Abstract
One prime feature of alcoholic liver disease (ALD) is iron accumulation in hepatic macrophages/Kupffer cells (KC) associated with enhanced NF-κB activation. Our recent work demonstrates a peroxynitrite-mediated transient rise in intracellular labile iron (ILI) as novel signaling for endotoxin-induced IKK and NF-κB activation in rodent KC. The present study investigated the mechanism of KC iron accumulation and its effects on ILI response in experimental ALD. We also tested ILI response in human blood monocytes. Chronic alcohol feeding in rats results in increased expression of transferrin (Tf) receptor-1 and hemochromatosis gene (HFE), enhanced iron uptake, an increase in nonheme iron content, and accentuated ILI response for NF-κB activation in KC. Ex vivo treatment of these KC with an iron chelator abrogates the increment of iron content, ILI response, and NF-κB activation. The ILI response is evident in macrophages derived from human blood monocytes by PMA treatment but not in vehicle-treated monocytes, and this differentiation-associated phenomenon is essential for maximal TNF-α release. PMA-induced macrophages load iron dextran and enhance ILI response and TNF-α release. These effects are reproduced in KC selectively loaded in vivo with iron dextran in mice and more importantly aggravate experimental ALD. Our results suggest enhanced iron uptake as a mechanism of KC iron loading in ALD and demonstrate the ILI response as a function acquired by differentiated macrophages in humans and as a priming mechanism for ALD.
Keywords: IKK, NF-κB, labile iron, TNF-α, transferrin receptor-1, HFE
enhanced tnf-α expression by hepatic macrophages/Kupffer cells (KC) underlies chronic inflammation and cytotoxicity in alcoholic liver disease (ALD) (5, 11, 13, 21). In search for the mechanism responsible for abnormal TNF-α upregulation, we discovered that an increased chelatable pool of nonheme iron is causally associated with enhanced NF-κB activation and TNF-α expression by KC in experimental ALD (32). As we further dissected the mechanistic link, we identified the novel signaling for activation of IKK involving a transient rise in the intracellular level of labile iron (ILI) in LPS or TNF-α-stimulated KC (37). The ILI signaling takes place at or within 2 min after agonist stimulation and is transient much like the well-known intracellular calcium concentration ([Ca2+]i) response (37). This study also identifies peroxynitrite as an upstream effector that evokes the ILI response (37). Exogenous iron is also rapidly taken up by macrophages within 2 min (1) and substitutes for the endogenous ILI response to activate IKK in cultured KC (28) in a manner requiring activation and protein-protein interactions of TGF-β-activated kinase 1, p21ras, and phosphatidylinositol 3-kinase in caveolae (4). In fact, we propose that this activation of IKK by extracellular ferrous iron is an important and endotoxin-independent mode of NF-κB activation and proinflammatory gene expression in chronic liver disease.
Our findings also demonstrate that the ILI response is upregulated by an increased nonheme and chelatable pool of iron resulting from genetic or environmental manipulations. For instance, a deficiency in natural resistance-associated macrophage protein-1 (Nramp1), the macrophage-specific iron transporter responsible for iron efflux from late endosomes, results in an increased chelatable pool of nonheme iron, accentuated LPS-induced ILI response, and consequent enhancement of NF-κB activation and TNF-α expression (37). In experimental ALD, a similar association between the increased iron pool and NF-κB activation is noted (32). However, why nonheme iron content in KC is increased in ALD and whether this results in augmented ILI response are yet to be determined. Furthermore, it is unknown whether the ILI response is relevant in humans. In this study, we examined the mechanism and consequence of the increased nonheme iron content in KC in the rodent models of ALD. We also tested whether the ILI response is evident in human macrophages and enhanced after iron loading.
MATERIALS AND METHODS
Materials.
PMA, LPS (Escherichia coli 055:B5), iron dextran, and dextran 500 were purchased from Sigma Chemical (St. Louis, MO). NycoPrep (AXIS-SHIELD POS AS) was obtained from Accurate Chem & Sci (Westbury, NY). TNF-α enzyme-linked immunosorbent assay kits for human and mouse were purchased from R&D Systems (Minneapolis, MN). Anti-human CD68-PE, control IgG-PE, and anti-p65 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), Fe59Cl3 were from PerkinElmer Life Sciences (Boston, MA), and peroxynitrite (ONOO−) from Calbiochem (San Diego, CA). Ultrafree-MC centrifugal filters (size-exclusion column) were purchased from Millipore (Bedford, MA) and SYBR GREEN real-time PCR master mixture from Applied Biosystems (Foster City, CA). Anti-mouse hepcidin and anti-mouse ferroportin-1 (FPN1) antibodies were purchased from Alpha Diagnostics International (San Antonio, TX), anti-human transferrin receptor (TfR) from Zymed Laboratories (South San Francisco, CA), and anti-mouse α-smooth muscle actin from Sigma Chemical.
Cell isolation and culture.
The use of animals for this study was approved by the Institutional Animal Care and Use Committee of the University of Southern California. KC were isolated from normal Wistar rats, alcohol-fed, pair-fed control rats, and iron dextran-injected mice by the Non-Parenchymal Liver Cell Core of Research Center for Alcoholic Liver and Pancreatic Diseases and Cirrhosis by in situ sequential digestion of the liver with pronase and collagenase followed by arabinogalactan gradient ultracentrifugation and adherence purification method as previously described (28, 32, 37). The cells were routinely cultured in DMEM containing 5% fetal calf serum (FCS) on plastic dishes for 2 days before experiments. KC isolated from alcohol-fed, pair-fed, and iron dextran-injected animals were cultured for 3 h or overnight in 2% FCS and studied immediately thereafter. For assessment of ILI signaling and TNF-α release, the cells were incubated in serum-free PBS and stimulated with peroxynitrite (10 μM) or LPS (500 ng/ml). For isolation of peripheral blood monocytes (PBMs), venous blood samples (40 ml) were withdrawn from healthy volunteer subjects who signed informed consent forms; and monocytes were isolated using NycoPrep 1.068 according to the manufacturer's directions. Briefly, the blood was first diluted 1:10 with a solution of dextran 500 (8.0%, wt/vol, ∼300 mOsm/kg) in 0.9% (wt/vol) NaCl and left at room temperature for 30–45 min. Leukocyte-rich plasma was removed and layered over 3 ml NycoPrep in a 15-ml Falcon tube and centrifuged at 600 g at 20°C for 15 min. PBMs were collected at the interface and washed twice by mixing with a solution of 0.9% (wt/vol) NaCl containing 0.13% (wt/vol) EDTA and 1% (vol/vol) FCS and centrifuging at 600 g for 7 min. Isolated PBMs were cultured in DMEM containing 10% FCS and antibiotics. The cells were treated with 100 nM PMA for 16 h at 37°C for induction of macrophage differentiation as determined by macrophage-specific gene expression by real-time PCR and fluorescence-activated cell sorting (FACS) described below.
