Abstract
Exposure to tobacco smoke is associated with increased DNA methylation at certain genes in both lung and bladder tumors. We sought to identify interactions in bladder cancer between DNA methylation and a history of smoking, along with any possible effect of aging. We measured DNA methylation in 342 transitional cell carcinoma (TCC) tumors at BCL2, PTGS2 (COX2), DAPK, CDH1 (ECAD), EDNRB, RASSF1A, RUNX3, TERT, and TIMP3. The prevalence of methylation at RUNX3, a polycomb target gene, increased as a function of age at diagnosis (p=0.031) and a history of smoking (p=0.015). RUNX3 methylation also preceded methylation at the other 8 genes (p<0.001). It has been proposed that DNA methylation patterns constitute a “molecular clock” and can be used to determine the “age” of normal tissues, i.e., the number of times the cells have divided. Since RUNX3 methylation increases with age, is not present in normal urothelium, and occurs early in tumorigenesis, it can be used for the first time as a molecular clock in order to determine the age of a bladder tumor. Doing so reveals that tumors from smokers are “older” than tumors from nonsmokers (p=0.009) either due to tumors in smokers initiating earlier or undergoing more rapid cell divisions. Since RUNX3 methylation is acquired early on in tumorigenesis then its detection in biopsy or urine specimens could provide a marker to screen cigarette smokers long before any symptoms of bladder cancer are present.
Keywords: RUNX3, methylation, bladder cancer, tobacco smoking, age
Introduction
Cancer etiology involves interactions between the environment, the genome, and the epigenome (1, 2). The epigenome consists of several layers of heritable transcriptional regulation imposed upon the genome, including DNA methylation, histone modifications, and nucleosome positioning (3). DNA becomes methylated at CpG dinucleotides and is associated with transcriptional repression when it occurs in CpG rich regions (CpG islands) located in gene promoters (4). During carcinogenesis, CpG islands become aberrantly hypermethylated (5). Alterations in the epigenome also accumulate during the aging process (6). For instance, normal epithelium in the colon acquires methylation in an age-dependent manner at genes that subsequently become hypermethylated in colon cancer (7). Therefore, it has been proposed that DNA methylation patterns constitute a molecular clock and can be used to determine the “age” of normal tissues, i.e., the number of times the cells have divided (8). It may also be possible to use methylation as a molecular clock to determine the age of tumor tissues by using a locus that is specifically methylated in tumor cells and does not acquire methylation in normal tissues as a function of aging. Information about the age of a tumor may give insight into when tumorigenesis initiated and have important implications for early detection.
Numerous types of environmental exposures, such as tobacco smoke, arsenic, cadmium, and nickel, are associated with aberrant DNA methylation (2). Specifically, tobacco-derived carcinogens are associated with DNA methylation at p16 (9), CYP1A1 (10), and RASSF1A (11) in lung tumors and p16 in bladder tumors (12). Marsit et al. (13) recently measured promoter hypermethylation of 16 different genes in bladder tumors and found a correlation between overall methylation and age, gender, and smoking history. However, their analysis combined the methylation at all the genes into one measure based on the premise that hypermethylation of promoters is not a targeted event. While the precise mechanism resulting in de novo methylation of specific CpG islands in cancer is currently unknown, recent work has demonstrated that genes targeted by polycomb complexes in embryonic stem (ES) cells are more likely than other genes to undergo promoter hypermethylation during carcinogenesis (14-16).
Bladder cancer is the fifth most commonly diagnosed cancer in the United States, where the majority of tumors are transitional cell carcinoma (TCC) (17). Long-term cigarette smokers are 2.5 times more likely to develop bladder cancer than nonsmokers (18). In this study, we took advantage of a large number of TCC samples from patients with known age, gender, and exposure to tobacco smoke in order to address the possibility that these factors are associated with epigenetic alterations. We utilized the real-time based methylation sensitive PCR assay MethyLight, developed by Eads et al. (19), to specifically and sensitively detect DNA methylation at nine different genes (RUNX3, BCL2, PTGS2 [COX2], DAPK, CDH1 [ECAD], EDNRB, RASSF1A, TERT, and TIMP3) in bladder tumor DNA samples from 342 patients. RUNX3 is a tumor suppressor involved in apoptosis and is both frequently silenced by methylation in bladder cancer (20, 21) and a confirmed polycomb target (22-24). RUNX3 methylation is associated with bladder tumor grade, invasiveness (20), stage, recurrence, and progression (21). Our results revealed interactions between environmental exposure, aging, and the epigenome. In particular, we have shown for the first time that DNA methylation, in addition to being used as a molecular clock to determine the age of normal tissues, can determine the age of tumors and our RUNX3 methylation data suggests that bladder tumors from smokers are older than from nonsmokers. Therefore, bladder tumorigenesis may initiate early in a smoker's life, while tumors from nonsmokers initiate at a later age.
