reactivity was detected with horseradish peroxidase-conjugated secondary antibodies and chemiluminescence (GE Healthcare Biosciences, Piscataway, NJ).
In vitro caspase-8 activity assay
Caspase-8 activity was assayed using the BD ApoAlert Caspase-8 Colorimetric Assay Kit and IETD-pNA(p-nitroaniline) as substrate (BD Bioscience). Caspase-8 activity was monitored at 405 nm using a spectrophotometer.
In vitro binding assays
Purified GST, GST-MUC1-CD(1-72), GST-MUC1-CD deletion mutants, GST-caspase-8, GST-caspase-8 deletion mutants, GST-FADD, GST-N-FADD and GST-C-FADD (36) were purified from E. coli (BL21 DE3) and immobilized on glutathione beads (Pierce). His-MUC1-CD(1-72) and His-caspase-8 deletion mutants were purified on Ni-NTA beads (Qiagen, Valencia, CA). GST and GST fusion proteins on glutathione beads were incubated with purified soluble proteins for 2 h at 4°C, washed and the adsorbates analyzed by immunoblotting. For competition studies, GST-caspase-8 DED bound to FADD was incubated with increasing amounts of MUC1-CD. Precipitated proteins were subjected to immunoblot analysis. In certain studies, binding was performed in the presence of a synthetic peptide (CQCRRKNYGQLDIFPARDTY) derived from amino acids 1-20 from MUC1-CD (Molecular Biology Core Facility, Dana-Farber Cancer Institute).
Apoptosis assays
Cells were fixed in 70% ethanol and incubated in PBS containing 50 μg/ml RNase and 2.5 μg/ml propidium iodide. DNA content was analyzed by flow cytometry. The percentage of cells with sub-G1 DNA was determined by the MODFIT LT Program (Verity Software, Topsham, ME). Cells were also suspended in Annexin-V-FLOUS containing propidium iodide (Roche) and analyzed by flow cytometry.
Results
MUC1 attenuates death receptor-induced activation of caspase-8
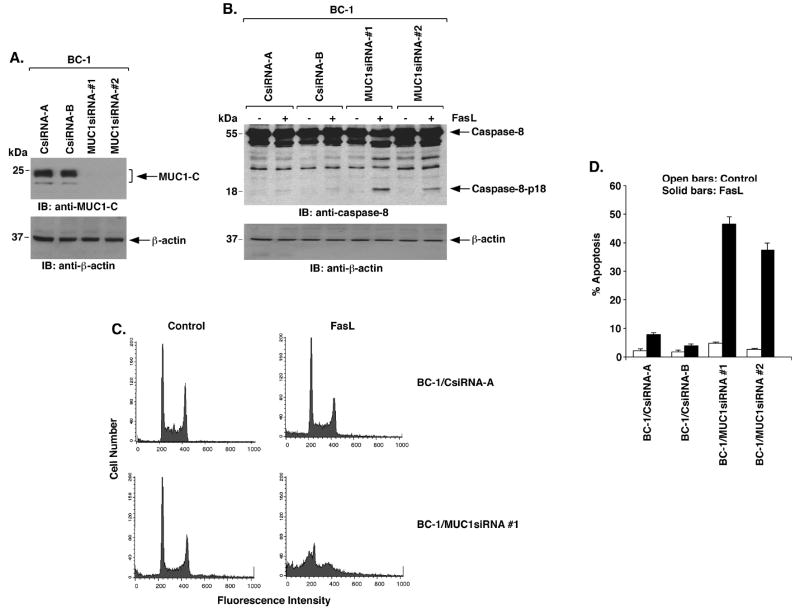
To determine whether MUC1 regulates death receptor signaling, we infected BC-1 cells with a lentivirus expressing a siRNA that targets the MUC1 sequence 5′-AAGGTACCATCAATGTCCACG-3′ encoding amino acids in MUC1-C (MUC1siRNA#1). As compared to BC-1 cells infected with a lentivirus expressing a control siRNA (CsiRNA), MUC1-C expression was stably downregulated by the MUC1siRNA#1 (Fig. 1A). To avoid potential off-target effects, we also infected the BC-1 cells with a lentivirus expressing a siRNA that targets a different MUC1 sequence encoding amino acids in MUC1-N (5′-AAGTTCAGTGCCCAGCTCTAC-3′; MUC1siRNA#2). Downregulation of MUC1-C was similar with the two MUC1siRNAs (Fig. 1A). Treatment of the BC-1/CsiRNA cells with FasL had little effect on activation of caspase-8 (Fig. 1B). By contrast, FasL treatment of BC-1 cells silenced for MUC1 was associated with cleavage of caspase-8 to the p18 fragment (Fig. 1B). To determine if silencing MUC1 affects death receptor-induced apoptosis, BC-1/CsiRNA and BC-1/MUC1siRNA cells were treated with FasL and then monitored for sub-G1 DNA content. In concert with the inhibitory effects of MUC1 on caspase-8 activation, silencing MUC1 sensitized BC-1 cells to FasL-induced apoptosis (Fig. 1C). The inhibitory effect of MUC1 on the apoptotic response to FasL was confirmed in repeated experiments with BC-1 cells expressing MUC1siRNA#1 and MUC1siRNA#2 (Fig. 1D) and in cells stained with Annexin-V and propidium iodide (Supplemental Fig. S1). Previous work showed that MUC1 blocks TRAIL-induced apoptosis of HCT116 cancer cells (22). Consistent with the results obtained using BC-1 cells, TRAIL-induced activation of caspase-8 in HCT116 cells was blocked by a MUC1-dependent mechanism (Supplemental Fig. S2). These findings indicate that MUC1 attenuates death receptor-induced activation of caspase-8 and apoptosis.
