Abstract
Tristetraprolin (TTP) is the prototype for a family of RNA binding proteins that bind the tumor necrosis factor (TNF) messenger RNA AU-rich element (ARE), causing deadenylation of the TNF poly(A) tail, RNA decay, and silencing of TNF protein production. Using mass spectrometry sequencing we identified poly(A) binding proteins-1 and -4 (PABP1 and PABP4) in high abundance and good protein coverage from TTP immunoprecipitates. PABP1 significantly enhanced TNF ARE binding by RNA EMSA and prevented TTP-initiated deadenylation in an in vitro macrophage assay of TNF poly(A) stability. Neomycin inhibited TTP-promoted deadenylation at concentrations shown to inhibit the deadenylases poly(A) ribonuclease and CCR4. Stably transfected RAW264.7 macrophages overexpressing PABP1 do not oversecrete TNF; instead they upregulate TTP protein without increasing TNF protein production. The PABP1 inhibition of deadenylation initiated by TTP does not require the poly(A) binding regions in RRM1 and RRM2, suggesting a more complicated interaction than simple masking of the poly(A) tail from a 3′-exonuclease. Like TTP, PABP1 is a substrate for p38 MAP kinase. Finally, PABP1 stabilizes cotransfected TTP in 293T cells and prevents the decrease in TTP levels seen with p38 MAP kinase inhibition. These findings suggest several levels of functional antagonism between TTP and PABP1 that have implications for regulation of unstable mRNAs like TNF.
Keywords: RNA, immunology, tumor necrosis factor
tristetraprolin (TTP) has been shown to be constitutively expressed in the liver (32) and to be upregulated in liver regeneration after partial hepatectomy (16), in the postmitotic intestinal crypt (15), and in the adaptive hyperplasia that occurs in a surgical model of short gut syndrome (17). TTP functions to regulate mRNAs by binding to the AU-rich elements (AREs) of unstable mRNAs, promoting their degradation. AREs are a feature of labile cytokine and immediate early response mRNAs that promote enzymatic removal of the poly(A) tail (deadenylation) and resultant mRNA decay in mRNA processing bodies (18, 57). In tristetraprolin knockout mice, absence of the protein prolongs macrophage TNF and granulocyte-macrophage colony stimulating factor (GM-CSF) mRNA half-life and enhances protein production, both of which contain well-characterized AREs in their 3′-UTRs (8, 9). The resultant TNF overexpression and tissue damage can be prevented with repeated injections of a neutralizing anti-TNF antibody (7).
TTP is the prototype of a family of RNA-binding proteins with unusual zinc binding domains that interact with RNA in a unique conformation (2, 20, 52). We found by RNA selection that the preferred TTP RNA binding site is UUAUUUAUU (53), the canonical destabilizing ARE motif found in cytokines and proto-oncogenes (29, 57). TTP-mediated deadenylation requires TTP binding to the ARE (30). TTP is multiply phosphorylated by MAPKAP kinase 2 (MK2) (10), a kinase downstream of p38 MAP kinase, and a pathway critical for TNF protein production (13, 26). Furthermore, mice lacking MK2 make TNF mRNA but are incapable of effectively translating it (26). TTP is also phosphorylated by p38 directly (10, 56), which inhibits the gene inhibitory effects of TTP (56).
We identified the MK2 and p38 phosphorylation sites on TTP using nanoflow HPLC electrospray ionization (nHPLC-μESI) mass spectrometry of a tryptic digest of heterologously expressed and immunoprecipitated TTP protein from mammalian cells (10). The design of these experiments also allowed detection of peptides from a number of coprecipitating proteins. One protein recovered in high abundance with good peptide coverage from all six sequencing runs (and absent when TTP was not present) was poly(A) binding protein-1 (PABP1). PABP1 interacts with the 5′ cap complex (35), has ARE-binding activity (45), and is best known as an enhancer of mRNA stability by binding to the poly(A) mRNA tail and protecting it from deadenylation (50, 51). Since the function of TTP is to promote deadenylation and subsequent mRNA decay, the isolation of PABP in high abundance raised the possibility of direct antagonism of TTP by PABP. PABP1 is also a reported target for MK2 on the GM-CSF ARE (5), although the mechanistic consequences of MK2 phosphorylation of PABP are unknown. Efficient translation of proteins requires PABP1 to bind both the 3′ end of the mRNA and the 5′ cap binding factor eIF4G. Deadenylation creates translationally incompetent mRNAs and silences protein expression even before the mRNA body can be destroyed (22, 24, 28, 49).