Materials and Methods
Study population
From 1987 to 1996 a population-based case-control study of bladder cancer was conducted in Los Angeles County as described previously (18). Eligibility criteria for cases included histologically confirmed transitional cell carcinoma diagnosed between January 1, 1987 and April 30, 1996 among non-Asian patients aged 25 to 65 years. In total 2,098 cases were identified through the Los Angeles County Cancer Surveillance Program, and of these 1,582 patients were included as part of a case-control pair.
Tissue collection
Hospitals and pathology laboratories provided tumor tissue blocks to the Los Angeles County SEER Registry Slide Retrieval Program, a component of the Tissue Procurement Core Resource of the USC/Norris Comprehensive Cancer Center. Of the specimens retrieved, 342 cases had sufficient tumor available to permit the analysis of DNA methylation (Table 1). Frozen blocks of matched bladder tumor and corresponding mucosa have been described previously (25) and urothelium from cancer-free bladders was obtained from age-matched patients undergoing prostatectomies. All study subjects had signed informed consent forms approved by the Human Subjects Committee at the University of Southern California Keck School of Medicine.
Table 1.
Clinicopathological characteristics of study population
| Variables | N | % |
|---|---|---|
| No. of patients | 342 | |
| TNM Stage | ||
| Ta | 154 | 45% |
| CIS | 9 | 3% |
| I | 93 | 27% |
| II | 54 | 16% |
| III | 18 | 5% |
| IV | 13 | 4% |
| Grade | ||
| 1 | 109 | 32% |
| 2 | 134 | 39% |
| 3 | 87 | 25% |
| 4 | 11 | 3% |
| Sex | ||
| Male | 259 | 76% |
| Female | 83 | 24% |
| Age at diagnosis | ||
| ≤40 | 22 | 6% |
| 41-50 | 60 | 18% |
| 51-60 | 175 | 51% |
| 61-65 | 84 | 25% |
| Median (range) | 58 (25-65) | |
| Smoking History | ||
| Never/Irregular | 45 | 13% |
| Former | 137 | 40% |
| Current | 159 | 46% |
| Duration of Smoking (years) | ||
| 0 | 46 | 13% |
| 1 to 15 | 36 | 11% |
| 16 to 29 | 79 | 23% |
| 30 to 39 | 95 | 28% |
| ≥ 40 | 86 | 25% |
| Smoking in Pack-Years | ||
| 0 | 46 | 13% |
| 1 to 30 | 103 | 30% |
| 31 to 60 | 105 | 31% |
| 61 to 85 | 53 | 15% |
| ≥ 86 | 35 | 10% |
DNA isolation and sodium bisulfite modification
DNA extraction from frozen tissues has been described previously (25). Microdissection was performed on one to three consecutive 5-micron sections (H&E stained) composed mainly of tumor tissue from 342 TCC patients. DNA was extracted and bisulfite converted as previously described (26, 27).
Quantitative methylation-sensitive real-time polymerase chain reaction
Methylation analysis was performed as described previously (26). Briefly, the proportion of bisulfite-converted DNA in each sample was controlled for by a collagen IIA (COL2A1) reaction, which only amplifies bisulfite-converted DNA and is located region with no CpG sites in order to be independent of the methylation status. TNFRSF25 (DR3) was used as a positive control and FADD as a negative control for methylation. Genomic DNA (Promega, Madison, WI) was treated with SssI DNA methyltransferase (New England Biolabs) and used as a fully methylated reference to which all samples were compared to yield the percentage of fully methylated DNA (PMR) (26). Primer sequences and locations have been previously described for BCL2, DAPK, EDNRB, FADD, RASSF1A, TERT, TNFRSF25 (DR3), COL2A1 (26); CDH1 (ECAD) (28); PTGS2 (COX2), and TIMP3 (29). RUNX3 (NM_004350, -326/-258) forward primer, probe, and reverse primer sequences were as follows: 5′-CGTTTTAGCGTTAGGGAGTTACG'3′, 6FAM5′-TTTGAGAGAGGGCGGTAAGGGCG-3′BHQ1, 5′-AACGTCCGAATCCCACGA-3′.
