Abstract
HIV encodes several proteins, including Tat that have been demonstrated to modulate the expression of receptors critical for innate immunity including MHC class I, mannose receptor and β2-microglobulin. We demonstrate that Tat targets the receptor tyrosine kinase RON, which negatively regulates inflammation and HIV transcription, for proteosome degradation. Tat decreases cell surface RON expression in HIV-infected monocytic cells and Tat-mediated degradation of RON protein is blocked by inhibitors of proteosome activity. Tat specifically induced down regulation of RON and not other cell surface receptors such as the transferrin receptor, the receptor tyrosine kinase TrkA or monocytic markers CD14 and ICAM-1. The Tat transactivation domain is required for RON degradation and this down regulation is dependent on the integrity of the kinase domain of RON receptor. We propose that Tat mediates degradation of RON through a ubiquitin-proteosome pathway and suggest that by targeting signals that modulate inflammation, Tat creates a microenvironment that is optimal for HIV replication and progression of AIDS associated diseases.
Keywords: HIV, Tat, RON Receptor Tyrosine Kinase, Proteosome-Dependent Degradation
Introduction
Macrophages, in addition to CD4+ T cells, are a major target for human immunodeficiency virus (HIV) and are potential long term cellular reservoir for virus dissemination (1, 2). Furthermore, dysregulated proinflammatory activities of macrophages associated with HIV infection directly contributes to AIDS and the development of associated pathologies in the central nervous system, lung, lymph nodes and skin (3, 4). Successful eradication of HIV infection will depend on a better understanding of the biochemical mechanisms that regulate HIV replication in macrophages.
HIV, like all viruses, has employed a variety of strategies such as down regulation of immune receptors on the surface of cells, to avoid detection and elimination by immune responses (5). The HIV encoded proteins gp120, Tat and Nef have been shown to influence the expression of a variety of cell surface molecules (6). For example, the viral protein Tat has been demonstrated to modulate the expression of cell surface proteins such MHC class I (7, 8), mannose receptor (9) and β2-microglobulin (10). Despite a large set of data regarding Tat functions, the exact molecular mechanism by which Tat affects cellular gene expression still remains elusive.
Tat is essential for efficient HIV replication and is required for provirus transcription initiation and elongation. Tat binds to the transactivation response element (TAR), which forms an RNA stem-loop structure present at the 5’-end of HIV transcripts (11-16). In addition, Tat has been reported to have a variety of other activities not associated with its ability to regulate HIV transcription including neurotoxicity (17), immunosuppression and inhibition of antigen-induced lymphocyte activation (18, 19). Furthermore, Tat contributes directly to the dysregulation of cytokine expression inducing the production of interleukin-1 (IL-1), IL-6, tumor necrosis alpha (TNF-α), TNF-β, transforming growth factor β1 (TGF-β1) and the IL-4 receptor (20-25). Tat has also been shown to regulate the expression of several molecules that could impact the immune response to HIV infection including cell surface proteins such as MHC class I (7, 8), mannose receptor (9), HIV coreceptors CXCR4 and CCR5 (26, 27), macrophage –inflammatory protein 1-α (MIP-1α) (28), β2-microglobulin (10), cytokine receptors (29, 30), vascular endothelial growth factor (VEGF) (31) and integrins (32). The ability of Tat to modulate the expression of many genes with no TAR element implies that Tat acts by interacting with other cellular regulatory proteins (33). These functions of Tat contribute to viral persistence and dissemination by directly or indirectly modulating the host anti-HIV response.
The receptor tyrosine kinase RON (Recepteur d'Origine Nantais), which belongs to the Met proto-oncogene family (34), is a critical regulator of macrophage function and inflammation (35). RON is expressed mostly on tissue resident macrophages and regulates multiple cellular responses including proliferation, differentiation, apoptosis, phagocytosis and cell motility (35-37). Macrophage-stimulating protein (MSP), the ligand for RON, is a potent inhibitor of proinflammatory mediators such as nitric oxide (NO), IL-12 and TNF-α (38-41), whereas it induces the expression of genes associated with wound healing and the resolution of inflammation including scavenger receptor A, IL-1Rα and arginase (42). Activated macrophages from RON knockout mice produce elevated levels of NO in vitro and in vivo and have demonstrated increased susceptibility to endotoxic shock as well as to autoimmune diseases mediated by inflammation (43-45). RON knockout mice also have compromised cell-mediated immunity as demonstrated by an increased susceptibility to Listeria monocytogenes infection (46). We have previously shown that RON inhibits HIV transcription (47, 48) and replication in macrophages and that the expression of this receptor tyrosine kinase is decreased in brains of AIDS patients (47).
In this study we demonstrate that Tat decreases cell surface RON expression by targeting this receptor tyrosine kinase for ubiquitin-mediated proteosome degradation. Considering the role of RON in regulating tissue resident macrophage activities and inflammation, these findings suggest an additional mechanism by which HIV escapes the immune system and creates a microenvironment that is favorable for virus replication and progression of AIDS.
Materials and Methods
Cells
The U937 promonocytic cell line (ATCC, Manassas, VA) was cultured in RPMI 1640 medium supplemented with 5% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin and 0.2 M l-glutamine. The 293T human embryonic kidney cell line (ATCC, Manassas, VA) was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.2 M l-glutamine. U937-RON cells were generated by transducing a RON retroviral vector, MSCV-RON, as previously described (47). Peripheral blood macrophages were isolated from whole blood obtained from healthy HIV-1-seronegative donors. Mononuclear cells were obtained by differential centrifugation using a Ficol/Hystopaque (Sigma-Aldrich) gradient and adherence to plastic culture flasks as previously described (49). Macrophages were separated from all blood cells by an initial adherence to plastic culture flasks overnight. After removing non-adherent cells, monocytes were cultured for 5−7 days to mature into monocyte-derived macrophages (MDM). Mouse peritoneal macrophages were obtained by peritoneal lavage with 10 ml of RPMI 1640 containing 10% FCS. Cells were incubated overnight and washed with PBS to eliminate all non-adherent cells. Cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Plasmids, transient transfections and luciferase activity assay
Pvuless-hRON was kindly provided by Dr. R. Breathnach (Institut de Biologie, Nantes, France). MSCV-RON construct (47) and the RON mutant constructs were generated as previously described (50). HXB.2, HIV-1 GST-Tat expression vectors (86R TK, GST-Tat 1 72R, GST-Tat 1 72R P18lS and GST-Tat 1 48 D TK) (51), Mtat30 (52), Nef (53) were obtained from the AIDS Research Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The JRFL expression construct was provided by Dr. R.I. Connor (Dartmouth, NH) (54). DNA for transfections was prepared using plasmid purification systems from Marligen Biosciences (Ijamsville, MD) following protocols provided by the manufacturer. Transfection of 293T cells was performed using TransIT-293 Transfection reagent from Mirus (Madison, WI). For transfection of U937 cells, DNA was added to 0.4 ml of U937 cells resuspended in serum free RPMI 1640 media. The cell suspensions were electroporated using a T820 square electroporation system (BTX, San Diego, CA) with one pulse for 20 mSec at 240 V in 4-mm cuvettes. Cells were resuspended in 5 ml of RPMI 1640 supplemented with 5% fetal calf serum. Transfection efficiency was assessed by co-transfecting pEGEP-N3 (Clonetech, Palo Alto, CA) and monitoring EGFP expression by fluorescence microscopy. Cell viability was confirmed by trypan blue staining. RON overexpression did not influence the transfection efficiencies of the different cell lines as previously described (47). CAT ELISA assay (Roche) was performed to determine the promoter activity of the RON gene in cells transfected with and without Tat DNA. Luciferase assay has been performed using a commercial luciferase assay kit (Promega, Madison WI) and a TD-20/20 luminometer (Turner BioSystems, Sunnyvale, CA).
Generation of HIV infectious titers, infections and transduction of primary macrophages
Replication competent HXB.2 and replication incompetent Mtat30 viruses were generated by cotransfecting 293T cells with 15 μg of cDNA for either HXB.2 or Mtat30, 3 μg of vesicular stomatitis virus-glycoprotein (VSV-G) and 3 μg RSV-Rev by CaPO4 transfection (55). Transfection efficiency was assessed by measuring p24 levels (p24 ELISA, Perkin Elmer, Wellesley, MA). We consistently generated titers of 1.0 ×106 infectious particles/ml. Supernatants were collected and filtered with 0.45 μm syringe filter (Whatman, Clifton, NJ). One milliliter of undiluted virus stocks was added to 1.0×106 cells for 24 h and then replaced with fresh media. MSCV-RON retroviral construct was packaged in 293T cells by transfecting 15 μg of the viral DNA, 3μg of VSV-G envelope, 3 μg of pECO and 3 μg of Tat by CaPO4 transfection. Supernatants were collected and filtered through a 0.45μm syringe filter (Whatman, Clifton, NJ). For some experiments, cells were treated with purified recombinant HIV-1 Tat (obtained from the AIDS Research Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health) (56) for 24 h in 1% serum media.