Figure 1. MUC1 blocks death receptor-induced activation of caspase-8 and apoptosis.
A. BC-1 cells were infected with lentiviruses expressing the CsiRNA, MUC1siRNA#1 or MUC1siRNA#2. Lysates from the indicated clones were immunoblotted with anti-MUC1-C and anti-β-actin. B. The indicated BC-1 cell clones were treated with 50 ng/ml FasL for 8 h. Lysates were immunoblotted with the indicated antibodies. C. The indicated BC-1 cells were treated with 50 ng/ml FasL for 24 h and then analyzed by FACS for sub-G1 DNA content. D. The results obtained with the indicated BC-1 cells left untreated (open bars) or treated with FasL (solid bars) are expressed as the percentage apoptosis (mean±SD for three experiments).
MUC1-C is sufficient to block death receptor-induced signaling
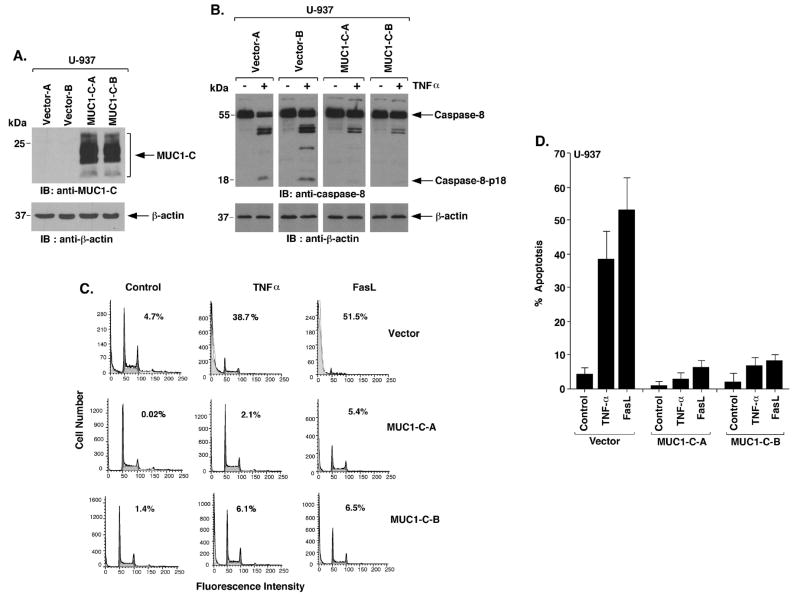
To determine if the MUC1-C subunit is sufficient to block activation of caspase-8, we stably transfected MUC1-negative U-937 cells with an empty vector or MUC1-C (Fig. 2A). Treatment of the U-937/vector cells with TNFα was associated with activation of caspase-8 (Fig. 2B). By contrast, this response to TNFα was attenuated in U-937/MUC1-C cells (Fig. 2B). Similar results were obtained when the U-937/vector and U-937/MUC1-C cells were stimulated with FasL (Supplemental Fig. S3). Moreover, expression of MUC1-C abrogated sensitivity to TNFα- and FasL-induced apoptosis (Fig. 2C). These responses were confirmed with both U-937/MUC1-C clones and in repeated experiments (Fig. 2D), and using Annexin-V and propidium iodide (Supplemental Fig. S4). These findings indicate that MUC1-C is sufficient to block death receptor-induced activation of caspase-8 and apoptosis.
Figure 2. MUC1-C is sufficient for inhibition of caspase-8 activation and apoptosis.
A. U-937 cells were infected with retroviruses expressing the control pLXIN vector or pLXIN-MUC1-C. Lysates were immunoblotted with the indicated antibodies. B. Lysates from the indicated U-937 cells treated with 10 ng/ml TNFα were immunoblotted with the indicated antibodies. C. The indicated U-937 cells were treated with 10 ng/ml TNFα or 50 ng/ml FasL for 24 h and monitored for sub-G1 content. D. The results are expressed as the percentage apoptosis (mean±SD of three experiments).