Quantification of DNA Methylation by Methylation-Sensitive Single Nucleotide Primer Extension
Quantification of methylation at specific CpG sites using methylation-sensitive single nucleotide primer extension, a method developed in our lab, has been described previously (30). The promoter region of RUNX3 was amplified with primers specific for bisulfite-converted DNA and with an annealing temperature of 58°C (forward 5′-GGGGTTGTAGAAGTTATAGGT-3′ and reverse 5′-CCAATACCACAACCCAAAAC-3′) and two specific CpG sites were assayed using SNuPE primers with an annealing temperature of 56°C (5′-GGGGTTGTAGAAGTTATAGGTT-3′ and 5′-TAGTAAGAGTTGGGGAAGTT-3′). RUNX3 methylation was calculated as the average percent methylation of two CpG sites.
Statistical analysis
For each gene (BCL2, COX2, DAPK, ECAD, EDNRB, RASSF1A, RUNX3, TERT, and TIMP3) the methylation, measured as the Percent of the fully Methylated Reference (PMR) (31), was scored two ways: as the rank among study samples of all PMR values for that gene and as methylated (PMR ≥ 10%) or unmethylated (PMR < 10%) using a biologically determined cutoff value. For each patient, the following information was also included in the analysis: age at diagnosis, sex, smoking history, tumor stage, and tumor grade (Table 1). To assess the overall association between each of these characteristics and the PMR ranking for each gene, patient and tumor characteristics were evaluated using either the Wilcoxon test for the dichotomous characteristics of sex (male and female), grade (low grade and high grade), and smoking history (never/irregular smokers and former/current smokers) or the Kruskal-Wallis test for the characteristics with 3 or more categories of tumor stage (Ta, CIS, T1, and T2-4) and patient's age at diagnosis (≤40, 41-50, 51-60, and 61-65) (Table 2).
Table 2.
Association of Demographics with Gene Methylation (PMR) (P values)
| Gene | N | % PMR ≥ 10% | Stage1,3 | Grade2,4 | Sex2,5 | Age1,6 | Smoked2,7 |
|---|---|---|---|---|---|---|---|
| RUNX3 | 304 | 39% | 0.008 | 0.015 | 0.38 | 0.027 | 0.004 |
| RASSF1A | 323 | 37% | <0.001 | <0.001 | 0.25 | 0.49 | 0.81 |
| EDNRB | 316 | 30% | 0.013 | 0.004 | 0.98 | 0.83 | 0.053 |
| BCL2 | 324 | 28% | 0.51 | <0.001 | 0.10 | 0.50 | 0.12 |
| COX2 | 239 | 13% | 0.39 | 0.08 | 0.72 | 0.52 | 0.19 |
| DAPK | 253 | 4% | 0.08 | 0.017 | 0.66 | 0.18 | 0.63 |
| TERT | 262 | 3% | 0.10 | 0.42 | 0.43 | 0.35 | 0.13 |
| TIMP3 | 222 | 1% | 0.047 | 0.017 | 0.47 | 0.69 | 0.81 |
| ECAD | 241 | < 1% | 0.33 | 0.034 | 0.72 | 0.82 | 0.49 |
P values based on Kruskal-Wallis test.
P values based on Wilcoxon test.
Data grouped into the categories of Ta, CIS, T1, and T2-T4.
Data grouped into the categories of male and female.
Data grouped into the categories of ≤40, 41-50, 51-60, and 61-65 years at diagnosis.
Data grouped into the categories of never/irregular smokers and former/current smokers.
A logistic regression model was used to evaluate the association between RUNX3 methylation, classified as positive (PMR ≥ 10%) or negative, and clinicopathological characteristics (Table 3). The model used in the logistic regression included the following variables: age at diagnosis (1-year increase), smoking history (never/irregular vs. former/current), sex, and tumor stage and grade (Ta low grade; Ta high grade, CIS, T1; and T2-T4). The interaction term for smoking status and age was not significant (p=0.23) and was omitted from the final model; similarly a quadratic term for age, to capture a nonlinear relationship between age and the log odds of methylation, was also not significant and was omitted in the final model. P-values were based on a likelihood ratio test. The curves in Figure 3 display the association between age and probability of methylation for smokers and non-smokers separately; the raw data were smoothed using a LOESS procedure (32) and the estimated probability of methylation was based on the logistic regression model with age as the only covariate. All p-values were reported as two-sided.
Table 3.