Animal experiments.
For chronic alcohol administration, male Wistar rats (14 wk old) and C57BL/6 mice (10 wk old) were aseptically implanted with gastrostomy catheters as described (33). A high-fat diet (35% calories as corn oil) was infused along with an increasing dose of ethanol (9–14.5 g/kg per day for rats and 22.7–35 g/kg per day for mice) or isocaloric dextrose solutions for 9 wk (rats) and 4 wk (mice). Iron dextran (25 or 75 mg/kg) was injected subcutaneously to C57BL/6 mice 2 wk before 4-wk alcohol feeding to selectively load iron in macrophages as previously described (23).
Flow cytometry.
Following the treatment with PMA (100 nM), PBMs were washed three times with cold PBS centrifuged at 400 g for 10 min, resuspended in PBS containing 1% (wt/vol) BSA, and incubated with anti-CD68 antibody (PE-labeled) for 30 min at room temperature. The cells were washed twice and resuspended with cold PBS before flow cytometry. A PE-labeled nonimmune mouse IgG was used as a control.
Real-time PCR.
Isolated cells were cultured in 0.5% FCS for 3 h, washed with PBS, and subjected to RNA extraction using TRIzol reagent following the manufacturer's instruction. Two micrograms of total RNA were reverse transcribed and amplified by using the SYBR GREEN PCR master mixture (Applied Biosystems) in a Stratagene MX3000 real-time PCR. Each Ct value from the treated sample was standardized to that of 36B4 housekeeping gene and compared with control samples. The PCR primers used for real-time PCR are shown in Table 1. A single peak of the dissociation curve was always confirmed for each PCR reaction.
Table 1.
Primer sequences for real–time PCR
| Gene | Sequences |
|---|---|
| TfR1 | 5′-CATGAGGGAAATCAATGATCGTA and 5′-GCCCCAGAAGATGTGTCGGAA |
| TfR2 | 5′-CTATCTGGTCCTGACCACCCT and 5′-TCAGAGTTGACATCTTCACCAA |
| HFE | 5′-CTGAAAGGGTGGGATTACATGTTC and 5′-AGGCACCACTCTCAACTTCGT |
| Zip14 | 5′-CTCAAAGGGGTTCGATATTCTG and 5′-ATCTCAGGGAACATATCGGCTA |
| IL-6 | 5′-GGAGAGGAGACTTCACAG and 5′-GCCATTGCACAACTCTTTTC |
| FPN-1 | 5′-TAGGGTCTACTGCGGC and 5′-GCACGGCTATAAGCAC |
| Hepcidin | 5′-GCACTAAGCACTCGGA and 5′-TGGGGAAGTTGGTGTC |
| Nramp1 | 5′-AGCGTTCAACATCTGTG and 5′-GCGTGCAAATCGAGAC |
| DMT-1 | 5′-GGTTAGCGTGGCTTATC and 5′-GCAACGGCACATACTT |
| TNF-α | 5′-CACGTCGTAGCAAACC and 5′-GCACATAGTCGGGGCA |
| CD68 | 5′-TGCTCAGTTGCCGGAC and 5′-CCGGGTAACGCAGAAG |
| CD14 | 5′-AGTCGGAGGCGTATAAC, and 5′-ACCCATTGAGCCATCT |
| 36B4 | 5′-TTCCCACTGGCTGAAAAGGT and 5′-CGCAGCCGCAAATGC |
TfR, transferrin receptor; HFE, hemochromatosis gene; FPN-1, ferroportin-1; Nramp1, natural resistance-associated macrophage protein-1; DMT-1, divalent metal transporter-1.
Immunoblot analysis and ELISA.
Release of TNF-α into the media by cultured PBMs or KC was determined by commercially available immunoassay kits for rodent and human TNF-α following the manufacturer's instruction (R&D Systems). Total KC lysates of control rats (Cont-KC) and alcohol-fed rats (Alc-KC) were prepared with an assay buffer (RIPA:PBS, pH 7.4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and complete protease inhibitor mixture). The protein extracts were resolved on a 10% SDS-PAGE, transferred onto a nitrocellulose membrane, and incubated with primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies. The antigen-antibody complexes were visualized by the enhanced chemiluminescence detection system (Pierce, IL). Plasma prohepcidin levels were determined by ELISA (DRG Instruments, Marburg, Germany).
ILI response assessment.
Cultured rat or mouse KC or PMA-primed PBM-derived macrophages were pretreated with or without iron dextran (1 μM) on a 60-mm dishes (1×106 cells per dish) for 16 h at 37°C and 5% CO2. The cells were then incubated with 5 μCi/ml FeCl3 for 14–16 h in 5 ml of DMEM containing 10% FCS. Labeled cells were sequentially washed with 5 ml of warm PBS once, PBS containing 100 μM bathophenanthroline sulfate once, and PBS twice. The washed cells were treated with LPS (500 ng/ml) or peroxynitrite (10 μM) in 5 ml of warm PBS for 2, 5, 10, or 20 min, and the incubation was stopped at the respective time points by removing PBS and adding 200 μl of lysis buffer (1.4 M NaCl, 0.1 M HEPES, pH 7.4, 1.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride). Peroxynitrite obtained from the supplier had the concentration of 170–200 mM in 4.7% NaOH. We took the midpoint (185 mM) for our use. For addition to cell culture, the stock was first diluted 1:10 using 4.7% NaOH and added to media (e.g., 5.4 μl into 10 ml) to achieve the final concentration of for 10 μM. For control, the same amount of 4.7% NaOH was added. A low molecular mass fraction (<5,000 Da) was prepared by a centrifugation of the lysate in a size-exclusion column (Millipore) at 8,600 g for 30 min at 4°C. Radioactivity of ultrafiltrate or total lysate was determined by a liquid scintillation counter as previously described (37). As an alternative method, ILI released by agonist stimulation was assessed by spectrophotometric detection at 430 nm of ILI chelated with desferrioxamine and added to culture of Alc-KC and Cont-KC as previously described (37).
EMSA.
EMSA for NF-κB was performed as previously described (28, 37). Briefly, nuclear proteins extracted from KC with and without prior treatment with L1 were incubated with P32-labeled double-strand κB consensus sequence and resolved on a 6% nondenaturing polyacrylamide gel for subsequent autoradiography. Two shifted bands detected by this method are previously shown to comprise p65/p50 heterodimer and p50/p50 homodimer by supershift assay (28).
Fe59 uptake by KC.
Freshly isolated Cont-KC and Alc-KC were plated onto 24-well plates, cultured in DMEM containing 0.5% FCS overnight. The cells were incubated with 59FeCl3 (5 μCi/ml in DMEM with 5% FCS) for 5, 10, 20, 30, 60, and 120 min, washed with PBS twice, and lysed in 2% (wt/vol) SDS. The KC lysate from triplicate wells were subjected to liquid scintillation counting for detection of 59Fe radioactivity.