MUC1-C inhibits caspase-8 in the response of nontransformed MCF-10A cells to death ligand stimulation
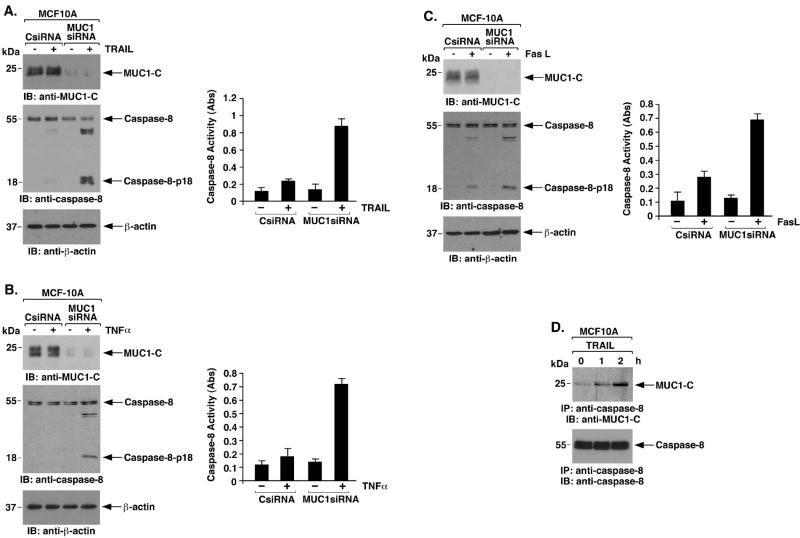
Inhibition of caspase-8 by MUC1-C could represent a response in nontransformed cells that is constitutively activated by overexpression of MUC1 in malignant cells. To address this possibility, studies were performed on the nontransformed MCF-10A cells (37, 38), which express endogenous MUC1 but at lower levels than that found in carcinoma cells (31). Treatment of MCF-10A cells with TRAIL had little if any effect on activation of caspase-8 (Fig. 3A, left). To assess the effects of MUC1 in the MCF-10A cells, we downregulated MUC1 with a MUC1siRNA pool, which for transient silencing was more effective than with the lentiviruses. TRAIL treatment of the MCF-10A cells with MUC1 silencing was associated with cleavage of caspase-8 to the p18 fragment (Fig. 3A, left). The inhibitory effects of MUC1 were confirmed by direct measurement of caspase-8 activity (Fig. 3A, right). Silencing MUC1 in MCF-10A cells was also associated with activation of caspase-8 in response to TNFα (Fig. 3B) and FasL (Fig. 3C) stimulation. Notably, the results from coimmunoprecipitation experiments demonstrated that MUC1-C and caspase-8 associate at a low level constitutively and that, this interaction is induced by death receptor stimulation (Fig. 3D). In addition, binding of MUC1-C and caspase-8 was detectable constitutively in BC-1, U-937 and HCT116 cells (Supplemental Fig. S5A-C), indicating that this interaction is found in diverse cell types. These findings indicated that MUC1 associates with caspase-8 and contributes to the physiologic regulation of caspase-8 activation.
Figure 3. MUC1-C attenuates caspase-8 activation in the response of MCF-10A cells to death receptor stimulation.
A-C. MCF-10A cells were transfected with control siRNA or MUC1 siRNA pools for 72 h and then stimulated with TRAIL (A), TNFα (B) or FasL (C). Lysates were immunoblotted with the indicated antibodies (left panels). Lysates were also assayed for caspase-8 activity using the BD ApoAlert kit (right panels). The results are expressed as the absorbance (Abs) at 405 nm. D. Lysates from MCF-10A cells left untreated or stimulated with 100 ng/ml TRAIL for the indicated times were immunoprecipitated with anti-caspase-8. The precipitates were immunoblotted with the indicated antibodies.
MUC1-CD binds directly to caspase-8
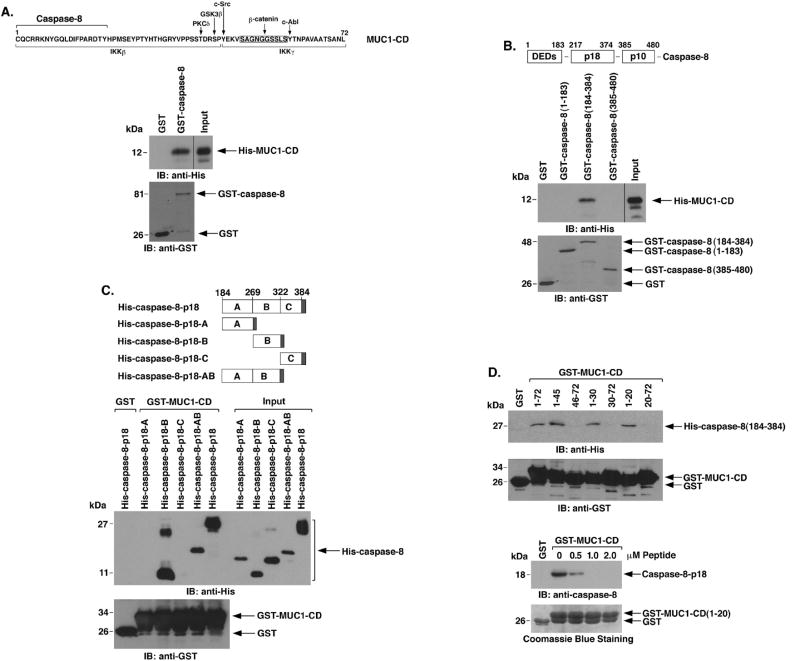
MUCl-C includes a 72 amino acid cytoplasmic domain (CD) that interacts with β-catenin, IKKs and multiple kinases (Fig. 4A, upper panel). To determine whether MUC1-CD interacts directly with caspase-8, we incubated purified His-tagged MUC1-CD with GST or GST-caspase-8 immobilized on glutathione beads. Immunoblot analysis of adsorbates with anti-His showed that MUC1-CD binds to caspase-8 (Fig. 4A, lower panels). Caspase-8 consists of an N-terminal region with two DEDs (amino acids 1-183), and the p18 (amino acids 217-374) and pl0 (amino acids 385-480) cleavage products (Fig. 4B, upper panel). To define the region of caspase-8 that interacts with MUCl-CD, we incubated GST fusion proteins of caspase-8 fragments with His-MUC1-CD. Analysis of the adsorbates demonstrated that MUC1-CD binds to the region (amino acids 184-384) that includes caspase-8-p18 (Fig. 4B, lower panels). To further define the sequences in caspase-8-p18 that are responsible for the interaction, we generated His-tagged fragments designated A (amino acids 184-269), B (amino acids 