Impact of Sex, Age, Tumor Stage, Tumor Grade, and Smoking History on RUNX3 Methylation (PMR ≥ 10%, N=304)
| OR (95% CI) | p-value | |
|---|---|---|
| Age at Diagnosis | 0.0311 | |
| 1-Year Increase | 1.04 (1.00, 1.08) | |
| Sex | 0.491 | |
| Female | 1.00 (reference) | |
| Male | 1.23 (0.68, 2.20) | |
| Tumor Stage & Grade | 0.0101 | |
| Ta - Low | 1.00 (reference) | |
| Ta – High/CIS/T1 | 1.18 (0.68, 2.07) | |
| T2+ | 2.16 (1.18, 3.94) | |
| Smoking | 0.0151 | |
| Never/Irregular | 1.00 (reference) | |
| Former/Current | 2.76 (1.15, 6.66) |
p-value based on likelihood ratio test.
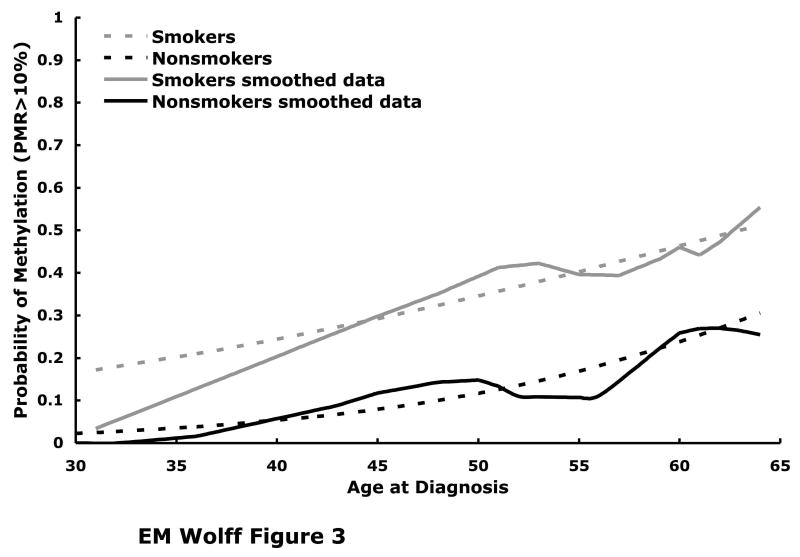
Fig. 3.
Estimated probability of RUNX3 methylation (PMR ≥ 10%) is plotted as a function of age at diagnosis for current or former smokers (N=265, dashed grey line) and for never or irregular smokers (N=38, dashed black line). These fitted curves are superimposed over the raw data that has been smoothed using a LOESS procedure, with the smokers represented by the solid grey line and the nonsmokers by the solid black line. The logistic regression analysis was performed on tumors from 304 patients and yielded a statistically significant association between RUNX3 methylation and smoking history (p=0.009, likelihood ratio test), adjusted by age at diagnosis, tumor grade, and tumor stage.
Results
RUNX3 is frequently and specifically methylated in bladder tumors
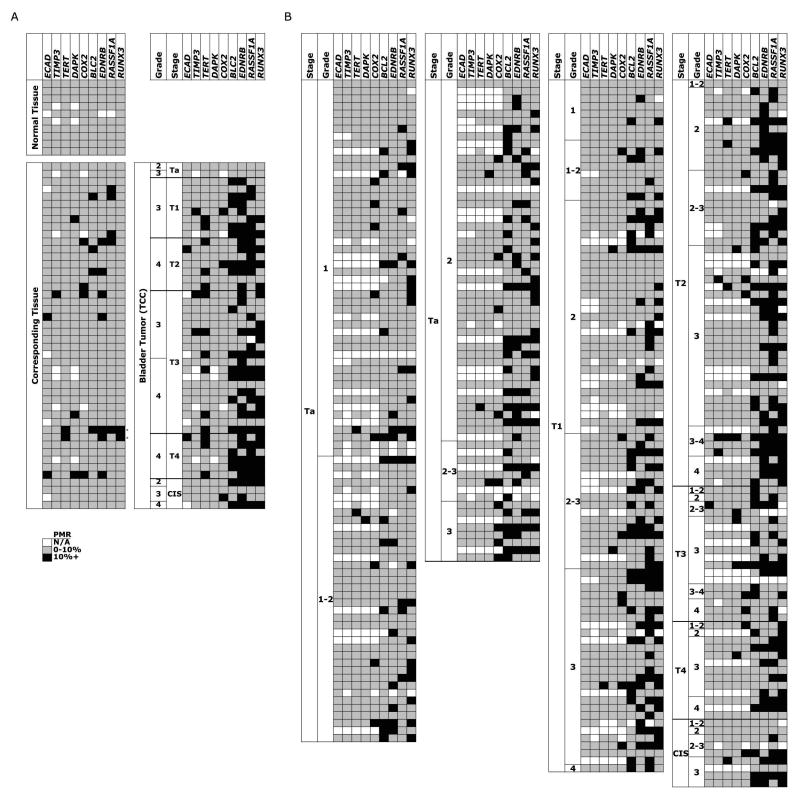
Since the methylation at nine genes in 10 urothelium samples from cancer-free bladders were all less than 10% PMR, this value was used as a biological cutoff for the presence or absence of methylation (Fig. 1A). In matched sets of bladder tumors and corresponding tissue EDNRB, RASSF1A, and BCL2 were frequently methylated in both the tumors (65%, 60%, and 39%) and the corresponding tissues (15%, 11%, and 9%) (Fig. 1A). While TERT was methylated in 30% of tumors and only 4% of corresponding tissues, RUNX3 was the most specific marker for bladder cancer, with frequent methylation in the tumor samples (23/41, 56%) and infrequent methylation in the corresponding tissues (2/46, 4%). The two corresponding tissues with RUNX3 methylation have nearly identical methylation patterns as their matched high grade and