RESULTS
Increased chelatable pool of iron causes enhanced ILI response in KC from ALD.
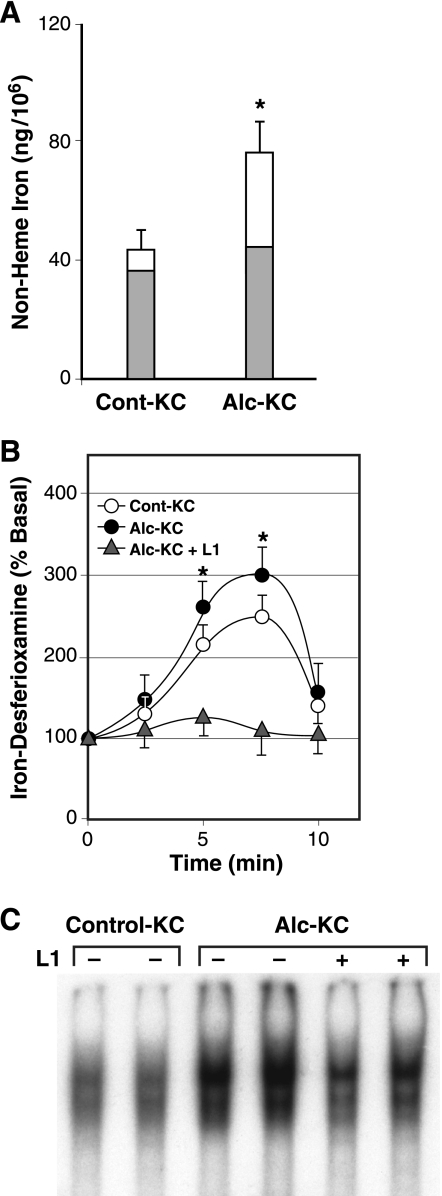
We have previously demonstrated that alcohol feeding to rats using the intragastric ethanol infusion model causes an increase in nonheme iron content, enhanced NF-κB activation, and cytokine expression in KC, all of which are abrogated by ex vivo treatment with the iron chelator L1 (1,2-dimethyl-3-hydroxypyridin-4-one) (32). Recently, we have also shown that agonist-stimulated IKK and NF-κB activation in KC requires novel ILI response as demonstrated as a peroxynitrite-mediated transient rise in ILI, which is positively regulated by increased iron content (37). Thus it is plausible that accentuated ILI response is a potential intermediary for the link between the increased nonheme iron content and enhanced NF-κB activation in KC in experimental ALD. To test this notion, we isolated KC from rats fed ethanol or control diet and assessed for the chelatable pool of nonheme iron and ILI signaling response. As previously shown, the nonheme iron content is increased 70% in Alc-KC compared with Cont-KC. After overnight treatment with the lipophilic iron chelator L1 (200 μM), the nonheme iron content in Alc-KC is normalized (a shaded portion of the bar), suggesting that the increment in the nonheme iron content in Alc-KC is due to an expansion of the chelatable pool of iron (Fig. 1A). These cells were also analyzed for the ILI response by the method involving chelation of liberated ILI with desferrioxamine and spectrophotometric determination of the iron chelator complex in cell suspension (37). This method was chosen over the Fe59-labeling technique because of a difficulty in performing a prolonged in vitro experiment of radioactive labeling for ILI analysis without altering in vivo changes of isolated cells. We have previously shown that a transient rise in ILI after LPS or peroxynitrite stimulation determined with Fe59 is also captured well by this spectrophotometric method (37). As shown in Fig. 1B, both Alc-KC and Cont-KC exhibit a transient peak of the iron chelator complex at 5–7 min after addition of peroxynitrite (10 μM), the effector molecule for the ILI signaling (37). Although the pattern of the ILI response is similar between the two groups, the peak for Alc-KC is significantly higher. Pretreatment of the cells with L1 completely abolishes the ILI response (Fig. 1B) and clearly attenuates enhanced NF-κB activation as assessed by EMSA (Fig. 1C) in Alc-KC. These results support the notion that enhanced ILI response constitutes the priming mechanism that bridges the increased chelatable pool of nonheme iron and enhanced NF-κB activation in Alc-KC.
Fig. 1.
A: nonheme iron content of Kupffer cells (KC) isolated from rats fed alcohol for 9 wk (Alc-KC) is increased compared with those from pair-fed controls (Cont-KC) as depicted by open bars. After ex vivo treatment with an iron chelator (L1, 100 μM) or vehicle (water) overnight, the nonheme iron level in Alc-KC and Cont-Alc were normalized (shaded bars). *P < 0.05 compared with Cont-KC. B: peroxynitrite-stimulated intracellular labile iron (ILI) response is augmented in Alc-KC. Alc-KC and Cont-KC were treated with peroxynitrite (10 μM) in PBS, and transiently liberated ILI was captured by desferrioxamine and measured spectrophotometrically. The iron chelator treatment (L1) abolishes the ILI response by Alc-KC. *P < 0.05 compared with Cont-KC. C: treatment of Alc-KC with L1 (100 μM) overnight attenuates enhanced NF-κB binding in Alc-KC as assessed by electrophoretic mobility shift assay.
Increased FPN1 expression by Alc-KC.
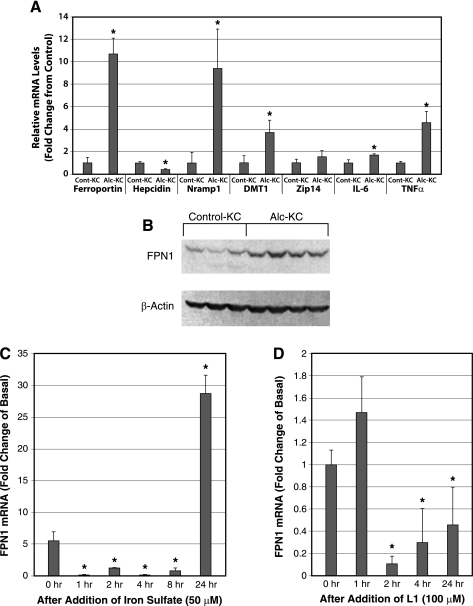
Next, we analyzed the expression of iron transporters Alc-KC and Cont-KC to help understand the mechanism of iron accumulation in Alc-KC. As shown in Fig. 2A, the expression of FPN1, Nramp1, and divalent metal transporter-1 are all increased in Alc-KC along with IL-6 and TNF-α, which serve as the positive controls for these cells (13). Zip14, which is a member of the SLC39A zinc transporter family, is also suggested to participate in nontransferrin-bound iron uptake (19). Expression of this transporter is unchanged by alcohol feeding. Hepcidin, which downregulates FPN1 expression via a posttranslational mechanism (7), is detected in Cont-KC and reduced in Alc-KC. Induction of FPN1 in Alc-KC is also confirmed at the protein level (Fig. 2B). Densitometric analysis reveals a significant 3.7-fold induction of FPN1 protein expression in Alc-KC (0.546 ± 0.144 vs. 0.147 ± 0.028, P < 0.05). These results suggest that iron accumulation in Alc-KC is not due to suppressed efflux of iron from endosomes and from the cells since Nramp1 and FPN1, which participate in these processes, respectively, are upregulated. Instead, our data suggest these iron transporters are induced as adaptive responses to iron loading. In particular, FPN1, a major iron exporter, is known to be positively regulated by iron loading and erythrophagocytosis in J774 macrophages (15). To test this notion, we treated cultured KC from normal rats with iron (iron sulfate) or the iron chelator L1 to examine their effects on FPN1 mRNA. As shown in Fig. 2C, FPN1 mRNA is reduced at 1–8 h after addition of iron but increased by 28-fold at 24 h. In contrast, L1 treatment suppresses FPN1 mRNA at 2–24 h (Fig. 2D). We next determined the plasma levels of hepcidin, the most important iron-regulatory hormone primarily expressed by hepatocytes by ELISA. Although other studies report suppressed hepcidin expression by alcohol feeding (2, 10), the prohepcidin levels were not different between alcohol-fed and pair-fed control animals in our study (195.0 ± 25.1 vs. 229.0 ± 15.1 ng/ml, P = 0.29).