270-322), C (amino acids 323-384) and AB (amino acids 184-322) (Fig. 4C, upper panel). Incubation of the His-caspase-8-p18 fragments with GST-MUC1-CD demonstrated that MUC1-CD binds to caspase-8-p18-B (amino acids 270-322) (Fig. 4C, lower panels and Supplemental Fig. S6). Additional binding studies with His-caspase-8(184-384) and GST-MUC1-CD fragments showed that MUC1-CD amino acids 1-20 confer the interaction with caspase-8 (Fig. 4D, upper). Moreover, binding of MUC1-CD(1-20) to caspase-8-p18 was attenuated by incubation in the presence of a synthetic MUC1-CD(1-20) peptide (Fig. 4D, lower). These findings demonstrate that MUC1-CD binds directly to caspase-8-p18.
Figure 4. MUC1-CD binds directly to caspase-8-p18.
A. Amino acid sequence of MUC1-CD with highlighting of phosphorylation sites and regions for β-catenin, IKKβ and IKKγ binding (upper panel). GST or GST-caspase-8 was incubated with purified His-MUC1-CD. The adsorbates and the input protein were immunoblotted with anti-His and anti-GST (lower panels). B. Schema of caspase-8 highlighting the N-terminal region containing the DEDs, and the p18 and p10 fragments (upper panel). GST and the indicated GST-caspase-8 fragments were incubated with His-MUC1-CD. The adsorbates and input protein were immunoblotted with anti-His and anti-GST (lower panels). C. Schema of the His-caspase-8-p18 fragment and the A, B, C and AB subfragments (upper panel). The shaded region denotes position of the His tag. GST or GST-MUC1-CD was incubated with the indicated His-caspase-8 proteins. The adsorbates and input proteins (1/10th that used in the reactions) were immunoblotted with anti-His and anti-GST (lower panels). D. GST or the indicated GST-MUC1-CD proteins were incubated with His-caspase-8-p18 (upper panel). The adsorbates were immunoblotted with anti-His and anti-GST. GST or GST-MUC1-CD(1-20) was incubated with His-caspase-8-p18 in the presence of increasing amounts of MUC1-CD(1-20) peptide (lower panel). The adsorbates were immunoblotted with anti-caspase-8. Input of the GST proteins was assessed by Coomassie blue staining.
MUC1-C blocks recruitment of caspase-8 to the DISC
Binding of TRAIL to DR4/5 results in recruitment of FADD to the DISC and, in turn, FADD recruits caspase-8 (39). To determine if MUC1-C affects DISC function, MCF-10A cells were treated with a complex of Flag-tagged TRAIL and anti-Flag to precipitate the DISC. Silencing MUC1 had no effect on DR4/5 levels in the DISC (Fig. 5A). Silencing MUC1 also had little effect on TRAIL-induced recruitment of FADD (Fig. 5A). Notably, however, TRAIL treatment was associated with recruitment of MUC1-C to the DISC (Fig. 5A). We also found that MUC1 attenuates recruitment of caspase-8 (Fig. 5A). These results thus indicated that MUC1-C is recruited to the DISC by a mechanism independent of its interaction with caspase-8. To determine whether the recruitment of MUC1-C is dependent on FADD, the MCF-10A cells were silenced for FADD and then treated with TRAIL. Recruitment of MUC1-C to the DISC was attenuated by silencing FADD (Fig. 5B). The results of coimmunoprecipitation studies further demonstrated that TRAIL induces the formation of MUC1-C-FADD complexes (Fig. 5C). Moreover, in MCF-7 cancer cells, we found that MUC1-C associates with FADD at high levels constitutively and that this interaction is modestly increased in the response to TRAIL (Fig. 5D). These findings indicated that MUC1-C interacts with FADD.
Figure 5. MUC1-C is recruited to the DISC and blocks recruitment of caspase-8.