invasive tumors, suggesting that these bladders either had epigenetic aberrancies throughout or that the invasive tumor had spread across the bladder. The pathological reports for these two cases indicated that the first case had multiple foci of carcinoma in situ and severe urothelial atypia and the second case had diffuse lesions throughout the bladder. Therefore the two corresponding tissue samples with RUNX3 methylation were atypical and are not considered “normal”. Of the other 44 cases, two had areas of urothelial hyperplasia and one had moderate chronic cystitis, although there was no detectable methylation in the corresponding tissues, and one case had areas of squamous metaplasia with methylation at TIMP3, COX2, and EDNRB in the corresponding tissue.
Fig. 1.
Methylation in bladder samples at nine different loci using quantitative methylation-sensitive real-time PCR and shown as percent of a fully methylated reference (PMR), with PMR > 10% in grey, PMR ≤ 10% in black, and unavailable data in white. A Ten normal urothelium samples from cancer-free bladders were obtained during prostatectomies from age-matched patients. Forty-six matched sets of bladder tumor tissue and corresponding tissue were obtained during cystectomies. B Methylation data for 342 bladder cancer samples from paraffin-embedded tissues.
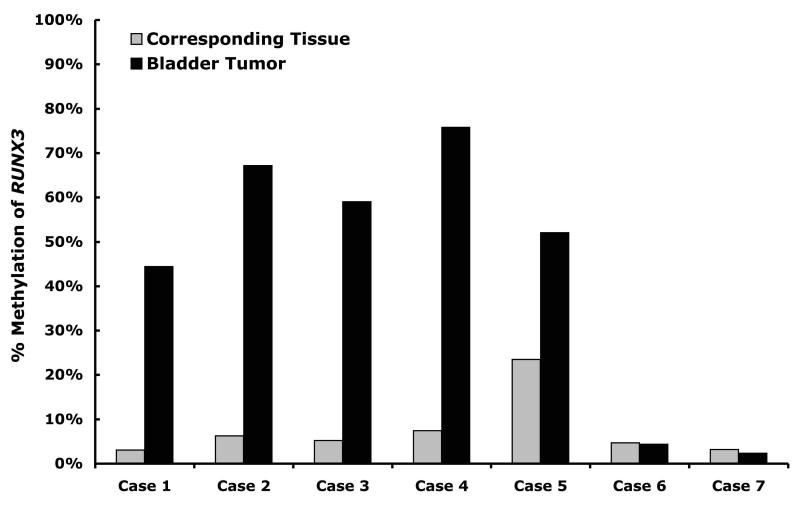
In order to confirm our methylation results we used another methylation assay, Ms-SNuPE, which was developed in our lab (30) and is able to quantify the methylation at specific CpG sites. When we examined two CpG sites just downstream of the region assayed using MethyLight we found that 5 out of 7 bladder tumors had high levels of RUNX3 methylation and only 1 out of 7 corresponding tissues had a high level of RUNX3 methylation in an independent cohort of 7 matched sets (Fig. 2). Upon examination of the pathological reports for these patients it was noted that in the patient with RUNX3 methylation in the corresponding tissue (Case 5) the entire bladder showed signs of cystitis cystica, a type of proliferative cystitis, with acute and chronic inflammation and had multifocal carcinoma in situ. Of the other six cases only one had reported areas of urothelial atypia.