Fig. 2.
A: expression of ferroportin-1 (FPN1), natural resistance-associated macrophage protein-1 (Nramp1), and divalent metal transporter-1 (DMT-1) mRNA are significantly increased in Alc-KC. RNA was extracted from Alc-KC and Cont-KC and analyzed by real-time PCR using the primers shown in Table 1. *P < 0.05 compared with Cont-KC. B: induction of FPN1 in Alc-KC is also confirmed at the protein level by immunoblot analysis. C: FPN1 mRNA level in cultured rat KC is markedly increased at 24 h after addition of iron sulfate following suppressive effects between 1 and 8 h. KC isolated from normal rats were cultured and treated with iron sulfate (50 μM) in serum-free media and RNA extracted at the indicated time points for real-time PCR analysis for FPN1. *P < 0.05 compared with the level at 0 h. D: treatment with the iron chelator L1 reduces FPN1 mRNA in cultured KC. Normal rat KC were isolated, cultured, and treated with L1 (100 μM), and RNA was extracted and analyzed as above. *P < 0.05 compared with the level at 0 h.
Enhanced iron uptake by Alc-KC.
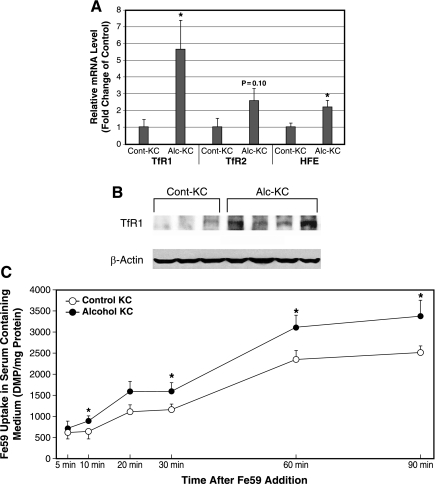
Since the observed changes in the iron efflux transporters cannot explain iron loading in Alc-KC, we next examined the mRNA expression of TfR1 and TfR2, as well as hemochromatosis gene (HFE) that is shown to regulate Tf-mediated iron uptake (22, 39). As shown in Fig. 3A, TfR1 mRNA but not TfR2 is significantly upregulated 5.7-fold in Alc-KC. HFE mRNA is also significantly increased twofold in Alc-KC. Increased TfR1 expression in Alc-KC is also confirmed at the protein level by immunoblot analysis (Fig. 3B). Densitometric analysis shows a significant 9.5-fold increase in TfR1 protein expression in Alc-KC (0.195 ± 0.075 vs. 0.020 ± 0.007, P < 0.05). We next determined uptake of iron by Alc-KC and Cont-KC using Fe59Cl3 in serum-containing media. Iron uptake by Alc-KC is significantly enhanced compared with Cont-KC over a 90-min period (Fig. 3C). These results suggest that iron uptake by Alc-KC is enhanced due to induction of TfR1, a dominant receptor type in KC and concomitant induction of HFE, which is suggested to promote Tf-dependent iron uptake in macrophages (22). Interestingly, a similar finding on alcohol-induced TfR1 induction has recently been reported for hepatocytes (16).
Fig. 3.
A: transferrin receptor-1 (TfR1) and hemochromatosis gene (HFE) mRNA levels are increased in Alc-KC. TfR1, TfR2, and HFE mRNA levels were analyzed for Alc-KC and Cont-KC as described for Fig. 2A. *P < 0.05 compared with Cont-KC. B: Alc-KC express higher levels of TfR1 protein as determined by immunoblot analysis. C: Alc-KC have significantly higher uptake of Fe59Cl3 compared with Cont-KC. The cells isolated from the animals were incubated with Fe59Cl3 in serum-containing media for the indicated periods, and the radioactivity in the cells was counted after washing. *P < 0.05 compared with Cont-KC.
ILI response is an acquired function of differentiated macrophages.
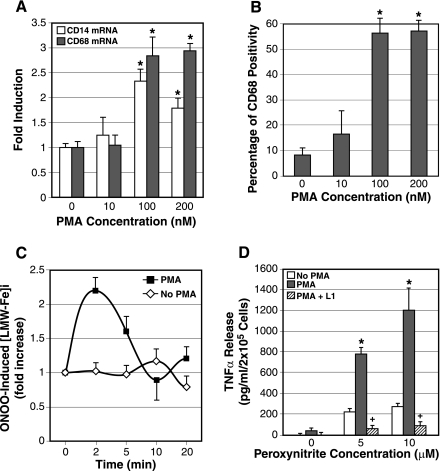
All experiments on ILI response described to date have utilized KC from rodents or murine macrophage cell line (RAW264.7) (37) but not macrophages from humans. Thus whether this phenomenon takes place in humans is an important question. To address this question, PBMs were isolated from human volunteers and tested for peroxynitrite-stimulated ILI response using Fe59. This experiment failed to detect ILI response in LPS or peroxynitrite-stimulated human PBMs. Next, we treated isolated PBMs with PMA (10–200 nM) overnight to force their differentiation to macrophages as assessed by mRNA expression of macrophage markers (CD14 and CD68) (Fig. 4A) and detection of CD68-positive cells by FACS (Fig. 4B). These results demonstrate that the PMA concentration of 100 nM achieves the maximal differentiation effect. Using this concentration, we repeated ILI analysis. The cells were washed after overnight PMA treatment and rested for 6 h before the treatment with peroxynitrite. As shown in Fig. 4C, PMA-treated cells but not vehicle (DMSO)-treated cells, exhibit ILI signaling at 2 min after peroxynitrite addition. Acquisition of this signaling event by differentiated macrophages is associated with a fourfold increase in the cells' ability to produce TNF-α in response to peroxynitrite compared with vehicle-treated cells (Fig. 4D). These results demonstrate that ILI response occurs in macrophages from humans and is a function acquired by macrophage differentiation. This acquired function confers that macrophage maximal cytokine release as both ILI response (data not shown) and TNF-α expression (Fig. 4D) is abrogated by pretreatment with the iron chelator L1.