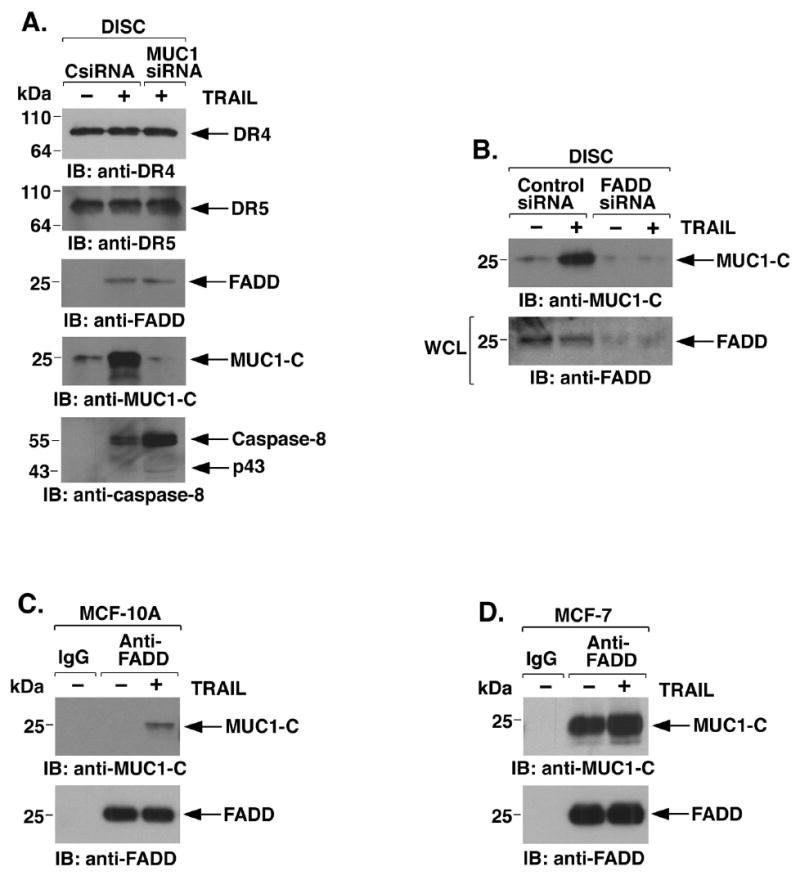
A. MCF-10A cells were transfected with the CsiRNA or MUC1siRNA pools for 72 h and then incubated with Flag-TRAIL. Anti-Flag immune complexes were precipitated with protein-G-sepharose to isolate the DISC and then immunoblotted with the indicated antibodies. B. MCF-10A cells were transfected with a control siRNA or FADDsiRNA for 72 h and then incubated with Flag-TRAIL to isolate the DISC. Anti-Flag precipitates were immunoblotted with anti-MUC1-C (upper panel). Whole cell lysates (WCL) were immunoblotted with anti-FADD to confirm FADD silencing (lower panel). C and D. MCF-10A (C) and MCF-7 (D) cells were incubated with TRAIL and then immunoprecipitated with a control IgG or anti-FADD. Immune complexes were immunoblotted with the indicated antibodies.
MUC1-CD binds directly to the FADD DED
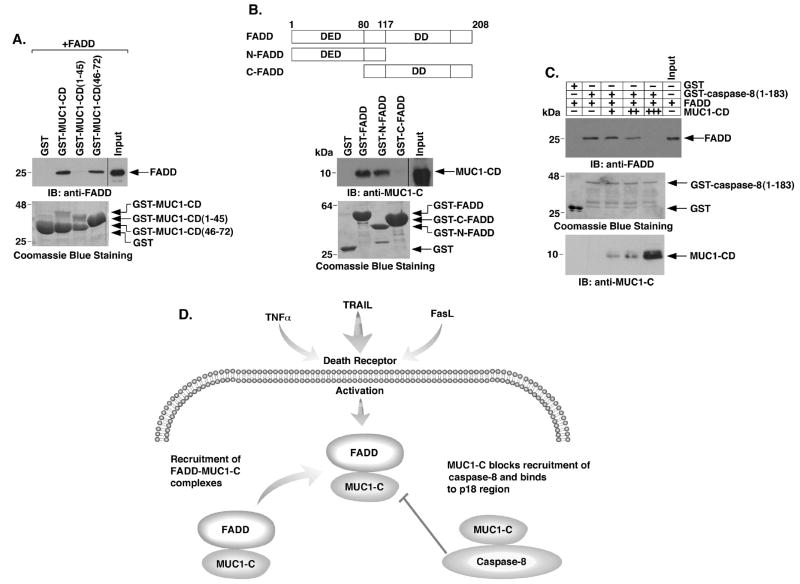
To determine whether MUC1-C binds to FADD, we incubated GST-MUC1-CD or GST-MUC1-CD deletion mutants with purified FADD. Analysis of the adsorbates demonstrated that MUC1-CD and FADD interact directly (Fig. 6A). We also found that MUC1-CD(46-72), and not MUC1-CD(1-45), binds to FADD (Fig. 6A). FADD consists of a DD that binds to DR4/5 and a DED that recruits caspase-8 to the DISC (Fig. 6B, upper panel). To define the region of FADD that confers the interaction, we incubated GST-FADD or GST-FADD deletion mutants (Fig. 6B, upper panel) with purified MUC1-CD. These binding studies confirmed the direct interaction between MUC1-CD and FADD and further demonstrated that, like caspase-8, MUC1-CD interacts with the FADD DED and not the DD (Fig. 6B, lower panel). Importantly, in competition experiments, incubation of GST-caspase-8(1-183) with FADD and then increasing amounts of MUC1-CD was associated with a progressive decrease in the interaction between FADD and caspase-8(1-183) (Fig. 6C), indicating that caspase-8 and MUC1-CD compete for binding to FADD. These findings indicate that FADD forms mutually exclusive complexes with MUC1-CD and caspase-8.