Pharmacological inhibitors
Proteosome activity was inhibited by treating the cells with the proteosome inhibitors MG-132 (Calbiochem) at a concentration of 5 μM at 37°C in 5% serum media for 30 min and Epoxomicin (Sigma-Aldrich) at a concentration of 1 μM at 37°C in 5% serum media for 1h. Lysosomal proteolysis pathway inhibitor Ammonium Chloride (NH4Cl) (Sigma-Aldrich) was applied to cells at a concentration of 20 μM for a duration of 30 min in 5% serum media. For inhibiting protein synthesis, cells were pretreated with 2 ng/ml of cycloheximide (Sigma-Aldrich) in 5% serum media for 1 h. For all experiments using inhibitors cell viability was monitored by trypan blue staining.
RNA extraction and RT-PCR
Total cellular RNA was prepared using Trizol Reagents (Invitrogen) according to the manufacturer's instructions. Tat expression was analyzed using Tat primers 5’GGA ATT CAC CAT GGA GCC AGT AGA TCC T 3’ and 5’ CGG GAT CCC TAT TCC TTC GGG CCT GT 3’. The PCR program consists of the following steps: 50°C for 30 min, 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, and one cycle of 72°C for 7 min. Murine RON mRNA was analyzed by RT-PCR using RON primers 5′ CAG CAG TGG ACA GCC TGT TCA 3′ and 5′ATG CCT TCC ACT CGG AAG TGC 3′. The PCR program consists of the following steps: 94°C for 3min followed by 35 cycles of 93°C for 30sec, 60°C for 1min and at the end 72°C for 2min.
Immunoprecipitation and Immunoblots
Cells were washed twice with phosphate-buffered saline and protein extracts were prepared by treating cells with lysis buffer (10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1.0 mM EDTA (pH 8.0), 2.0 mM sodium vanadate, 10 mM sodium fluoride, 10 mM sodium pyrophosphate, 1% Nonidet P-40, 1.0 mM phenylmethylsufonyl fluoride, 1.0 mM pepstain) at 4°C for 30 min. Lysates were mixed with 2×SDS loading buffer containing dithiothreitol and heated at 100°C for 5 min before resolving by SDS-PAGE with 8% polyacrylamide. Proteins were transferred to nitrocellulose membrane (Pall Corporation) and blocked with 5% non-fat dry milk in PBS with 0.02% v/v Tween-20 (J. T. Baker chemical corporation, Phillipsburg, NJ). Rabbit-anti-RON polyclonal antibody (Santa Cruz Biotechnologies) was used to probe for RON. Horseradish peroxidase conjugated goat anti-rabbit IgG (Sigma-Aldrich) was used as the secondary antibody. For detection of β-actin, membranes were stripped with 100 mM β-mercaptoethanol, 62.5 mM Tris-HCl (pH 6.7), 2% SDS for 30 min at 55°C and reprobed with mouse anti-β-actin antibody (Sigma-Aldrich) for 1 h at 25°C, which was detected with horseradish peroxidase-conjugated goat-anti-mouse (Sigma-Aldrich) secondary antibody. Goat-anti-CD71 antibody (Santa Cruz Biotechnologies) was used to detect transferrin receptor expression. Horseradish peroxidase conjugated anti-goat IgG (Sigma-Aldrich) was used as the secondary antibody. TrkA was detected with anti-TrkA (Millipore) and the secondary anti-rabbit IgG conjugated with horseradish peroxidase conjugated (Sigma-Aldrich). MHC I antibody (Santa Cruz Biotechnologies) was used to probe for MHC I expression. Horseradish peroxidase conjugated anti-rabbit IgG (Sigma-Aldrich) was used as the secondary antibody. Nef protein expression was detected with HIV-1 Nef antiserum (Shugars et al., 1993) and gp120 (JRFL) envelope protein with antiserum to HIV-1 gp120 (Chiron Corporation) obtained from NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Anti-serum against Ron was raised against the last 15 amino acids of the carboxyl tail. Anti-ubiquitin antibody (Pierce) has been used to detect ubiquitinated form of proteins and anti-rabbit IgG was used as the secondary antibody. Blots were developed using an ECL-plus kit (Amersham Biosciences, Piscataway, NJ) and exposed to films. For some blots, densitometry with a personal ImageQuant analysis tool (Amersham Biosciences) was performed to determine signal intensities.
For immunoprecipitation, cell lysates were precleared for 30 min with protein A/G (Santa Cruz Biotechnologies). RON was immunoprecipitated with 1.0 μg anti-RON antibody (Santa Cruz Biotechnologies) and protein A/G agarose beads (Santa Cruz Biotechnologies). Protein-bound beads were washed four times with PBS and resuspended in 1×SDS buffer (50 mM Tris-HCl (pH 6.8), 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mM DTT) and resolved by SDS-PAGE.
Cell surface protein biotinylation and ubiquitination
Cell surface protein biotinylation and isolation was performed using Pinpoint cell surface protein isolation kit from Pierce. U937-RON cells were labeled with EZ-Link Sulfo-NHS-SS-Biotin and then lysed and isolated with immobilized NeutrAvidin gel. The bound proteins were released by incubating with SDS-PAGE sample buffer containing 50 mM DTT. The flow-through and elution were kept for analysis. Western blot was performed using an anti-ubiquitin antisera (included with kit). Ubiquitination of RON receptor at the cell surface is performed by using ubiquitin enrichment kit from Pierce. Cell lysates were incubated with ubiquitin affinity resin overnight. Ubiquitinated forms of RON were eluated with SDS-PAGE loading buffer and western analysis was performed using anti-ubiquitin antisera monoclonal antibody (included with the kit).
Results
Tat decreases RON protein
We previously observed a correlation between HIV infection and decreased RON expression in brain tissue (47). One possible explanation for diminished RON expression is that HIV directly targets this receptor. Therefore, to address whether HIV down regulates RON expression, U937-RON cells, a U937 monocytic cell line transduced with RON, were transfected with HIV HXB.2 cDNA plasmid and RON expression was monitored by immunoblotting (Fig. 1A). RON is expressed as a 160 kDa precursor protein and 140 kDa proteolytic product, although the detection of this doublet depends upon the resolution of the gel and the quality of the anti-RON antibody. We consistently observed a decrease in RON protein by greater than 75% when cells were transfected with the HIV plasmid.
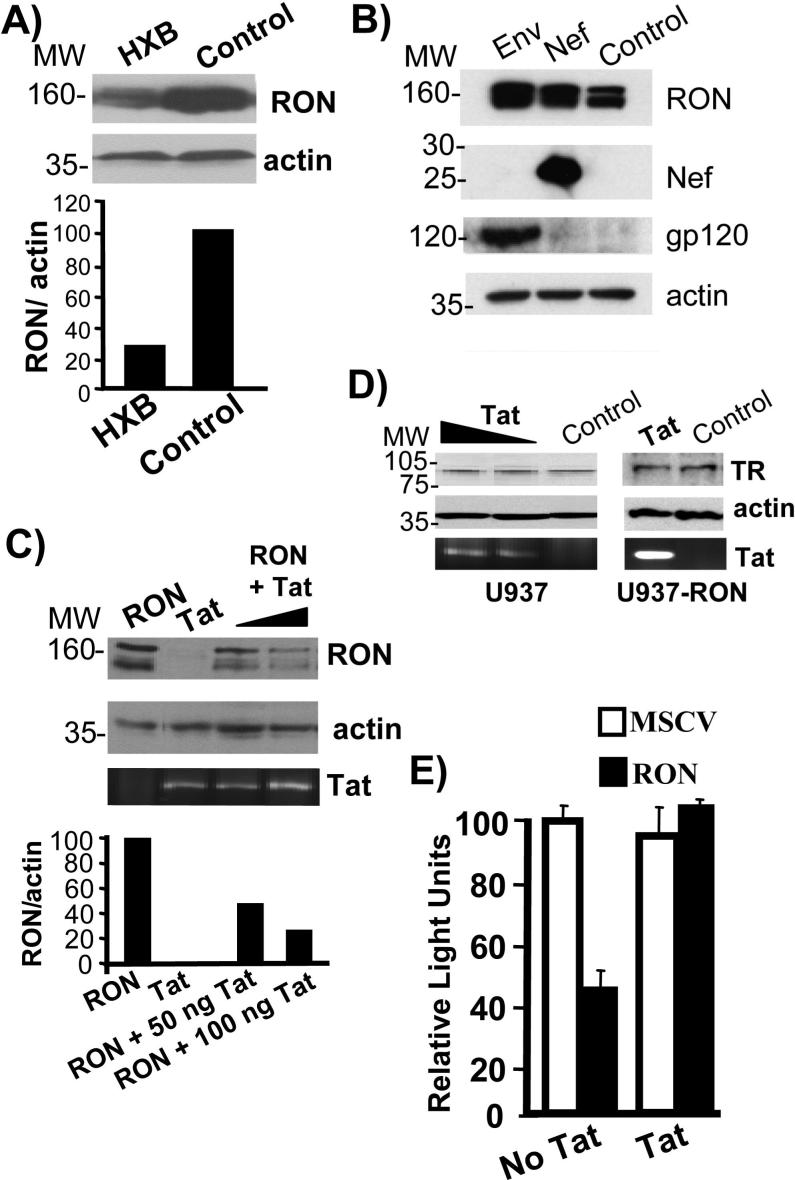
Figure 1.