Fig. 4.
A: PMA treatment overnight induces macrophage marker genes (CD14 and CD68) in peripheral blood monocytes (PBMs) isolated from healthy volunteers as determined by real-time PCR. PBMs were cultured overnight with the indicated concentrations of PMA and RNA extracted for CD14 and CD68 real-time PCR. *P < 0.05 compared with vehicle-treated cells. B: CD68-positive macrophages increase in number with PMA treatment at 100 and 200 nM. PBMs treated with PMA as above were analyzed for CD68 expression by fluorescence-activated cell sorting (FACS). *P < 0.05 compared with vehicle-treated cells. C: in both PMA-induced macrophages and vehicle (DMSO)-treated monocytes, the ILI response was determined by assessment of intracellular low molecular weight-Fe59 complexes ([LMW-Fe]i) in response to peroxynitrite (10 μM) following Fe59Cl3 labeling. Note PMA-induced macrophages but not monocytes exhibit the ILI response. D: peroxynitrite-induced TNF-α release by PMA-induced macrophages derived from PBMs is fourfold greater than vehicle-treated PBMs. L1 treatment abrogates this differentiation-associated, maximal TNF-α expression. *P < 0.05 compared with vehicle-treated PBMs (no PMA). +P < 0.05 compared with PMA-treated PBMs without L1 treatment.
Iron dextran increases macrophage iron content and promotes ILI signaling and TNF production.
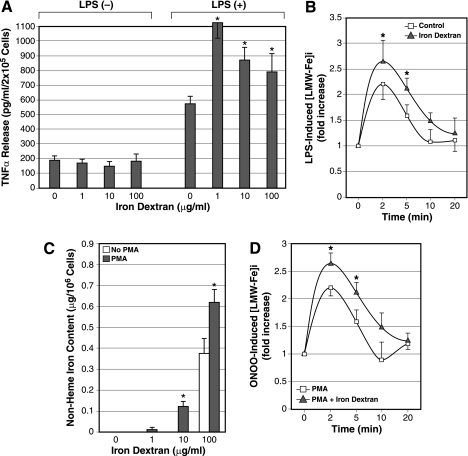
Next, we increased nonheme iron content in cultured rat KC by treating the cells with iron dextran to determine how this manipulation influences peroxynitrite-induced ILI response. Iron in this large complex with dextran is readily and preferentially taken up by macrophages and is therefore used to selectively induce iron loading in macrophages (23). Cultured rat KC were treated with iron dextran (1, 10, and 100 μg/ml) overnight, washed extensively, and stimulated with LPS (500 ng/ml) in serum-free medium for 6 h. Iron dextran treatment at 1, 10, and 100 μg/ml increases the nonheme iron content 2.2-, 5.4-, and 11.1-fold, respectively (data not shown). These treatments do not affect basal TNF-α release (Fig. 5A, left) but enhance LPS-stimulated TNF-α release 190, 151, and 138%, respectively (Fig. 5A, right). Using the 1-μg/ml concentration, LPS-stimulated ILI response was determined using Fe59. The Fe59-specific activity was not different in both untreated and iron dextran-treated cells after Fe59 labeling. As predicted, LPS-induced ILI response is significantly accentuated in the cells pretreated with iron dextran (Fig. 5B). Next we tested iron dextran treatment for PBMs from rats. Iron dextran treatment does not increase nonheme iron content in vehicle-treated PBMs until the concentration is raised to 100 μg/ml (Fig. 5C). On the other hand, PMA-induced, PBM-derived macrophages dose dependently increase their nonheme iron content in response to iron dextran (Fig. 5C), suggesting their enhanced pinocytotic activity. PMA-treated PBMs with or without subsequent iron dextran treatment (1 μg/ml) were tested for peroxynitrite-induced ILI response. As shown for KC, iron dextran-treated, PBM-derived macrophages show an accentuated ILI response to the agonist stimulation (Fig. 5D), suggesting that the priming with iron dextran occurs in macrophages in general.
Fig. 5.
A: normal rat cultured KC were pretreated with the different concentrations of iron dextran and stimulated with LPS to determine the effects of iron loading on TNF-α release. Note iron dextran treatment alone does not affect the basal TNF-α release (left) but enhances LPS-stimulated TNF-α release twofold at 1 μg/ml concentration and 40–50% at 10–100 μg/ml of iron dextran (right). *P < 0.05 compared with the cells without the iron dextran treatment. B: iron loading by incubation with 1 μg/ml iron dextran overnight accentuates the ILI response in cultured KC as determined by LPS-induced [LMW-Fe]i. *P < 0.05 compared with the cells without prior iron dextran treatment (control). C: PMA-induced macrophages from rat blood monocytes take up iron dextran to increase iron storage more efficiently than vehicle-treated monocytes (no PMA). Normal rat blood monocytes were cultured with or without PMA (100 nM) and subsequently treated with iron dextran for 24 h before the cells were collected for determination of nonheme iron content. *P < 0.05 compared with vehicle-treated rat blood monocytes (no PMA). D: iron dextran loading enhances peroxynitrite-induced ILI response in rat blood monocyte-derived macrophages by PMA treatment. PMA-induced rat macrophages with or without iron dextran treatment (1 μg/ml) were analyzed for peroxynitrite-induced [LMW-Fe]i using Fe59Cl. *P < 0.05 compared with the cells without iron dextran treatment.
Iron dextran administration in mice promotes KC TNF-α production and aggravates experimental ALD.