Figure 6. MUC1-CD competes with caspase-8 for direct binding to the FADD DED.
A. GST and the indicated GST-MUC1-CD proteins were incubated with purified FADD. The adsorbates and input FADD were immunoblotted with anti-FADD. The GST and GST-MUC1-CD proteins were stained with Coomassie blue. B. Schematic representation of full length FADD, N-FADD and C-FADD with positioning of the DED and DD (upper panel). The indicated GST or GST-FADD proteins were incubated with purified MUC1-CD. The adsorbates and input MUC1-CD were immunoblotted with anti-MUC1-C (lower panel). Input of the GST and GST-FADD proteins was assessed with Coomassie blue staining. C. GST or GST-caspase-8(1-183) containing the DEDs was incubated with FADD in the absence or presence of increasing amounts of purified MUC1-CD. The adsorbates and input FADD were immunoblotted with anti-FADD. Input of GST and GST-caspase-8(1-183) was determined by Coomassie blue staining. Input of MUC1-CD was determined by immunoblotting. D. Proposed interactions of MUC1-C with FADD and caspase-8 in blocking death receptor signaling.
Discussion
MUC1 blocks death receptor-induced activation of caspase-8
Previous work had shown that overexpression of MUC1 in HCT116 cells inhibits TRAIL-induced apoptosis (22). These findings were attributed to localization of MUC1-C to the mitochondrial outer membrane with a block in Bid-induced release of apoptogenic factors and thereby the extrinsic pathway (22). However, the present results demonstrate that MUC1 blocks TRAIL-induced signaling upstream to Bid by inhibiting caspase-8 activation. MUC1 also blocked TNFα- and FasL-induced activation of caspase-8 to the p18 fragment. In concert with these results, MUC1 blocked death receptor-induced apoptosis in diverse cell models. In addition, MUC1-C was sufficient for blocking caspase-8 activation and the apoptotic response, indicating that the MUC1-N mucin component is dispensable for inhibition of the extrinsic pathway. Previous studies of COS7 cells expressing a CD8/MUC1 fusion protein and CHO cells expressing a hamster Muc1 showed an augmentation of FasL-induced activation of caspase-8 and apoptosis (40). The basis for the discrepancy of those and the present results are not clear, but may relate to the expression of a MUC1 fusion protein that affects MUC1 function or the upregulation of Fas in these models. In this regard, MUC1 had little if any effect on Fas expression in the present studies. Moreover, our findings obtained with human malignant cells with and without MUC1 expression clearly show that MUC1 suppresses induction of both caspase-8 activity and apoptosis in the response to activation of diverse death receptors with FasL, TRAIL and TNFα.
MUC1-C binds directly to caspase-8
The long form of the caspase-8 (FLICE)-like inhibitory protein (cFLIPL) has two DEDs that confer binding to the caspase-8-p18 region (amino acids 217-374) (41). The present results demonstrate that, like cFLIPL, MUC1-C interacts directly with caspase-8-p18 (amino acids 270-322) and that this interaction is conferred by amino acids 1-20 of MUC1-CD (Fig. 6D). Mutation of the two cysteines in MUC1-CD had no effect on binding of to caspase-8-p18, indicating that the interaction is not mediated by disulfide bonds. Caspase-8-p18 contains the catalytic Cys-360 residue that is directly inhibited by the p35 protein from baculovirus (42). Our results indicate that MUC1-CD binds upstream to Cys-360. Moreover, unlike p35, incubation of active recombinant caspase-8 with stoichiometric concentrations of MUC1-CD in vitro had little effect on caspase-8 activity (data not shown). Homodimerization of caspase-8 results in interdimer processing to the p18 and p10 fragments that form active caspase-8 (43). Binding of cFLIP to caspase-8 blocks caspase-8 dimerization and the processing of caspase-8 to the p18 and p10 fragments (44). Binding of MUC1-C to the p18 region of caspase-8 could thus also interfere with interdimer processing and thereby prevent caspase-8 activation. In other studies, caspase-8 has been linked to activation of the IKK complex and NF-κB (44-47). In this context, recent work has shown that MUC1-C interacts with IKKβ/IKKγ and activates NF-κB signaling (31). However, it is presently not known if the interaction between MUC1-C and caspase-8 contributes to MUC1-C-mediated activation of the IKK->NF-κB pathway.