Tat decreases RON expression and circumvents RON-dependent repression of HIV transcription. A) U937-RON cells were transfected with HXB.2 or PCI DNA (empty vector control). Whole cell extracts were prepared 24 h post-transfections and immunoblotted to determine RON expression. RON is expressed as a 160 kDa precursor protein and 140 kDa proteolytic product, although the detection of this doublet depends upon the resolution of the gel and the quality of the anti-RON antibody. Blots were stripped and reprobed for β-actin. RON expression relative to β-actin was assessed by densitometry. B) 293T cells were cotransfected with MSCV-RON and 100 ng PCI control plus 100 ng Nef or JRFL Env expression constructs. 16 h post-transfection, cells were lysed and immunoblots were performed to determine RON, Nef and gp120 expression. In control experiments, Nef was able to down regulate CD4 in Jurkat T cells (data not shown). C) 293T cells were cotransfected with MSCV-RON and either PCI or 50 ng and 100 ng of wild-type Tat expression construct (86R TK). Cells were also transfected with 50 ng of wild-type Tat expression construct alone as a control. 16 h post-transfection, cells were lysed and RON immunoblots were performed. Blots were stripped and reprobed for β-actin. RON expression relative to β-actin was assessed by densitometry. Since there are no reliable Tat antibodies available for detecting Tat protein expression, Tat expression was analyzed by RT-PCR of the total RNA using Tat-specific primers. PCR products were analyzed on 1% agarose gel. D) U937 and U937-RON cells were transfected with 0.5 and 2 μg of Tat expression plasmid. 48 h post-transfection cells were lysed and the expression of transferrin receptor (TR) was detected by immunoblotting. Blots were stripped and reprobed for β-actin. RT-PCR was used to detect Tat expression using Tat primers. PCR products were analyzed on 1% agarose gel. E) U937-MSCV (unfilled bars) or U937-RON (filled bars) cells were transfected with 10 μg LTR-Luc and 1 μg Tat or PCI empty vector. Cells were harvested 48 h post-transfection and assayed for luciferase activity. Following transfections trypan blue staining was performed to control for cell viability. Transfections were performed in triplicate and data are presented as the percentage of luciferase, with luciferase activity in the cells lacking RON being set at 100%. All experiments were performed a minimum of three times.
HIV encodes six accessory proteins that potentially influence cell function and HIV replication. In particular, Nef and Tat have been shown to have activities that alter gene expression, receptor turnover, cell growth and differentiation (6). To determine which HIV protein(s) were potentially involved in down regulating RON expression, 293T cells were cotransfected with expression vectors encoding RON and different HIV proteins. As shown in Fig. 1B and 1C, Nef and the envelope protein gp120 did not affect RON expression, whereas transfected Tat decreased RON expression by 50 to 70%. Tat did not mediate a general decrease in protein expression, since no changes were observed in the expression of transferrin receptor in U937 or U937-RON cells transfected with Tat (Fig. 1D). To determine if Tat has the ability to overcome the suppressive effects of RON on HIV transcription, U937-RON and U937-MSCV (cells transduced with the empty vector) were co-transfected with Tat and a HIV LTR-luciferase reporter (LTR-Luc) and assayed for luciferase activity. The presence of RON inhibited LTR activity by more than 2-fold compared with LTR activity in Tat transfected U937-MSCV cell line. Moreover, Tat induces LTR activity by more than 2-fold in the presence of RON suggesting that Tat is able to at least partly circumvent RON-dependent inhibition of HIV transcription (Fig. 1E). The LTR-Luc reporter construct used in this experiment lacks the TAR element which is replaced by the luciferase gene, therefore, any Tat-induced transcription would reflect activities other than its ability to activate provirus transcription processivity.
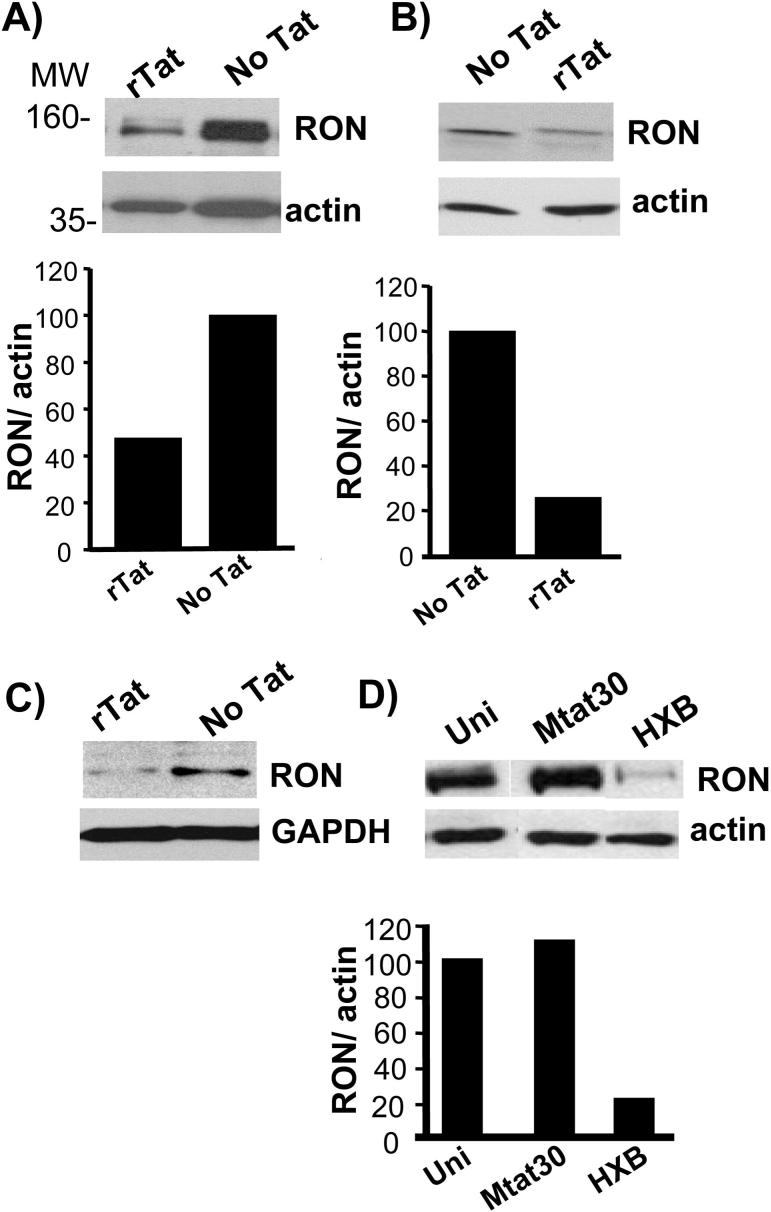
Tat decreased the expression of RON in the context of both U937 monocytic cells and primary macrophages. For these experiments cells were treated with recombinant Tat, which is rapidly taken up by cells, as an alternative to transfection (57, 58). Recombinant Tat was capable of transactivating a transfected HIV-LTR-luciferase reporter plasmid and did not induce cell death as determined by exclusion of trypan blue or propidium iodide exclusion and flow cytometry (data not shown). As shown in figure 2A, recombinant Tat significantly decreased RON expression in U937-RON cells by more than 50% compared with cells that were not treated with Tat. The decrease in RON protein was rapid, occuring within two hours upon addition of recombinant Tat to U937-RON cells and extending up to at least 24 h (data not shown). Similar results were observed in primary monocyte-derived macrophages transduced with RON following Tat treatment (Fig. 2B). It is important to note that the addition of Tat did not alter the activity of the MSCV retroviral promoter used to express RON since no differences were observed in RON mRNA levels in cells transduced with RON retroviral vector cultured in the presence and absence of Tat (data not shown). In addition, the expression of GFP from a MSCV-EGFP retroviral vector was not altered by the presence or absence of Tat (data not shown). The above experiments depend on overexpression of RON, in part because human primary tissue resident macrophages that express RON are not readily accessible, therefore, we tested whether Tat influenced the expression of RON in primary murine peritoneal macrophages. Addition of recombinant Tat to these cells diminished the expression of RON protein by more than 75 % (Fig. 2C). The ability of Tat to down modulate RON in the context of primary mouse macrophages is independent of its ability to increase transcription processivity, since Tat is unable to interact with P-TEFb in the context of murine cells (59). Diminished RON expression was also observed following infection with HIV IIIB, whereas infection with HIV clone Mtat30, which expresses a missense Tat, did not alter RON protein levels (Fig. 2D) indicating that the levels of Tat associated with an infection are sufficient to down regulate RON.