Fig. 2.
Average percent methylation of two CpG sites located in the promoter of RUNX3 in 7 cases of matched bladder tumors (black bars) and corresponding tissues (grey bars) measured by Ms-SNuPE.
Since the matched tumor and corresponding tissue sets were taken from patients undergoing cystectomies, most of the tumors were highly invasive. Therefore we extended our study to include more noninvasive and low-grade tumor samples obtained by transurethral resections. When we assayed the methylation status of nine genes in these additional 342 bladder tumor samples, RUNX3 was still the most frequently methylated gene (39%), with the next most frequently methylated genes being RASSF1A (37%), EDNRB (30%), and BCL2 (28%) (Fig. 1B, Table 2).
RUNX3 methylation precedes methylation at the other 8 genes
Methylation at all genes except COX2 and TERT was significantly associated with tumor grade and methylation at EDNRB, RASSF1A, RUNX3, and TIMP3 was associated with tumor stage, revealing that methylation was more likely in tumors of higher grade or stage (Table 2). Based on the assumption that tumors of the lowest stage and grade are the precursors of tumors of higher stage and grade, we compared the methylation patterns of the four most frequently methylated genes in Ta low grade tumors to tumors of higher stage and grade in order to determine whether RUNX3 methylation precedes methylation at the other genes (Fig. 1B). An analysis of the discordance of RUNX3 methylation revealed that of the 20 tumors that had only RUNX3 methylated, 17 cases were Ta low stage tumors (85%) compared to 32 out of 89 cases (36%) with methylation at any of the other genes besides RUNX3 (p<0.001, Fisher's exact test). In contrast, methylation of EDNRB (35% compared to 31%, p=0.80) and BCL2 (33% compared to 28%, p=0.67) showed no discordance of methylation while RASSF1A showed the opposite pattern of RUNX3, with significantly more methylation in tumors of higher grade and stage (24% compared to 44%, p=0.042). The observed discordance is compatible with the hypothesis that RUNX3 methylation occurs early in tumorigenesis and before methylation of any of the other genes examined.
RUNX3 methylation increases with age and a history of smoking
Only methylation at RUNX3 was significantly associated with the age at which diagnosis of TCC was made (p=0.027) and a history of tobacco smoking (p=0.004) based on univariable analyses (Table 2). Using a logistic regression model controlling for age, sex, tumor stage, and grade, we found methylation of RUNX3 more frequently in tumors of smokers versus nonsmokers (p=0.015, OR=2.76, 95% CI: 1.15, 6.66) (Table 3). There was no effect of the duration of smoking when added to the above logistic regression model (p=0.88). Methylation at RUNX3 was significantly associated with the age at diagnosis (p=0.031) based on a logistic regression adjusting for sex, smoking history, tumor stage and grade.
In order to examine the joint association between smoking, age, and RUNX3 methylation we used a logistic regression model with age, smoking history, tumor stage and grade (Fig. 3). We found a statistically significant association between RUNX3 methylation and both age (p=0.004) and smoking status (p=0.009). The probability of RUNX3 methylation is higher in smokers and increases as a function of age in both nonsmokers and smokers (Fig. 3). Since RUNX3 methylation increases with age, is not present in the normal urothelium, and occurs early in tumorigenesis, it can be used as a molecular clock in order to determine the “age” of a bladder tumor and doing so reveals that tumors from smokers are “older” than tumors nonsmokers.
Discussion
In this study we evaluated the methylation of nine genes in transitional cell carcinoma samples from 342 patients about whom detailed demographic, clinicopathological, and smoking information had been previously collected (18). We found several genes to be more frequently methylated in tumors of higher stage and grade. The four most commonly methylated genes in our cohort of bladder tumors are involved in apoptotic pathways, including RUNX3, and BCL2, EDNRB, RASSF1A (Fig. 1), consistent with our previous study (26). In addition, we found that RUNX3 is not methylated in normal-appearing tissue corresponding to tumors or tissue from cancer-free bladders. However, since quantitative methylation-sensitive real-time PCR detects individual strands of DNA that are simultaneously methylated at all sites a cutoff value of 10% PMR cannot rule out the presence of sporadic sites of methylation. Therefore, we measured methylation of two specific CpG sites in the RUNX3 promoter in a small independent cohort of matched tumor and corresponding tissue. We found significant levels of methylation in 5 of the 7 tumors and only one of the corresponding tissues (Fig. 2). However, this specific corresponding tissue is not considered to be non-neoplastic, as it was taken from a bladder with cystitis cystica with acute and chronic inflammation and multifocal carcinoma in situ. In addition, other studies have also found that RUNX3 is not methylated in normal bladder (21), prostate (20, 33, 34), lung (20), breast (20), and colon (34) tissues using a variety of methods and also never or infrequently methylated in non-neoplastic tissue corresponding to bladder tumors (20), gastric tumors (34, 35), breast tumors (36), non-small cell lung tumors (37), and hepatocellular carcinomas (38). Since RUNX3 is not methylated in normal urothelium or normal-appearing tissue corresponding to tumors, we can assume that RUNX3 methylation was not initially present in the normal urothelium of these patients, is specific to bladder tumors, and is not acquired as a function of aging.