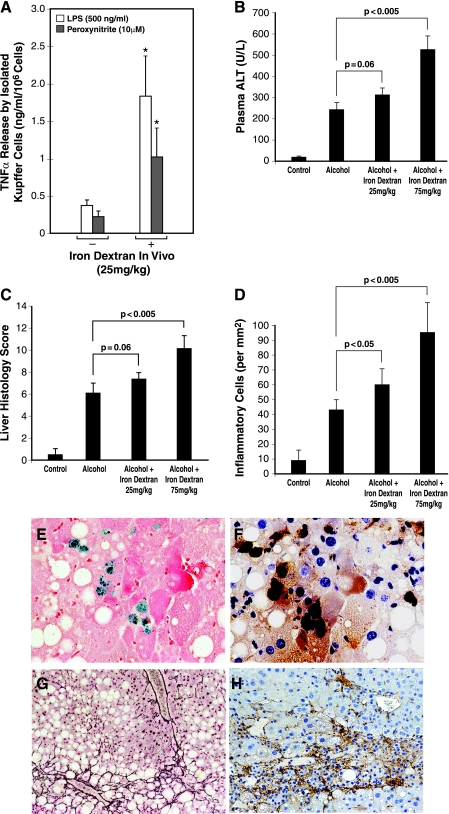
Next, we extended the manipulation with iron dextran in vivo by injecting it subcutaneously into mice at different doses (10–100 mg/kg). A single subcutaneous injection of iron dextran achieves a gradual uptake by macrophages over several weeks (23). After testing various doses, the doses of 25 and 75 mg/kg were shown to increase 1.7- and 3.8-fold the nonheme iron content of KC isolated from mice at 2 wk after the injection, respectively. KC isolated from the mice given 25 mg/kg were tested for TNF-α release without or with peroxynitrite (10 μM) or LPS (500 ng/ml) stimulation. As shown in Fig. 6A, KC isolated from iron dextran-injected mice release fourfold more TNF-α regardless of whether the cells are stimulated with LPS or peroxynitrite. This result demonstrates that in vivo KC iron loading with iron dextran indeed primes the cells for agonist-induced TNF-α expression.
Fig. 6.
A: KC were isolated from mice at 2 wk after a single subcutaneous injection of iron dextran (25 mg/kg) to assess LPS- or peroxynitrite-stimulated TNF-α expression. Note KC isolated from iron dextran-injected mice release 4–5 times more TNF-α regardless of whether they are stimulated with LPS or peroxynitrite. *P < 0.05 compared with the cells isolated from mice without iron dextran injection. B: plasma alanine aminotransferase (ALT) levels of mice fed intragastrically with alcohol and high fat diet or pair-fed with isocaloric diet for 4 wk without or with prior iron dextran injection at 25 mg/kg or 75 mg/kg. Note iron dextran injection at 75 mg/kg significantly aggravates plasma ALT elevation compared with alcohol-fed mice injected with vehicle (dextran). C: liver histological score of mice fed alcohol for 4 wk without or with iron dextran injection. Note iron dextran injection at 75 mg/kg significantly worsen the liver histological score compared with alcohol-fed mice with vehicle treatment (dextran). D: morphometric analysis of inflammatory cells in the liver of the different experimental groups. Note iron dextran injection at 25 mg/kg or 75 mg/kg significantly increases inflammatory cells in the liver compared with alcohol-fed mice with vehicle treatment. E: representative microphotographs of aggravated alcoholic liver injury in iron dextran-injected mice. In the liver of alcohol-fed mice given 75 mg/kg iron dextran, hepatic macrophages stained for iron (Prussian blue reaction) are noted, ×436 magnification. F: these iron staining-positive macrophages are also stained positively for active p65, ×436 magnification. G: pericellular liver fibrosis is evident in the livers of alcohol-fed mice with 75 mg/kg iron dextran treatment as demonstrated by reticulin staining, ×109 magnification. H: numerous activated hepatic stellate cells are present in a focus of liver necrosis and inflammation as shown by α-smooth muscle actin staining, ×109 magnification.
Finally, we tested the iron dextran priming method in the mouse intragastric ethanol infusion model to determine its effects on experimental ALD. One single injection of iron dextran (25 or 75 mg/kg sc) was given 2 wk before the initiation of 4-wk ethanol administration. Iron dextran administration increases plasma alanine aminotransferase (ALT) levels (an index of liver damage) from 241 ± 33.7 (U/l) in alcohol-fed mice without iron dextran treatment to 315.5 ± 30.1 and 522 ± 71.4 in alcohol-fed mice given the two respective doses (Fig. 6B). Blind histological analysis and scoring of the liver specimens were also performed, and the results show similar trends of aggravated liver pathology (Fig. 6C). Furthermore, a morphometric analysis reveals 40 and 220% increases in the number of inflammatory cells (both mononuclear and polymorphonuclear cells) in the livers of alcohol-fed mice given 25 mg/kg and 75 mg/kg iron dextran, respectively, compared with alcohol-fed mice without iron dextran treatment (Fig. 6D). In mice given alcohol and 75 mg/kg of iron dextran, liver necrosis and accompanying inflammation are evident. In these necrotic and inflammatory areas, KC stained for iron are noted (Fig. 6E), and these cells are also stained positively for active p65 (Fig. 6F), suggesting that KC with iron accumulation have enhanced NF-κB activation. Liver fibrosis is also evident in these animals as demonstrated by reticulin staining (Fig. 6G) and numerous activated hepatic stellate cells identified by α-smooth muscle actin immunostaining (Fig. 6H). Iron dextran administration to pair-fed control mice only slightly affect plasma ALT levels and liver histology in pair-fed control mice (data now shown). This absence of drastic effects is consistent with the lack of stimulation for basal TNF-α expression in cultured KC exposed to iron dextran (Fig. 5A, left). In summary, these results demonstrate that KC iron loading and consequent NF-κB priming by iron dextran administration accentuate alcoholic liver damage and further suggest the importance of the primed ILI signaling in the pathogenesis of ALD.
DISCUSSION
ALD is a chronic inflammatory disease much like atherosclerosis, pulmonary fibrosis, glomerulosclerosis, and neurodegenerative diseases, and its pathogenesis is largely predicated by gene and environment interactions (30). Among several key environmental risk factors identified for ALD, iron deserves particular attention. This transition metal is long known to aggravate ALD regardless of whether its excess in the liver is achieved by intake or a genetic mechanism (9, 24, 31). To add to the complexity, alcohol consumption itself increases hepatic iron content (16, 31, 35). Chronic inflammation also leads to increased iron storage. This effect may be mediated through induction of hepcidin by inflammatory cytokines and the ability of this iron regulatory hormone to reduce the expression of the iron exporter FPN (7). However, in our rat model of ALD, FPN1 expression is induced in KC whose nonheme iron content is increased. Thus reduced iron export is an unlikely cause of iron accumulation in these cells, and FPN1 induction may reflect a consequence of iron loading (15). Indeed, our results from cultured KC exposed to iron or iron chelator support this notion of a positive relationship between iron and FPN (Fig. 2, C and D). Instead, our research has identified induced expression of TfR1 and enhanced iron uptake as the most probable mechanism of KC iron loading in alcohol-fed rats. TfR1 is a predominant receptor type expressed by KC (40) and is increased nearly sixfold in Alc-KC (Fig. 3A). We also observed twofold induction of HFE Alc-KC. Mechanisms by which HFE controls iron homeostasis is poorly understood. HFE appears to have different effects depending on the cell types studied. For instance, HFE expression causes reduced intracellular levels in HeLa and H1299 cell lines associated with decreased Tf-mediated iron uptake (26, 36). Although this inhibition is thought to be mediated by the ability of HFE to compete with Tf for binding to TfR1 (17), this effect is also evident in the cells lacking endogenous TfR expression (3). In contrast, in macrophage and colonic intestinal cell lines, HFE increases cellular iron content via inhibition of iron export (6, 8). Consistent with this notion, reconstitution of HFE-deficient mice with wild-type bone marrow results in increased splenic iron storage, whereas the reverse reconstitution (wild-type mice with HFE-deficient bone marrow) causes decreased splenic iron content (20). Furthermore, transduction of wild-type HFE in PBMs from patients with HFE C282Y mutation normalizes Tf-dependent iron uptake and ferritin iron accumulation (22). Thus HFE appears to facilitate iron accumulation in macrophages, and this effect might have also contributed to increased iron content in Alc-KC via HFE induction.