Figure 2.
Tat diminishes RON expression in monocyte/macrophages. A) U937-RON cells were treated without and with 50 ng/ml of recombinant Tat for 24 h. Cells were lysed and immunoblots were performed using anti-RON polyclonal antibody. B) Primary monocyte-derived-macrophages transduced with MSCV-RON were incubated with 100 ng/ml recombinant Tat for 24 h. Cells were lysed and immunoblotting was performed using anti-RON polyclonal antibody. C) Primary mouse peritoneal macrophages were treated with 100 ng/ml recombinant Tat for 24 h. Cells were lysed and immunoblots were performed using anti-Ron (RON) antibody. D) U937-RON cells were infected with HIV clones Mtat30 or HXB.2. Controls included uninfected cells (Uni). 24 h post-infection, immunoblots were performed on whole cell extract to determine RON expression. For these experiments the immunoblots were stripped and reprobed for β-actin. RON expression relative to β-actin was assessed by densitometry. All experiments were performed a minimum of three times. All composite figures were taken from the same gel and a single exposure.
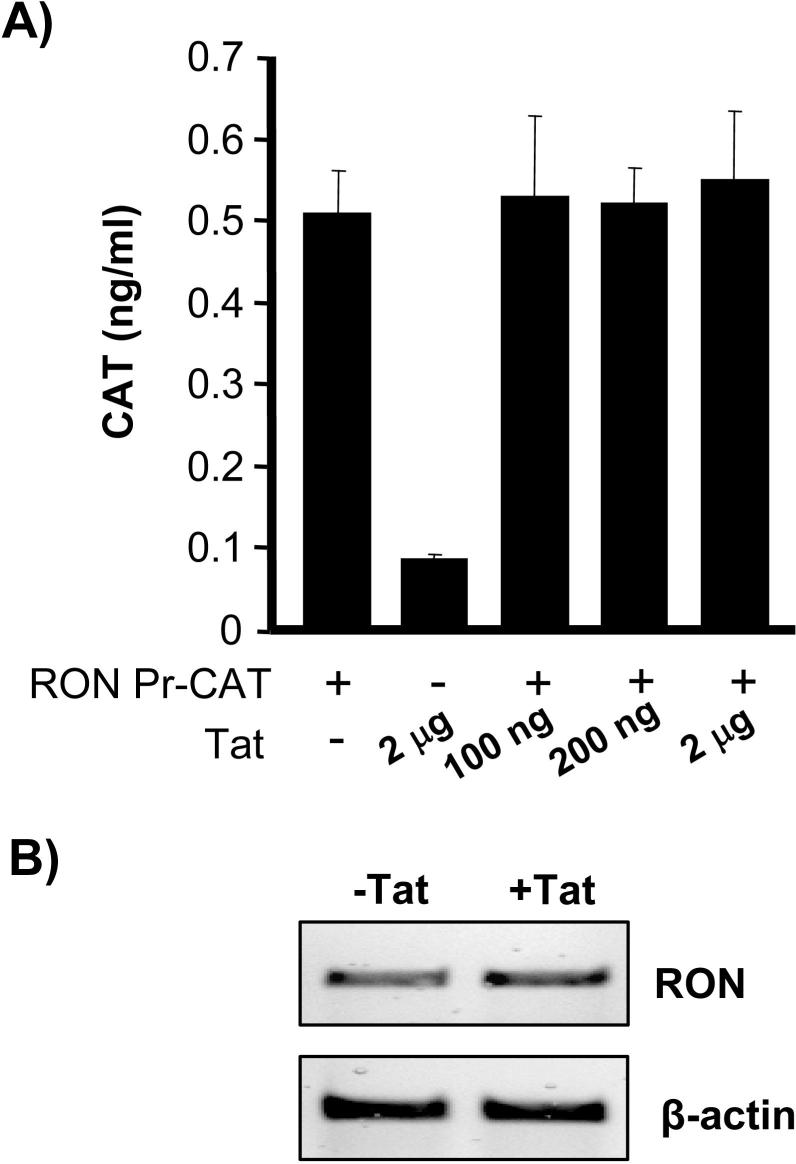
Tat does not appear to regulate RON transcription. In cotransfection experiments Tat was unable to transactivate the RON promoter (Fig 3A). Furthermore, treating primary mouse peritoneal macrophages with recombinant Tat did not alter RON mRNA levels (Fig. 3B). Taken together, these data suggest that Tat is down-regulating RON independent of transcriptional regulation.
Figure 3.
Tat does not affect RON transcription. A) 293T cells were cotransfected with RON-CAT, a plasmid in which the CAT reporter is regulated by the RON promoter and the indicated amount of Tat expression plasmid. 48 h post-transfection, CAT activity was measured by ELISA (Roche Applied Science). As a positive control for Tat activity, 293T cells were cotransfected with Tat expression plasmid and an HIV-LTR luciferase reporter construct. 100 ng of Tat DNA induced HIV-LTR activity by greater than 10 fold and 2 μg of Tat expression plasmid induced HIV-LTR activity by greater than 200 fold (data not shown). These data are representative of at least three experiments. B) Primary mouse peritoneal macrophages were treated with (100 ng/ml) recombinant Tat for 24 h. mRNA was extracted from these cells and RT-PCR was performed for Ron (RON)expression.
Tat increases ubiquitin-dependent proteosome degradation of RON
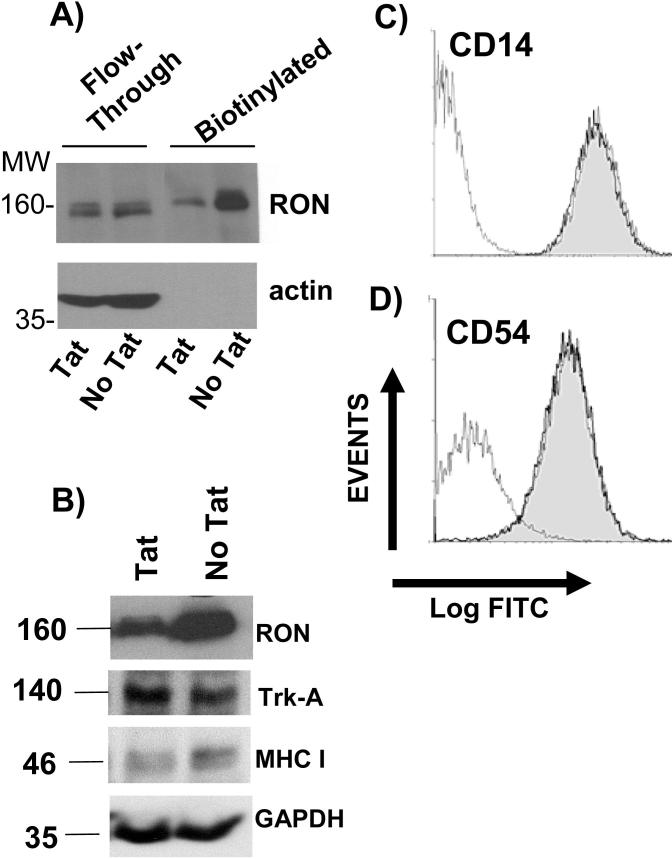
To gain insight into the mechanism by which Tat is targeting RON, we examined whether RON was being expressed at the cell surface in the absence and presence of Tat. Since available antibodies to RON poorly detect cell surface staining, we employed a strategy in which cell surface proteins were biotinylated and subsequently immunoprecipitated and purified using NeutrAvidin Gel. In Tat treated U937-RON cells, reduced biotinylated RON was present compared to U937-RON cells that were not treated with Tat, suggesting that Tat is inducing the internalization and subsequent degradation of the receptor (Fig. 4A). There was no significant change in the expression of the surface receptor tyrosine kinase TrkA (Fig. 4B) or the monocytic markers CD14, CD54 (ICAM-1) on U937-RON cells in the presence and absence of Tat (Fig. 4C and 4D) indicating that Tat is not leading to a general turnover of surface proteins. A modest decrease was observed for MHC I (Fig 4B) consistent with previous studies which have shown that Tat inhibits MHC I transcription (7, 8).