We have shown that RUNX3 methylation precedes methylation at the other 8 genes we examined, indicating that it is an early event in bladder tumorigenesis. RUNX3 methylation appears to occur early in tumorigenesis in a variety of cancer types. Methylation of RUNX3 has been found in invasive ductal breast carcinoma (IDC) and its precursor lesion of ductal carcinoma in situ (DCIS) (39), prostate cancer and its precursor prostatic intraepithelial neoplasia (33), and gastric cancer and its precursor lesions of chronic gastritis and intestinal metaplasia (34). RUNX3 methylation also occurs in Barrett's esophagus (BE), a precursor of esophageal adenocarcinoma (EAC), and is associated with progression to EAC (40). In addition, we found methylation in the corresponding tissue from three bladders, two of which had multifocal carcinoma in situ and acute and chronic inflammation with cystitis cystica, a proliferative cystitis, or severe urothelial atypia, and one of which with diffuse lesions throughout the bladder. Other groups have shown that similar inflammatory lesions in the bladder express telomerase (41, 42), indicating that these lesions are likely to be undergoing immortalization or malignant transformation. RUNX3 is a polycomb target (24) and several groups have found that polycomb targets become preferentially methylated during cancer (14-16), lending support to the theory that cancer is derived from stem cells. Several preinitiation events occur in stem cells before they are initiated and then clonally expand during promotion (43). Initiating events are irreversible and if they occur in stem cells then that event will remain in the asymmetrically dividing stem cell. Therefore, our results showing that corresponding tissue from a bladder with widespread inflammatory lesions has elevated RUNX3 methylation indicates that RUNX3 methylation occurs earlier than tumorigenesis and accumulates in the bladder stem cells that will eventually develop into a tumor.
According to a multivariable logistic regression model, the probability of RUNX3 methylation increases with age in both smokers and non-smokers. Biological age is a surrogate marker for the number of times a cell has divided and the more times a cell divides the more opportunities for aberrant methylation to accumulate (44). Because the degree of hypermethylation in normal colonic epithelium is related to age (7), it has been suggested that methylation can be used as a molecular “clock” to predict the age of a tissue (8). Therefore, we are able to use RUNX3 methylation to show for the first time that methylation in tumor cells might be useful as a molecular clock. We have shown that RUNX3 methylation is present early in tumorigenesis and that the level increases with age (Fig. 3). When we apply our RUNX3 molecular clock to tumors from smokers versus nonsmokers, our data suggests bladder tumors in smokers are “older” than in nonsmokers, i.e. have undergone more cell divisions before diagnosis, since RUNX3 is more prevalent in tumors from smokers. There are at least two possible explanations for such an observation. Either tumors in smokers “age” or divide more quickly than in nonsmokers, resulting in faster accumulation of methylation errors over a similar amount of time, or tumors in smokers were initiated at an earlier age compared with tumors of nonsmokers, and therefore have had more time to divide and accumulate RUNX3 methylation. Since RUNX3 methylation appears to be an early event in bladder tumorigenesis and increases over time, then detection of RUNX3 methylation in biopsy or urine specimens could provide a marker to screen the at risk population of cigarette smokers long before any symptoms are present.