The second key finding of the present study is the relevance of the ILI response in humans. Using PBMs from healthy volunteers, we have shown PMA-treated, PBM-derived macrophages, but not vehicle-treated monocytes, exhibit peroxynitrite-provoked ILI response. A functional significance of this ILI response is suggested by a fourfold enhancement in peroxynitrite-induced TNF-α production by macrophages that have acquired the ILI response. This notion is supported by the concomitant abrogation of the ILI response and TNF-α production by the iron chelator treatment. These results reveal that the ILI response is relevant in humans and is an acquired function of macrophage differentiation. This conclusion is not surprising since macrophages are already known to utilize iron as a vital tool for their host defense mechanisms. Such examples include a bacteriostatic effect on intracellular pathogens by iron deprivation of late endosomes achieved by the transporter Nramp1, unique in macrophages and an antimicrobial action of iron released by macrophages against extracellular microbes (12).
Finally, our study demonstrates that an increase in nonheme iron content augments the ILI response. This finding has a significant implication in understanding the link between KC iron accumulation and chronic liver disease such as ALD at a mechanistic level. Alc-KC have increased nonheme iron content and enhanced ILI response and NF-κB activation, and the treatment of these cells with a lipophilic iron chelator (L1) coordinately suppresses all these three parameters. This delayed ILI response at 5–7 min detected by the chelator method vs. <2 min by tracing with Fe59 appears to result from the fact that the chelator is not reaching liberated ILI immediately. Nevertheless, this method accurately detects and validates agonist-stimulated ILI response demonstrated by the Fe59 method (37). Iron dextran is a convenient tool to validate our notion because it is more readily taken up by macrophages and increases KC iron content. This manipulation was used in vitro and in vivo in our study to confirm the positive relationship between iron content, ILI signaling, and TNF-α expression in KC. Furthermore, this priming technique was extended to the mouse alcohol model to demonstrate that KC iron loading aggravates experimental ALD. The iron chelator treatment has previously been shown to reduce the severity of liver injury in the rat intragastric ethanol infusion model (27) and KC NF-κB activation and liver damage in a rat model of cholestatic liver injury (18). In patients with viral hepatitis (14, 38) or nonalcoholic steatohepatitis (NASH) (29, 34), iron reduction therapy by phlebotomy improves liver pathology. In a recent elegant study on the rabbit model of NASH, erythrophagocytosis by KC was shown to promote liver inflammation and fibrosis (25). In addition to increased iron uptake demonstrated in the present study, this mechanism of KC iron loading may also contribute to KC iron overload in ALD in which abnormalities of red blood cells are common and enhanced erythrophagocytosis by KC is expected. We propose that the link between accentuated ILI signaling and enhanced NF-κB activation revealed in the present study provides a cellular and molecular explanation for the role of KC iron in the pathogenesis of chronic liver disease.
GRANTS
This work was supported by National Institutes of Health Grants, P50 AA011999, Research Center for Alcoholic Liver and Pancreatic Diseases (H. Tsukamoto), and its Animal and Morphology Core facilities, R24 AA012885, Nonparenchymal Liver Cell Core (H. Tsukamoto), U56 AA014643, Collaborative Alcohol Research Center (V. Gordeuk), Suntory Ltd., and by the Medical Research Service of the Department of Veterans Affairs.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Atkinson PG, Barton CH. High level expression of Nramp1G169 in RAW264.7 cell transfectants: analysis of intracellular iron transport. Immunology 96: 656–662, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bridle K, Cheung TK, Murphy T, Walters M, Anderson G, Crawford DG, Fletcher LM. Hepcidin is down-regulated in alcoholic liver injury: implications for the pathogenesis of alcoholic liver disease. Alcohol Clin Exp Res 30: 106–112, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Carlson H, Zhang AS, Fleming WH, Enns CA. The hereditary hemochromatosis protein, HFE, lowers intracellular iron levels independently of transferrin receptor 1 in TRVb cells. Blood 105: 2564–2570, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Xiong S, She H, Lin SW, Wang J, Tsukamoto H. Iron causes interactions of TAK1, p21ras, and phosphatidylinositol 3-kinase in caveolae to activate IkappaB kinase in hepatic macrophages. J Biol Chem 282: 5582–5588, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Colell A, Garcia-Ruiz C, Miranda M, Ardite E, Mari M, Morales A, Corrales F, Kaplowitz N, Fernandez-Checa JC. Selective glutathione depletion of mitochondria by ethanol sensitizes hepatocytes to tumor necrosis factor. Gastroenterology 115: 1541–1551, 1998. [DOI] [PubMed] [Google Scholar]
- 6.Davies PS, Enns CA. Expression of the hereditary hemochromatosis protein HFE increases ferritin levels by inhibiting iron export in HT29 cells. J Biol Chem 279: 25085–25092, 2004. [DOI] [PubMed] [Google Scholar]
- 7.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell 18: 2569–2578, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drakesmith H, Sweetland E, Schimanski L, Edwards J, Cowley D, Ashraf M, Bastin J, Townsend AR. The hemochromatosis protein HFE inhibits iron export from macrophages. Proc Natl Acad Sci USA 99: 15602–15607, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher LM, Dixon JL, Purdie DM, Powell LW, Crawford DH. Excess alcohol greatly increases the prevalence of cirrhosis in hereditary hemochromatosis. Gastroenterology 122: 281–289, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Harrison-Findik DD, Schafer D, Klein E, Timchenko NA, Kulaksiz H, Clemens D, Fein E, Andriopoulos B, Pantopoulos K, Gollan J. Alcohol metabolism-mediated oxidative stress down-regulates hepcidin transcription and leads to increased duodenal iron transporter expression. J Biol Chem 281: 22974–22982, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Iimuro Y, Gallucci RM, Luster MI, Kono H, Thurman RG. Antibodies to tumor necrosis factor-alpha attenuate hepatic necrosis and inflammation caused by chronic exposure to ethanol in the rat. Hepatology 26: 1530–1537, 1997. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Baldwin CL. Iron augments macrophage-mediated killing of Brucella abortus alone and in conjunction with interferon-gamma. Cell Immunol 148: 397–407, 1993. [DOI] [PubMed] [Google Scholar]