Figure 4.
Tat diminishes the expression of cell surface RON. A) U937-RON cells were treated with 100ng/ml recombinant Tat and then surface proteins were subjected to biotinylatation with Sulfo-NHS-SS-Biotin. Biotinylated proteins were enriched using immobilized NeutrAvidin Gel. The elution and flow-through fractions were analyzed for RON by immunoblot. Blots were reprobed for β-actin as a control. B) U937-RON cells were transfected with Tat or PCI DNA (empty vector control). Whole cell extracts were prepared 24 h post-transfections and immunoblotted to determine RON, TrkA and MHC I expression. C) and D) Flow cytometry histograms of U937 cells treated with or without recombinant Tat (100ng/ml) for 24 h. Cells were stained for C) CD14 and D) CD54 (ICAM-1) and analyzed by flow-cytometry. The histograms are: isotype control (thin line); treated with Tat (shaded light gray) and untreated with Tat (thick line).
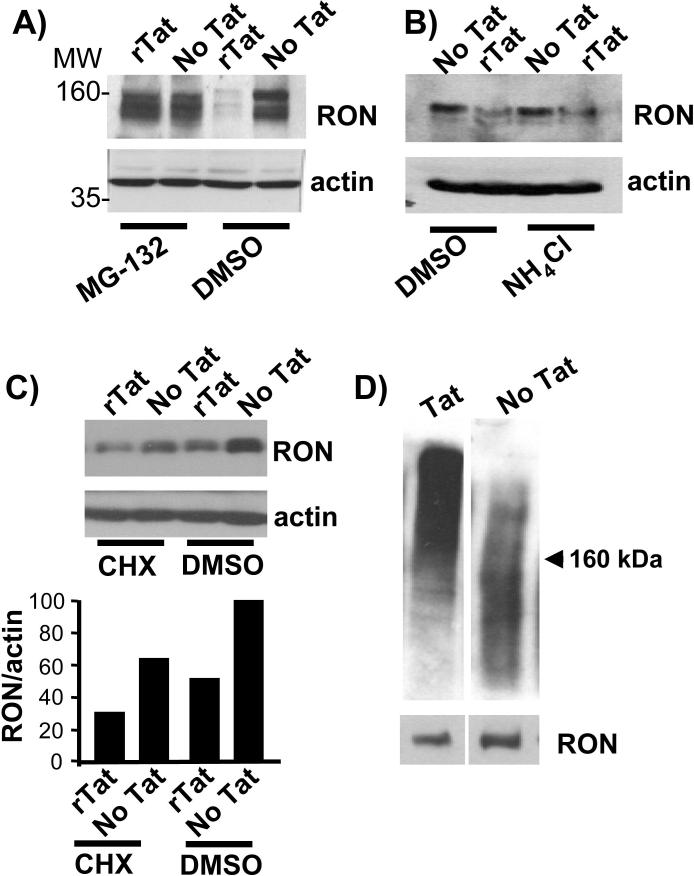
Previous studies have shown that MET and RON expression is regulated by proteosome-dependent degradation (60-62). To investigate whether Tat-mediated down regulation of RON requires the proteosome pathway, we treated U937-RON cells with recombinant Tat protein in the presence and absence of the proteosome inhibitor MG-132. MG-132 blocked the ability of Tat to mediate RON turnover (Fig. 5A). We observed similar results with Epoxomicin, a selective proteosome inhibitor (data not shown). Ammonium chloride (NH4Cl), an inhibitor of the lysosomal proteolysis pathway in macrophages (63) or cycloheximide, a inhibitor of protein synthesis, had no effect on the RON degradation in the presence of Tat (Fig. 5B & C).
Figure 5.
Tat-mediated degradation of RON is ubiquitin proteosome-dependent. A) U937-RON cells were treated with recombinant Tat (100 ng/ml) for 24 h in the presence of proteosome inhibitor MG-132 (5 μM) or DMSO as a vehicle control for 1 h. Immunoblots were probed with an anti-RON antibody. B) U937-RON cells were treated with recombinant Tat (100 ng/ml) for 24 h in the presence of NH4Cl (20 μM) or DMSO as a vehicle control for 30 min. RON expression was monitored by immunoblotting. C) U937-RON cells were incubated with cycloheximide (CHX; 2 ng) or DMSO for 1 h and then were treated with 100 ng/ml recombinant Tat for 24 h. Whole cell extracts were prepared and probed for RON. All immunoblots were stripped and reprobed for β-actin. RON expression relative to β-actin was assessed by densitometry. D) Lysates from 293T cells cotransfected with RON (3 μg) and Tat (6 μg) were subjected to immunoprecipitation with RON antibody followed by enrichment for ubiquitinated protein. Immunoblots were performed using anti-ubiquitin antibody. The blot then was stripped and reprobed for RON protein. All data in composite figure were taken from the same gel and exposure. Data are representative of at least three independent experiments.
Proteins are directed to the 26S proteosome for degradation by polyubiquitination, a post-translational modification in which ubiquitin polypeptides are covalently linked to lysines on the targeted proteins. In order to determine whether Tat promotes RON polyubiquitination, 293T cells were cotransfected with RON and Tat plasmids and subsequently RON was immunoprecipitated and examined for polyubiquitynation by immunoblotting with an anti-ubiquitin antibody. RON from the Tat transfected cells resolved in a high molecular weight smear, consistent with polyubiquitination, whereas, less ubiquitinated RON was observed in cells that lack Tat (Fig. 5D). Taken together our data show that Tat mediated down regulation of RON occurs through a ubiquitin-proteosome pathway.
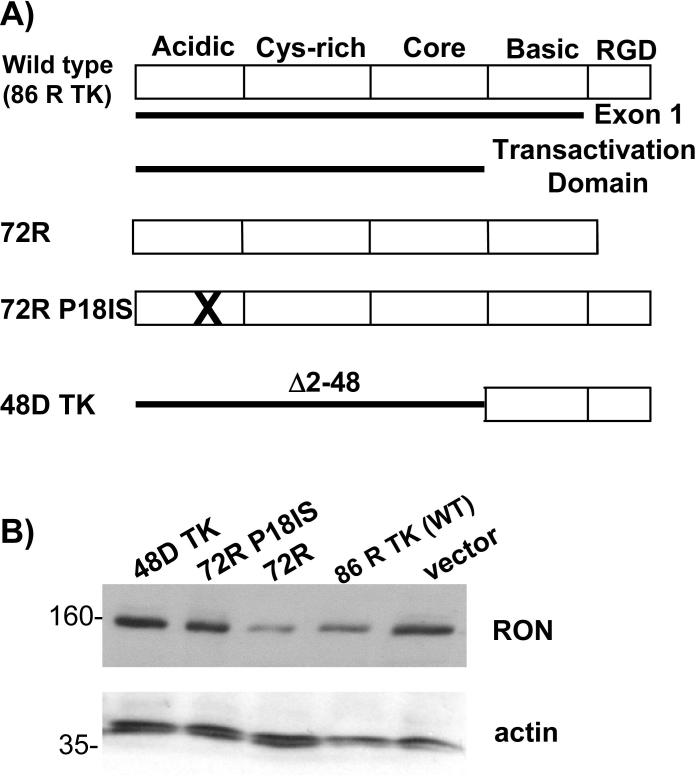
The Tat transactivation domain is required for degradation of RON
Tat protein is composed of five functional domains (Fig. 6A) (64). The transactivation domain includes the amino terminal acidic region, the adjacent cysteine-rich domain, the core domain and the basic domain which are encoded by the first exon (1−72 aa) (51) and are able to fully activate the HIV-LTR (65). To determine which domain(s) of Tat is mediating the degradation of RON protein, we cotransfected various Tat mutants with a RON expression construct. The first 72 amino acids (Tat 72R) were sufficient to mediate down regulation of RON and disrupting the transactivation domain by either inserting a glycine and phenylalanine between amino acid residues 18 and 19 (72R P181S) or deleting amino acids 2−48 (48D TK) abolished the ability of Tat to regulate RON expression. Furthermore, the basic region, which mediates RNA binding and the RGD domain, was unable to alter RON expression (48D TK; Fig 6B). These data suggest that the transactivation domain of HIV Tat is required for regulating RON expression (Fig. 6B).
Figure 6.
The Tat transactivation domain is required for degradation of RON. A) Schematic presentation of the domains of wild type Tat and mutations. B) U937-RON cells were transfected with 5 μg PCI (control empty vector) and expression vector for wild type Tat or Tat mutants. 24 h post transfection, cells were lysed and RON was detected by immunoblotting. Blots were stripped and reprobed for β-actin, a control for protein loading.