Acknowledgments
Grant support: NCI P01 CA 86871
References
- 1.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–32. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 8.Kim JY, Siegmund KD, Tavare S, Shibata D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 2005;3:10. doi: 10.1186/1741-7015-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Nelson HH, Wiencke JK, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–24. [PubMed] [Google Scholar]
- 10.Anttila S, Hakkola J, Tuominen P, et al. Methylation of cytochrome P4501A1 promoter in the lung is associated with tobacco smoking. Cancer Res. 2003;63:8623–8. [PubMed] [Google Scholar]
- 11.Kim DH, Kim JS, Ji YI, et al. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003;63:3743–6. [PubMed] [Google Scholar]
- 12.Marsit CJ, Karagas MR, Danaee H, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–6. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 13.Marsit CJ, Houseman EA, Schned AR, Karagas MR, Kelsey KT. Promoter Hypermethylation is Associated with Current Smoking, Age, Gender, and Survival in Bladder Cancer. Carcinogenesis. 2007 doi: 10.1093/carcin/bgm116. [DOI] [PubMed] [Google Scholar]
- 14.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 15.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 16.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff EM, Liang G, Jones PA. Mechanisms of Disease: genetic and epigenetic alterations that drive bladder cancer. Nat Clin Pract Urol. 2005;2:502–10. doi: 10.1038/ncpuro0318. [DOI] [PubMed] [Google Scholar]
- 18.Castelao JE, Yuan JM, Skipper PL, et al. Gender- and smoking-related bladder cancer risk. J Natl Cancer Inst. 2001;93:538–45. doi: 10.1093/jnci/93.7.538. [DOI] [PubMed] [Google Scholar]
- 19.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M, Shigematsu H, Shames DS, et al. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93:1029–37. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kim WJ, Kim EJ, Jeong P, et al. RUNX3 inactivation by point mutations and aberrant DNA methylation in bladder tumors. Cancer Res. 2005;65:9347–54. doi: 10.1158/0008-5472.CAN-05-1647. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz YB, Kahn TG, Nix DA, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 23.Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–7. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008 doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–8. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 26.Friedrich MG, Weisenberger DJ, Cheng JC, et al. Detection of methylated apoptosis-associated genes in urine sediments of bladder cancer patients. Clin Cancer Res. 2004;10:7457–65. doi: 10.1158/1078-0432.CCR-04-0930. [DOI] [PubMed] [Google Scholar]
- 27.Widschwendter M, Siegmund KD, Muller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–13. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 28.Eads CA, Lord RV, Kurumboor SK, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–6. [PubMed] [Google Scholar]
- 29.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–8. [PubMed] [Google Scholar]
- 30.Gonzalgo ML, Liang G. Methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) for quantitative measurement of DNA methylation. Nat Protoc. 2007;2:1931–6. doi: 10.1038/nprot.2007.271. [DOI] [PubMed] [Google Scholar]
- 31.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 32.SAS Institute Inc. SAS/STAT User's Guide, Version 8. Cary, NC: SAS Publishing; 2000. The Loess Procedure; pp. 1853–900. [Google Scholar]
- 33.Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–40. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 34.Kim TY, Lee HJ, Hwang KS, et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004;84:479–84. doi: 10.1038/labinvest.3700060. [DOI] [PubMed] [Google Scholar]
- 35.Waki T, Tamura G, Sato M, Terashima M, Nishizuka S, Motoyama T. Promoter methylation status of DAP-kinase and RUNX3 genes in neoplastic and non-neoplastic gastric epithelia. Cancer Sci. 2003;94:360–4. doi: 10.1111/j.1349-7006.2003.tb01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau QC, Raja E, Salto-Tellez M, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–20. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- 37.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–92. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park WS, Cho YG, Kim CJ, et al. Hypermethylation of the RUNX3 gene in hepatocellular carcinoma. Exp Mol Med. 2005;37:276–81. doi: 10.1038/emm.2005.37. [DOI] [PubMed] [Google Scholar]
- 39.Subramaniam MM, Chan JY, Soong R, et al. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9917-4. [DOI] [PubMed] [Google Scholar]
- 40.Schulmann K, Sterian A, Berki A, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–48. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 41.Lancelin F, Anidjar M, Villette JM, et al. Telomerase activity as a potential marker in preneoplastic bladder lesions. BJU Int. 2000;85:526–31. doi: 10.1046/j.1464-410x.2000.00466.x. [DOI] [PubMed] [Google Scholar]
- 42.Kavaler E, Landman J, Chang Y, Droller MJ, Liu BC. Detecting human bladder carcinoma cells in voided urine samples by assaying for the presence of telomerase activity. Cancer. 1998;82:708–14. doi: 10.1002/(sici)1097-0142(19980215)82:4<708::aid-cncr14>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 43.Luebeck EG, Moolgavkar SH. Multistage carcinogenesis and the incidence of colorectal cancer. Proc Natl Acad Sci U S A. 2002;99:15095–100. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preston-Martin S, Pike MC, Ross RK, Jones PA, Henderson BE. Increased cell division as a cause of human cancer. Cancer Res. 1990;50:7415–21. [PubMed] [Google Scholar]