MUC1-C blocks recruitment of caspase-8 to the DISC
Activation of TNF-R1 results in binding to TRADD and then the recruitment of FADD (2). The activated Fas and DR4/5 receptors directly bind FADD through their DDs (9). Significantly, the present results demonstrate that MUC1-C is recruited to the DISC and that this response is dependent on FADD (Fig. 6D). We also found that MUC1-CD binds directly to the FADD DED. In concert with these results, MUC1-C blocks recruitment of caspase-8 to the DISC and MUC1-CD competes with caspase-8 for binding to the FADD DED in vitro. Direct binding studies further showed that MUC1-CD(46-72) confers the interaction with FADD. cFLIP competes with caspase-8 for recruitment to the DISC (41, 48-50). Thus, the demonstration that MUC1-C behaves like cFLIP by interacting with the FADD DED and blocking caspase-8 recruitment indicates that MUC1-C and cFLIP have redundant functions (Fig. 6D). However, MUC1-C is devoid of a DED and the same region of MUC1-CD that binds to FADD also interacts with β-catenin (30), IKKγ (31) and the HSP90/HSP70 complex (23, 24). These findings and the present results with FADD indicate that MUC1-C has multiple unrelated binding partners and therefore may have a chaperone-like function. MUC1-C also functions as a transmembrane receptor and thus would potentially represent a novel class of chaperone.
Binding of MUC1-C to FADD and caspase-8 is a physiologic response
Malignant cells could exploit a physiologic function of MUC1 through aberrant expression of this protein. For example, studies in MCF-10A cells demonstrated little constitutive interaction between MUC1-C and FADD or caspase-8. However, death receptor stimulation was associated with increased binding of MUC1-C to both FADD and caspase-8. Moreover, MUC1 attenuated death receptor-induced activation of caspase-8 in the MCF-10A cells. Thus, expression of MUC1 by normal epithelial cells may activate a physiological response involving inhibition of caspase-8 activation as a protective mechanism. In this regard, epithelial cells that are exposed to inflammatory conditions could utilize MUC1-dependent attenuation of caspase-8 to transiently protect against death in response to local release of TNFα, TRAIL or FasL by immune effector cells. In contrast to nonmalignant cells, MUC1 is aberrantly overexpressed by most human carcinomas and certain hematologic malignancies (10, 51, 52). The present findings indicate that overexpression of MUC1 as found in human tumors is of importance to attenuation of caspase-8 activation in response to death receptor stimulation and that MUC1-mediated inhibition of this pathway may have been exploited by malignant cells for survival under adverse conditions.
Supplementary Material
Acknowledgments
This work was supported by Grants CA100707, CA98628 and CA97098 awarded by the National Cancer Institute. The authors thank Dr. Marcus Peter (U. of Chicago) for the GST-FADD constructs. Mr. Kamal Chauhan is acknowledged for technical support. D.K. holds equity in Genus Oncology and is a consultant to the company.
The abbreviations used are
- MUC1
mucin 1
- MUC1-CD
MUC1 cytoplasmic domain
- TNFα
tumor necrosis factor α
- TNF-R1
TNF receptor 1
- TRAIL
TNF-related apoptosis-inducing ligand
- TRAIL-R1/DR4
TRAIL receptor 1
- TRAIL-R2/DR5
TRAIL receptor 2
- FasL
Fas ligand
- TRAF2
TNFR-associated factor 2
- DD
death domain
- DED
death effector domain
- RIP1
receptor interacting protein 1
- FADD
Fas associating protein with death domain
- DISC
death-inducing signaling complex
- cFLIP
caspase-8 (FLICE)-like inhibitory protein
- MUC1-N
MUC1 N-terminal subunit
- MUC1-C
MUC1 C-terminal subunit
References
- 1.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–8. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–90. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 3.Schneider-Brachert W, Tchikov V, Neumeyer J, et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–28. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Boldin M, Goncharov T, Goltsev Y, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell-death. Cell. 1996;85:803–15. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Chinnaiyan AM, Kischkel FC, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–27. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 6.Stennicke H, Jurgenmeier J, Shin H, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–90. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Zhu H, Xu C-j, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the fas pathway of apotosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 8.Luo X, Budiharjo H, Zou H, Slauhter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–90. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 9.Scaffidi C, Kirchhoff S, Krammer PH, Peter ME. Apoptosis signaling in lymphocytes. Curr Opin Immunol. 1999;11:277–85. doi: 10.1016/s0952-7915(99)80045-4. [DOI] [PubMed] [Google Scholar]
- 10.Kufe D, Inghirami G, Abe M, et al. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–32. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 11.Ligtenberg MJ, Kruijshaar L, Buijs F, et al. Cell-associated episialin is a complex containing two proteins derived from a common precursor. J Biol Chem. 1992;267:6171–7. [PubMed] [Google Scholar]
- 12.Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–6. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 13.Levitin F, Stern O, Weiss M, et al. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–86. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 14.Gendler S, Taylor-Papadimitriou J, Duhig T, Rothbard J, Burchell JA. A highly immunogenic region of a human polymorphic epithelial mucin expressed by carcinomas is made up of tandem repeats. J Biol Chem. 1988;263:12820–3. [PubMed] [Google Scholar]