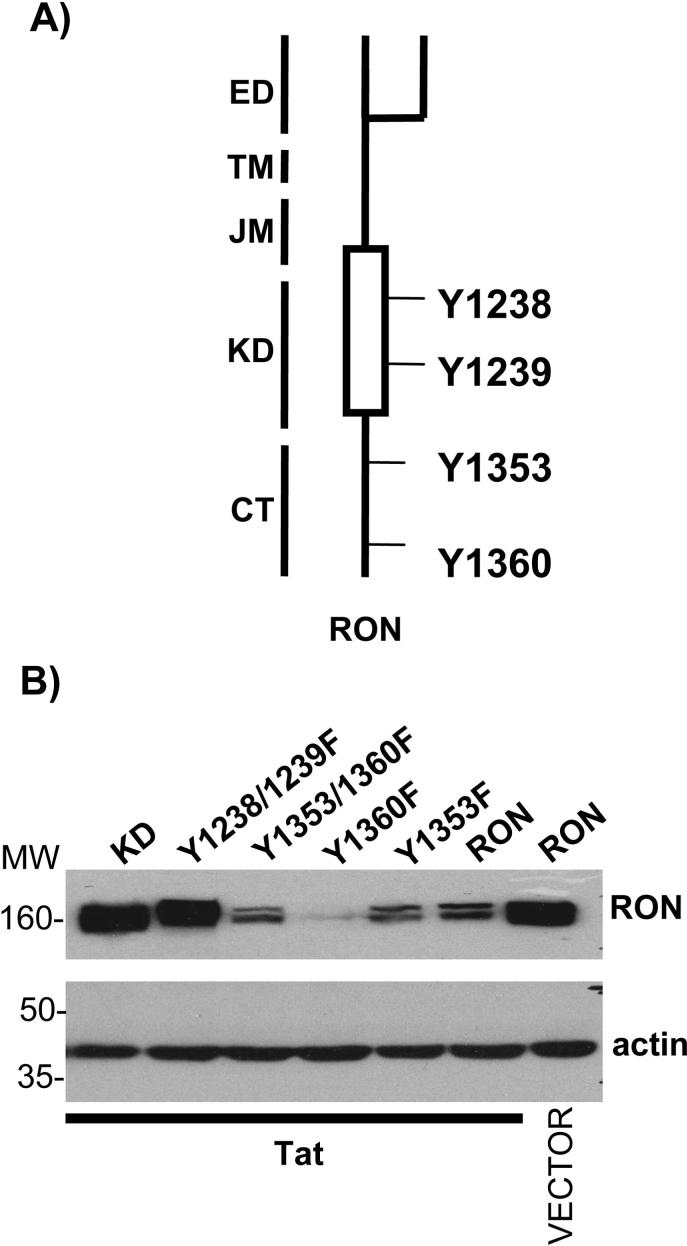
Tat mediated degradation requires the RON kinase domain but not the docking sites
To investigate which tyrosine residues of RON are involved in Tat mediated degradation of this receptor, we employed a series of RON mutations in which critical tyrosines were mutated to phenylalanines (Fig. 7A). Tyrosines 1353 and 1360 serve as a multifunctional docking site for several signaling molecules including Gab-1, Grb2, PI3K, phopholipase C and SHP-2 (35). Mutating these residues individually or in combination (Y1353F, Y1360F and Y 1353/1360F) did not affect the ability of Tat to down regulate RON protein. By contrast, the kinase defective RON mutants, RON KD and RON Y1238/1239F were not susceptible to Tat-dependent degradation of RON (Fig. 7B) indicating that Tat-mediated RON turnover is dependent on the integrity of the kinase domain.
Figure 7.
Tat-mediated degradation requires the kinase domain but not the docking site of RON. A) Schematic diagram of critical tyrosine residues in RON. ED, extracellular domain; TM, transmembrane domain; JM, juxtamembrane domain; KD, kinase domain; CT, C-terminal. B) 293T cells were transfected with 2 μg of RON, RON mutant expression vectors and Tat expression vector or PCI empty vector. 24 h post transfection, cells were lysed and RON was detected by immunoblotting. Blots were then stripped and reprobed for β-actin, a control for protein loading.
Discussion
HIV-1 targets critical receptors that participate in the regulation of the immune response. For example, Nef has been shown to interact with several signal transduction proteins and down regulates the cell surface expression of CD4, CD28, MHC class I molecules and FcγRs (66-68). Tat has also been shown to repress the expression of multiple cell surface proteins such a MHC class I (7, 8), mannose receptor proteins (9) and β2-microglobulin (10). In this study we demonstrate that Tat diminishes the expression of RON, a receptor tyrosine kinase that regulates inflammation and macrophage function, through an ubiquitin proteosome-dependent degradation.
Tat decreases RON expression in a variety of cell types including primary murine peritoneal macrophages indicating a lack of tissue and species specificity. However, the ability of Tat to target RON for degradation does not reflect a general turnover of cell surface receptors since TrkA, CD14 and ICAM-1 surface expression were not altered in presence and absence of Tat. Furthermore, RON expression was decreased by the addition of exogenous Tat. Although we do not know the exact mechanism by which recombinant Tat is mediating this activity, we have shown that the RGD domain, which is recognized by cell surface integrin receptors is not required for modulating RON expression, whereas the transactivation domain is sufficient for this RON degradation. We suspect that recombinant Tat is rapidly entering the cells, even though the levels of Tat associated with an in vitro infection were sufficient to modulate RON expression. Tat has been reported to be released from infected cells and taken up by neighboring uninfected cells (57, 69) providing a mechanism by which HIV can compromise immune function of uninfected macrophages. Whether the levels of Tat present in the serum or tissue are sufficient to have an impact on cell function in vivo is controversial.
Tat targets RON for ubiquitin proteosome degradation as demonstrated by polyubiquitination of RON in the presence of Tat and the observation that Tat-dependent degradation of RON was blocked with the proteosome inhibitors MG-132 and Epoxomicin. This ability of Tat to down regulate protein expression by ubiquitin-proteosome degradation is a novel function of Tat. A recent report also suggested that Tat mediated proteosome-dependent degradation of microtubule-associated protein 2 (MAP-2) accounted for the loss of MAP-2 and neuronal damage observed in the brain of AIDS patients with neurological dysfunctions (70). Tat has also been shown to interact with several subunits of the 26S proteosome and to modulate the 20S proteosome activity (71, 72). We have not observed a physical interaction between Tat and RON (data not shown) suggesting that Tat is targeting RON possibly by activating signal transduction pathways that negatively regulate RON. Consistent with this hypothesis is the observation that RON degradation requires a functional kinase domain. Previous studies have similarly shown that the degradation of the Met receptor tyrosine kinase requires kinase activity (73). RON and Met as well as other receptor tyrosine kinases have been shown to be degraded upon activation in part by the recruitment of the E3 ubiquitin ligase c-Cbl (60). The role of c-Cbl in Tat-dependent degradation of RON is currently being explored.
HIV Tat is a potent transactivator of transcription and is essential for viral replication (74, 75). This viral protein interacts with the TAR element and in cooperation with host cellular factors, in particular P-TEFb, functions to activate provirus transcription (76, 77). Furthermore, Tat has been shown to activate NF-κB signaling to transactivate gene expression in the absence of TAR (78-80). Several reports have also shown that Tat represses the transcription of multiple genes such as MHC class I (7, 8), mannose receptor (9) and β2-microglobulin (10). However, the ability of Tat to regulate RON expression is independent of P-TEFb since Tat was able to induce RON degradation in mouse peritoneal macrophages, despite the inability of Tat to interact with the murine CycT1 subunit of P-TEFb. Exon 1 of Tat, which includes the transactivation domain, is required for down modulation of RON indicating that the transactivation domain most likely mediates protein-protein interactions that facilitates targeting of RON to the proteosome. It is unlikely that Tat is inducing the expression of additional genes that indirectly down-modulate RON since cycloheximide does not influence Tat-dependent degradation of RON.
RON signaling has been shown to repress the transcription of iNOS, TNF-α and IL-12 (38-41). We have previously demonstrated that RON is also able to inhibit HIV transcription (47, 48). Since HIV transcription is tightly linked to proinflammatory signals and RON signaling has been shown to inhibit these signals, decreasing RON expression would be advantageous for virus replication. In addition, decreased RON expression would lead to dysregulation of the cytokine network and compromised innate immunity, increasing the susceptibility to opportunistic infection and facilitating disease progression. We have previously shown that RON expression is repressed in brain samples of a subset of HIV patients (47). Although this decrease in RON expression may be a consequence of multiple mechanisms associated with chronic infection and inflammation including changes in cell populations, we propose that HIV Tat, in part by down regulating RON, contributes to a microenvironment that favors HIV replication and further facilitates tissue damage and disease progression.