- 13.Kamimura S, Tsukamoto H. Cytokine gene expression by Kupffer cells in experimental alcoholic liver disease. Hepatology 22: 1304–1309, 1995. [PubMed] [Google Scholar]
- 14.Kato J, Miyanishi K, Kobune M, Nakamura T, Takada K, Takimoto R, Kawano Y, Takahashi S, Takahashi M, Sato Y, Takayama T, Niitsu Y. Long-term phlebotomy with low-iron diet therapy lowers risk of development of hepatocellular carcinoma from chronic hepatitis C. J Gastroenterol 42: 830–836, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood 102: 4191–4197, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Kohgo Y, Ohtake T, Ikuta K, Suzuki Y, Hosoki Y, Saito H, Kato J. Iron accumulation in alcoholic liver diseases. Alcohol Clin Exp Res 29: 189S–193S, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Lebron JA, West AP Jr, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol 294: 239–245, 1999. [DOI] [PubMed] [Google Scholar]
- 18.Lin M, Rippe RA, Niemela O, Brittenham G, Tsukamoto H. Role of iron in NF-κB activation and cytokine gene expression by rat hepatic macrophages. Am J Physiol Gastrointest Liver Physiol 272: G1355–G1364, 1997. [DOI] [PubMed] [Google Scholar]
- 19.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci USA 103: 13612–13617, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makui H, Soares RJ, Jiang W, Constante M, Santos MM. Contribution of Hfe expression in macrophages to the regulation of hepatic hepcidin levels and iron loading. Blood 106: 2189–2195, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClain CJ, Cohen DA. Increased tumor necrosis factor production by monocytes in alcoholic hepatitis. Hepatology 9: 349–351, 1989. [DOI] [PubMed] [Google Scholar]
- 22.Montosi G, Paglia P, Garuti C, Guzman CA, Bastin JM, Colombo MP, Pietrangelo A. Wild-type HFE protein normalizes transferrin iron accumulation in macrophages from subjects with hereditary hemochromatosis. Blood 96: 1125–1129, 2000. [PubMed] [Google Scholar]
- 23.Muir AR, Golberg L. Observations on subcutaneous macrophages. Phago-cytosis of iron-dextran and ferritin synthesis. Q J Exp Physiol Cogn Med Sci 46: 289–298, 1961. [DOI] [PubMed] [Google Scholar]
- 24.Nahon P, Sutton A, Rufat P, Ziol M, Thabut G, Schischmanoff PO, Vidaud D, Charnaux N, Couvert P, Ganne-Carrie N, Trinchet JC, Gattegno L, Beaugrand M. Liver iron, HFE gene mutations, and hepatocellular carcinoma occurrence in patients with cirrhosis. Gastroenterology 134: 102–110, 2008. [DOI] [PubMed] [Google Scholar]
- 25.Otogawa K, Kinoshita K, Fujii H, Sakabe M, Shiga R, Nakatani K, Ikeda K, Nakajima Y, Ikura Y, Ueda M, Arakawa T, Hato F, Kawada N. Erythrophagocytosis by liver macrophages (Kupffer cells) promotes oxidative stress, inflammation, and fibrosis in a rabbit model of steatohepatitis: implications for the pathogenesis of human nonalcoholic steatohepatitis. Am J Pathol 170: 967–980, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem 274: 9022–9028, 1999. [DOI] [PubMed] [Google Scholar]
- 27.Sadrzadeh SM, Nanji AA, Price PL. The oral iron chelator, 1,2-dimethyl-3-hydroxypyrid-4-one reduces hepatic-free iron, lipid peroxidation and fat accumulation in chronically ethanol-fed rats. J Pharmacol Exp Ther 269: 632–636, 1994. [PubMed] [Google Scholar]
- 28.She H, Xiong S, Lin M, Zandi E, Giulivi C, Tsukamoto H. Iron activates NF-κB in Kupffer cells. Am J Physiol Gastrointest Liver Physiol 283: G719–G726, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Sumida Y, Kanemasa K, Fukumoto K, Yoshida N, Sakai K, Nakashima T, Okanoue T. Effect of iron reduction by phlebotomy in Japanese patients with nonalcoholic steatohepatitis: a pilot study. Hepatol Res 36: 315–321, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Tsukamoto H Conceptual importance of identifying alcoholic liver disease as a lifestyle disease. J Gastroenterol 42: 603–609, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Tsukamoto H, Horne W, Kamimura S, Niemela O, Parkkila S, Yla-Herttuala S, Brittenham GM. Experimental liver cirrhosis induced by alcohol and iron. J Clin Invest 96: 620–630, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukamoto H, Lin M, Ohata M, Giulivi C, French SW, Brittenham G. Iron primes hepatic macrophages for NF-κB activation in alcoholic liver injury. Am J Physiol Gastrointest Liver Physiol 277: G1240–G1250, 1999. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamoto H, Mkrtchyan H, Dynnyk A. Intragastric ethanol infusion model in rodents. Methods Mol Biol 447: 33–48, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Valenti L, Fracanzani AL, Dongiovanni P, Bugianesi E, Marchesini G, Manzini P, Vanni E, Fargion S. Iron depletion by phlebotomy improves insulin resistance in patients with nonalcoholic fatty liver disease and hyperferritinemia: evidence from a case-control study. Am J Gastroenterol 102: 1251–1258, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Valerio LG Jr, Parks T, Petersen DR. Alcohol mediates increases in hepatic and serum nonheme iron stores in a rat model for alcohol-induced liver injury. Alcohol Clin Exp Res 20: 1352–1361, 1996. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Chen G, Pantopoulos K. The haemochromatosis protein HFE induces an apparent iron-deficient phenotype in H1299 cells that is not corrected by co-expression of beta 2-microglobulin. Biochem J 370: 891–899, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong S, She H, Takeuchi H, Han B, Engelhardt JF, Barton CH, Zandi E, Giulivi C, Tsukamoto H. Signaling role of intracellular iron in NF-kappaB activation. J Biol Chem 278: 17646–17654, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Yano M, Hayashi H, Yoshioka K, Kohgo Y, Saito H, Niitsu Y, Kato J, Iino S, Yotsuyanagi H, Kobayashi Y, Kawamura K, Kakumu S, Kaito M, Ikoma J, Wakusawa S, Okanoue T, Sumida Y, Kimura F, Kajiwara E, Sata M, Ogata K. A significant reduction in serum alanine aminotransferase levels after 3-month iron reduction therapy for chronic hepatitis C: a multicenter, prospective, randomized, controlled trial in Japan. J Gastroenterol 39: 570–574, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Zhang AS, Davies PS, Carlson HL, Enns CA. Mechanisms of HFE-induced regulation of iron homeostasis: insights from the W81A HFE mutation. Proc Natl Acad Sci USA 100: 9500–9505, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood 103: 1509–1514, 2004. [DOI] [PubMed] [Google Scholar]