- 15.Siddiqui J, Abe M, Hayes D, et al. Isolation and sequencing of a cDNA coding for the human DF3 breast carcinoma-associated antigen. Proc Natl Acad Sci USA. 1988;85:2320–3. doi: 10.1073/pnas.85.7.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–10. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Yu W-H, Ren J, et al. Heregulin targets γ-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 protein. Mol Cancer Res. 2003;1:765–75. [PubMed] [Google Scholar]
- 18.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–78. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Chen D, Liu D, et al. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 21.Wen Y, Caffrey T, Wheelock M, Johnson K, Hollingsworth M. Nuclear association of the cytoplasmic tail of MUC1 and β-catenin. J Biol Chem. 2003;278:38029–39. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 22.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren J, Bharti A, Raina D, et al. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 24.Ren J, Raina D, Chen W, et al. MUC1 oncoprotein functions in activation of fibroblast growth factor receptor signaling. Mol Cancer Res. 2006;4:873–83. doi: 10.1158/1541-7786.MCR-06-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlo G, Siddiqui J, Cropp C, et al. DF3 tumor-associated antigen gene is located in a region on chromosome 1q frequently altered in primary human breast cancer. Cancer Res. 1989;49:6966–71. [PubMed] [Google Scholar]
- 26.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3β and β-catenin. J Biol Chem. 2001;276:6061–4. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3β with the DF3/MUC1 carcinoma-associated antigen and β-catenin. Mol Cell Biol. 1998;18:7216–24. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren J, Li Y, Kufe D. Protein kinase C δ regulates function of the DF3/MUC1 carcinoma antigen in β-catenin signaling. J Biol Chem. 2002;277:17616–22. doi: 10.1074/jbc.M200436200. [DOI] [PubMed] [Google Scholar]
- 29.Raina D, Ahmad R, Kumar S, et al. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774–83. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and β-catenin in cell adhesion. J Biol Chem. 1997;272:12492–4. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–27. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–64. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 33.Yin L, Huang L, Kufe D. MUC1 oncoprotein activates the FOXO3a transcription factor in a survival response to oxidative stress. J Biol Chem. 2004;279:45721–7. doi: 10.1074/jbc.M408027200. [DOI] [PubMed] [Google Scholar]
- 34.Yin L, Kharbanda S, Kufe D. Mucin 1 oncoprotein blocks hypoxia-inducible factor 1alpha activation in a survival response to hypoxia. J Biol Chem. 2007;282:257–66. doi: 10.1074/jbc.M610156200. [DOI] [PubMed] [Google Scholar]
- 35.Bodmer JL, Holler N, Reynard S, et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–3. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 36.Scaffidi C, Volkland J, Blomberg I, et al. Phosphorylation of FADD/ MORT1 at serine 194 and association with a 70-kDa cell cycle-regulated protein kinase. J Immunol. 2000;164:1236–42. doi: 10.4049/jimmunol.164.3.1236. [DOI] [PubMed] [Google Scholar]
- 37.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 38.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–92. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–75. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Chaturvedi R, Srivastava RK, Hisatsune A, et al. Augmentation of Fas ligand-induced apoptosis by MUC1 mucin. Int J Oncol. 2005;26:1169–76. [PubMed] [Google Scholar]
- 41.Han DK, Chaudhary PM, Wright ME, et al. MRIT, a novel death-effector domain-containing protein, interacts with caspases and BclXL and initiates cell death. Proc Natl Acad Sci USA. 1997;94:11333–8. doi: 10.1073/pnas.94.21.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu G, Cirilli M, Huang Y, et al. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–7. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- 43.Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
- 45.Kreuz S, Siegmund D, Rumpf JJ, et al. NFkappaB activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. J Cell Biol. 2004;166:369–80. doi: 10.1083/jcb.200401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su H, Bidere N, Zheng L, et al. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–8. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 47.Park SM, Schickel R, Peter ME. Nonapoptotic functions of FADD-binding death receptors and their signaling molecules. Curr Opin Cell Biol. 2005;17:610–6. doi: 10.1016/j.ceb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Irmler M, Thome M, Hahne M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–5. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasula SM, Ahmad M, Ottilie S, et al. FLAME-1, a novel FADD-like anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. J Biol Chem. 1997;272:18542–5. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 50.Goltsev YV, Kovalenko AV, Arnold E, et al. CASH, a novel caspase homologue with death effector domains. J Biol Chem. 1997;272:19641–4. doi: 10.1074/jbc.272.32.19641. [DOI] [PubMed] [Google Scholar]
- 51.Dyomin VG, Palanisamy N, Lloyd KO, et al. MUC1 is activated in a B-cell lymphoma by the t(1;14)(q21;q32) translocation and is rearranged and amplified in B-cell lymphoma subsets. Blood. 2000;95:2666–71. [PubMed] [Google Scholar]
- 52.Brossart P, Schneider A, Dill P, et al. The epithelial tumor antigen MUC1 is expressed in hematological malignancies and is recognized by MUC1-specific cytotoxic T-lymphocytes. Cancer Res. 2001;29:6846–50. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.