Aknowledgements
We are grateful to Dr. Avery August for critical review of the manuscript and scientific discussions. Also we thank Elaine Kunze and Susan F. Magargee at Pennsylvania State University flow cytometry core facility for valuable assistance and Qingping Liu and Xin Wei for the RON mutant expression constructs. The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 GST-Tat expression vectors from Dr. Andrew Rice, pMtat30 from Dr. Reza Sadaie, Nef expression vector from Drs. Yingying Li, Feng Gao and Beatrice H. Hahn, HIV-1 Tat protein from Dr. John Brady, HIV-1 Nef antiserum from Dr. Ronald Swanstrom, and gp120 antibody from Chiron Corporation.
Footnotes
This project is supported by funds from the Penn State Tobacco Formula Funds and NIH grant AI46261 to A.J.H. and AHA predoctoral fellowship grant 0415425U to P.K.
References
- 1.Gendelman HE, Orenstein JM, Baca LM, Weiser B, Burger H, Kalter DC, Meltzer MS. The macrophage in the persistence and pathogenesis of HIV infection. AIDS. 1989;3:475–495. doi: 10.1097/00002030-198908000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 3.Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 4.Giulian D, Vaca K, Noonan CA. Secretion of neurotoxins by mononuclear phagocytes infected with HIV-1. Science. 1990;250:1593–1596. doi: 10.1126/science.2148832. [DOI] [PubMed] [Google Scholar]
- 5.Ploegh HL. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 6.Emerman M, Malim MH. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 7.Howcroft TK, Strebel K, Martin MA, Singer DS. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science. 1993;260:1320–1322. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 8.Weissman JD, Brown JA, Howcroft TK, Hwang J, Chawla A, Roche PA, Schiltz L, Nakatani Y, Singer DS. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc. Natl. Acad. Sci. USA. 1998;95:11601–11606. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell RL, Egan BS, Shepherd VL. HIV-1 Tat represses transcription from the mannose receptor promoter. J. Immunol. 2000;165:7035–7041. doi: 10.4049/jimmunol.165.12.7035. [DOI] [PubMed] [Google Scholar]
- 10.Carroll IR, Wang J, Howcroft TK, Singer DS. HIV Tat represses transcription of the beta 2-microglobulin promoter. Mol. Immunol. 1998;35:1171–1178. doi: 10.1016/s0161-5890(98)00107-2. [DOI] [PubMed] [Google Scholar]
- 11.Dayton AI, Terwilliger EF, Potz J, Kowalski M, Sodroski JG, Haseltine WA. Cis-acting sequences responsive to the rev gene product of the human immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 1988;1:441–452. [PubMed] [Google Scholar]
- 12.Fisher AG, Feinberg MB, Josephs SF, Harper ME, Marselle LM, Reyes G, Gonda MA, Aldovini A, Debouk C, Gallo RC, et al. The trans-activator gene of HTLV-III is essential for virus replication. Nature. 1986;320:367–371. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- 13.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 14.Berkhout B, Gatignol A, Rabson AB, Jeang KT. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabatier JM, Vives E, Mabrouk K, Benjouad A, Rochat H, Duval A, Hue B, Bahraoui E. Evidence for neurotoxic activity of tat from human immunodeficiency virus type 1. J. Virol. 1991;65:961–967. doi: 10.1128/jvi.65.2.961-967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanyam M, Gutheil WG, Bachovchin WW, Huber BT. Mechanism of HIV-1 Tat induced inhibition of antigen-specific T cell responsiveness. J. Immunol. 1993;150:2544–2553. [PubMed] [Google Scholar]
- 19.Viscidi RP, Mayur K, Lederman HM, Frankel AD. Inhibition of antigen-induced lymphocyte proliferation by Tat protein from HIV-1. Science. 1989;246:1606–1608. doi: 10.1126/science.2556795. [DOI] [PubMed] [Google Scholar]
- 20.Lafrenie RM, Wahl LM, Epstein JS, Yamada KM, Dhawan S. Activation of monocytes by HIV-Tat treatment is mediated by cytokine expression. J. Immunol. 1997;159:4077–4083. [PubMed] [Google Scholar]
- 21.Buonaguro L, Barillari G, Chang HK, Bohan CA, Kao V, Morgan R, Gallo RC, Ensoli B. Effects of the human immunodeficiency virus type 1 Tat protein on the expression of inflammatory cytokines. J. Virol. 1992;66:7159–7167. doi: 10.1128/jvi.66.12.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buonaguro L, Buonaguro FM, Giraldo G, Ensoli B. The human immunodeficiency virus type 1 Tat protein transactivates tumor necrosis factor beta gene expression through a TAR-like structure. J. Virol. 1994;68:2677–2682. doi: 10.1128/jvi.68.4.2677-2682.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puri RK, Aggarwal BB. Human immunodeficiency virus type 1 tat gene up-regulates interleukin 4 receptors on a human B-lymphoblastoid cell line. Cancer Res. 1992;52:3787–3790. [PubMed] [Google Scholar]
- 24.Sastry KJ, Reddy HR, Pandita R, Totpal K, Aggarwal BB. HIV-1 tat gene induces tumor necrosis factor-beta (lymphotoxin) in a human B-lymphoblastoid cell line. J. Biol. Chem. 1990;265:20091–20093. [PubMed] [Google Scholar]
- 25.Scala G, Ruocco MR, Ambrosino C, Mallardo M, Giordano V, Baldassarre F, Dragonetti E, Quinto I, Venuta S. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 TAT protein. J. Exp. Med. 1994;179:961–971. doi: 10.1084/jem.179.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss JM, Nath A, Major EO, Berman JW. HIV-1 Tat induces monocyte chemoattractant protein-1-mediated monocyte transmigration across a model of the human blood-brain barrier and up-regulates CCR5 expression on human monocytes. J. Immunol. 1999;163:2953–2959. [PubMed] [Google Scholar]
- 27.Secchiero P, Zella D, Capitani S, Gallo RC, Zauli G. Extracellular HIV-1 tat protein up-regulates the expression of surface CXC-chemokine receptor 4 in resting CD4+ T cells. J. Immunol. 1999;162:2427–2431. [PubMed] [Google Scholar]
- 28.Sharma V, Xu M, Ritter LM, Wilkie NM. HIV-1 tat induces the expression of a new hematopoietic cell-specific transcription factor and downregulates MIP-1 alpha gene expression in activated T-cells. Biochem. Biophys. Res. Commun. 1996;223:526–533. doi: 10.1006/bbrc.1996.0928. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Drenkow J, Lankford CS, Frucht DM, Rabin RL, Gingeras TR, Venkateshan C, Schwartzkopff F, Clouse KA, Dayton AI. HIV regulation of the IL-7R: a viral mechanism for enhancing HIV-1 replication in human macrophages in vitro. J. Leuk. Biol. 2006;79:1328–1338. doi: 10.1189/jlb.0704424. [DOI] [PubMed] [Google Scholar]
- 30.Husain SR, Leland P, Aggarwal BB, Puri RK. Transcriptional up-regulation of interleukin 4 receptors by human immunodeficiency virus type 1 tat gene. AIDS Res. Hum. Retroviruses. 1996;12:1349–1359. doi: 10.1089/aid.1996.12.1349. [DOI] [PubMed] [Google Scholar]
- 31.Albini A, Soldi R, Giunciuglio D, Giraudo E, Benelli R, Primo L, Noonan D, Salio M, Camussi G, Rockl W, Bussolino F. The angiogenesis induced by HIV-1 tat protein is mediated by the Flk-1/KDR receptor on vascular endothelial cells. Nat. Med. 1996;2:1371–1375. doi: 10.1038/nm1296-1371. [DOI] [PubMed] [Google Scholar]
- 32.Zocchi MR, Poggi A, Rubartelli A. The RGD-containing domain of exogenous HIV-1 Tat inhibits the engulfment of apoptotic bodies by dendritic cells. AIDS. 1997;11:1227–1235. doi: 10.1097/00002030-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Jones KA. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 34.Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–1202. [PubMed] [Google Scholar]
- 35.Correll PH, Morrison AC, Lutz MA. Receptor tyrosine kinases and the regulation of macrophage activation. J. Leuk. Biol. 2004;75:731–737. doi: 10.1189/jlb.0703347. [DOI] [PubMed] [Google Scholar]
- 36.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell Motility Is Controlled by SF2/ASF through Alternative Splicing of the Ron Protooncogene. Mol. Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Lutz MA, Correll PH. Activation of CR3-mediated phagocytosis by MSP requires the RON receptor, tyrosine kinase activity, phosphatidylinositol 3-kinase, and protein kinase C zeta. J. Leuk. Biol. 2003;73:802–814. doi: 10.1189/jlb.0602319. [DOI] [PubMed] [Google Scholar]
- 38.Wang MH, Cox GW, Yoshimura T, Sheffler LA, Skeel A, Leonard EJ. Macrophage-stimulating protein inhibits induction of nitric oxide production by endotoxin- or cytokine-stimulated mouse macrophages. J. Biol. Chem. 1994;269:14027–14031. [PubMed] [Google Scholar]
- 39.Chen YQ, Fisher JH, Wang MH. Activation of the RON receptor tyrosine kinase inhibits inducible nitric oxide synthase (iNOS) expression by murine peritoneal exudate macrophages: phosphatidylinositol-3 kinase is required for RON-mediated inhibition of iNOS expression. J. Immunol. 1998;161:4950–4959. [PubMed] [Google Scholar]
- 40.Liu QP, Fruit K, Ward J, Correll PH. Negative regulation of macrophage activation in response to IFN-gamma and lipopolysaccharide by the STK/RON receptor tyrosine kinase. J. Immunol. 1999;163:6606–6613. [PubMed] [Google Scholar]
- 41.Morrison AC, Wilson CB, Ray M, Correll PH. Macrophage-stimulating protein, the ligand for the stem cell-derived tyrosine kinase/RON receptor tyrosine kinase, inhibits IL-12 production by primary peritoneal macrophages stimulated with IFN-gamma and lipopolysaccharide. J. Immunol. 2004;172:1825–1832. doi: 10.4049/jimmunol.172.3.1825. [DOI] [PubMed] [Google Scholar]
- 42.Morrison AC, Correll PH. Activation of the stem cell-derived tyrosine kinase/RON receptor tyrosine kinase by macrophage-stimulating protein results in the induction of arginase activity in murine peritoneal macrophages. J. Immunol. 2002;168:853–860. doi: 10.4049/jimmunol.168.2.853. [DOI] [PubMed] [Google Scholar]
- 43.Correll PH, Iwama A, Tondat S, Mayrhofer G, Suda T, Bernstein A. Deregulated inflammatory response in mice lacking the STK/RON receptor tyrosine kinase. Genes Funct. 1997;1:69–83. doi: 10.1046/j.1365-4624.1997.00009.x. [DOI] [PubMed] [Google Scholar]
- 44.Leonis MA, Toney-Earley K, Degen SJ, Waltz SE. Deletion of the Ron receptor tyrosine kinase domain in mice provides protection from endotoxin-induced acute liver failure. Hepatology. 2002;36:1053–1060. doi: 10.1053/jhep.2002.36822. [DOI] [PubMed] [Google Scholar]
- 45.Tsutsui S, Noorbakhsh F, Sullivan A, Henderson AJ, Warren K, Toney-Earley K, Waltz SE, Power C. RON-regulated innate immunity is protective in an animal model of multiple sclerosis. Ann. Neurol. 2005;57:883–895. doi: 10.1002/ana.20502. [DOI] [PubMed] [Google Scholar]
- 46.Lutz MA, Gervais F, Bernstein A, Hattel AL, Correll PH. STK receptor tyrosine kinase regulates susceptibility to infection with Listeria monocytogenes. Infect. Immun. 2002;70:416–418. doi: 10.1128/IAI.70.1.416-418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee ES, Kalantari P, Tsutsui Section S, Klatt A, Holden J, Correll PH, Power Section C, Henderson AJ. RON receptor tyrosine kinase, a negative regulator of inflammation, inhibits HIV-1 transcription in monocytes/macrophages and is decreased in brain tissue from patients with AIDS. J. Immunol. 2004;173:6864–6872. doi: 10.4049/jimmunol.173.11.6864. [DOI] [PubMed] [Google Scholar]
- 48.Klatt A, Zhang Z, Kalantari P, Hankey PA, Gilmour DS, Henderson AJ. The receptor tyrosine kinase RON represses HIV-1 transcription by targeting RNA polymerase II processivity. J. Immunol. 2008;180:1670–1677. doi: 10.4049/jimmunol.180.3.1670. [DOI] [PubMed] [Google Scholar]
- 49.Henderson AJ, Calame KL. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4(+) T cells. Proc. Natl. Acad. Sci. USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei X, Hao L, Ni S, Liu Q, Xu J, Correll PH. Altered exon usage in the juxtamembrane domain of mouse and human RON regulates receptor activity and signaling specificity. J. Biol. Chem. 2005;280:40241–40251. doi: 10.1074/jbc.M506806200. [DOI] [PubMed] [Google Scholar]
- 51.Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadaie MR, Rappaport J, Benter T, Josephs SF, Willis R, Wong-Staal F. Missense mutations in an infectious human immunodeficiency viral genome: functional mapping of tat and identification of the rev splice acceptor. Proc. Natl. Acad. Sci. USA. 1988;85:9224–9228. doi: 10.1073/pnas.85.23.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, Shaw GM, Zajac AJ, Letvin N, Hahn BH. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses. 2003;19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- 54.Connor RI, Sheridan KE, Lai C, Zhang L, Ho DD. Characterization of the functional properties of env genes from long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 1996;70:5306–5311. doi: 10.1128/jvi.70.8.5306-5311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohan CA, Kashanchi F, Ensoli B, Buonaguro L, Boris-Lawrie KA, Brady JN. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 1992;2:391–407. [PMC free article] [PubMed] [Google Scholar]
- 57.Ensoli B, Barillari G, Salahuddin SZ, Gallo RC, Wong-Staal F. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature. 1990;345:84–86. doi: 10.1038/345084a0. [DOI] [PubMed] [Google Scholar]
- 58.Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 1993;67:277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penengo L, Rubin C, Yarden Y, Gaudino G. c-Cbl is a critical modulator of the Ron tyrosine kinase receptor. Oncogene. 2003;22:3669–3679. doi: 10.1038/sj.onc.1206585. [DOI] [PubMed] [Google Scholar]
- 61.Hammond DE, Urbe S, Vande Woude GF, Clague MJ. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene. 2001;20:2761–2770. doi: 10.1038/sj.onc.1204475. [DOI] [PubMed] [Google Scholar]
- 62.Hammond DE, Carter S, Clague MJ. Met receptor dynamics and signalling. Curr. Top. Microbiol. Immunol. 2004;286:21–24. doi: 10.1007/978-3-540-69494-6_2. [DOI] [PubMed] [Google Scholar]
- 63.Hart PD, Young MR. Ammonium chloride, an inhibitor of phagosomelysosome fusion in macrophages, concurrently induces phagosome-endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J. Exp. Med. 1991;174:881–889. doi: 10.1084/jem.174.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rice AP, Carlotti F. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J. Virol. 1990;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sodroski J, Rosen C, Wong-Staal F, Salahuddin SZ, Popovic M, Arya S, Gallo RC, Haseltine WA. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985;227:171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- 66.Swigut T, Shohdy N, Skowronski J. Mechanism for down-regulation of CD28 by Nef. EMBO J. 2001;20:1593–1604. doi: 10.1093/emboj/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blagoveshchenskaya AD, Hannah MJ, Allen S, Cutler DF. Selective and signal-dependent recruitment of membrane proteins to secretory granules formed by heterologously expressed von Willebrand factor. Mol. Biol. Cell. 2002;13:1582–1593. doi: 10.1091/mbc.01-09-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De SK, Venkateshan CN, Seth P, Gajdusek DC, Gibbs CJ. Adenovirus-mediated human immunodeficiency virus-1 Nef expression in human monocytes/macrophages and effect of Nef on downmodulation of Fcgamma receptors and expression of monokines. Blood. 1998;91:2108–2117. [PubMed] [Google Scholar]
- 69.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 70.Aprea S, Del Valle L, Mameli G, Sawaya BE, Khalili K, Peruzzi F. Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J. Neurosci. 2006;26:4054–4062. doi: 10.1523/JNEUROSCI.0603-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- 72.Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ, Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Letters. 2003;553:200–204. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- 73.Jeffers M, Taylor GA, Weidner KM, Omura S, Vande Woude GF. Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeang KT, Chang Y, Berkhout B, Hammarskjold ML, Rekosh D. Regulation of HIV expression: mechanisms of action of Tat and Rev. AIDS. 1991;5(Suppl 2):S3–14. [PubMed] [Google Scholar]
- 75.Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Ann. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 76.Dayton AI, Sodroski JG, Rosen CA, Goh WC, Haseltine WA. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986;44:941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- 77.Arya SK, Guo C, Josephs SF, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 78.Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bagasra O, Khalili K, Seshamma T, Taylor JP, Pomerantz RJ. TAR-independent replication of human immunodeficiency virus type 1 in glial cells. J. Virol. 1992;66:7522–7528. doi: 10.1128/jvi.66.12.7522-7528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor JP, Pomerantz R, Bagasra O, Chowdhury M, Rappaport J, Khalili K, Amini S. TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. EMBO J. 1992;11:3395–3403. doi: 10.1002/j.1460-2075.1992.tb05418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]