In this manuscript, we identify the interaction of TTP and PABP1, investigate the functional antagonism between PABP1 in an in vitro model of TTP function, and explore the consequences of this interaction in the context of TNF mRNA metabolism.
MATERIALS AND METHODS
Chemicals and reagents.
G418 was obtained from Invitrogen. Neomycin, phenol-extracted and ion exchange-purified Escherichia coli LPS (L4524), and other general chemicals were obtained from Sigma-Aldrich. The anti-glutathione-S-transferase (GST) antibody was obtained from Upstate Biotechnologies, the 9E10 myc epitope tag monoclonal antibody was obtained from the University of Virginia Lymphocyte Core, and the anti-PABP1 antibody was obtained from Abcam. The anti-poly(A) ribonuclease (anti-PARN) antibody was a kind gift from Michael Wormington and the rabbit anti-TTP antibody was a kind gift from Jiahuai Han. The GFP-PABP1 and mTTP tag plasmids were kind gifts from George Pavlakis (1) and Christoph Moroni (46), respectively. pEGFP-N1 was obtained from Clontech.
Cell culture and transfection.
RAW264.7 cells were obtained from the American Type Culture Collection and propagated in RPMI 1640 media with 10% fetal bovine serum (Invitrogen). Transfection was performed with FuGENE-6 reagent using the accompanying protocol (Roche). Selection to create stable cell lines was performed at 100 ng/ml G418 and LPS stimulation performed at 10 ng/ml. HEK 293T cells were obtained from the American Type Culture Collection, propagated in the recommended media, and transfected with FuGENE-6.
Cloning and plasmids.
All plasmid preparations were performed with endotoxin-free plasmid DNA (EndoFree; Qiagen) The Balb/c TNF 3′-untranslated region (UTR) was amplified from random primed Balb/c cDNA from LPS-stimulated buffy coat cells using the primers GGA CTC ATC TAG ACT TTC CGA ATT CAC TGG AGC CTC and CGT TTA TTC TAG AAG CGA TCT TTA TTT CTC TC. This was cloned via XbaI (New England Biolabs) into similarly cut pSEAP Control and pGL3Control. The HindIII/BamH1 fragment was shuttled into Bluescript KS(-) to create templates for capped synthetic mRNA production using Message Machine kit (Ambion). Human PABP1 was subcloned into the pET28 (EMD Biosciences) and pGEX (Amersham Pharmacia) by standard methods. Subcloning of RRM1+2 and RRM3+4-COOH-terminal PABP fragments was done by using the native EcoR1 site in human PABP1. Further details of subcloning are available on request. All cloning was verified with DNA sequencing using BigDye Terminator chemistry (Applied Biosystems) on both strands.
Protein methods.
On-bead digestion and identification of TTP interacting proteins were performed by previously described methods (10). The ECL-Plus Western blotting detection system was used for all immunoblotting (Amersham Biosciences) using the appropriate secondary antibodies. GST-TTP, His-PABP1, GST-PABP1, and GST-PABP1 fragments were expressed and purified by the manufacturer's methods, then dialyzed and concentrated as described in our laboratory's previous work (53). RNAse contamination was assayed with the RNaseALERT (Ambion). Ponceau S staining of all Western blot transfers was performed to verify even loading and transfer (55). Active recombinant p38 was prepared as previously described (10). kinase assays were performed at 30°C in 25 μl reactions containing (final concentrations) 25 mM HEPES, pH 7.4, 10 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 0.1 mg/ml substrate, 10 μg/ml p38, 10 mM MgCl2, 50 μM [γ-32P]ATP (∼4,000 cpm/pmol). Phosphate incorporation was quantified by use of a phosphoimager and equal protein loading by Coomassie staining.
Synthesis of mRNAs and RNA EMSAs.
Short radioactive TNF ARE RNA probes were expressed as previously described (53). Larger mRNAs were generated by using linearized plasmid DNAs with the Message Machine kit (which includes a final DNase I digestion to remove plasmid DNA; Ambion), followed by tailing with the Poly(A) Tailing kit (Ambion), which typically added at least 200 poly(A) residues. All Message Machine mRNAs are capped with the accompanying cap analog. RNA EMSAs were performed as previously described (53).
In vitro deadenylation reactions.
RAW264.7 cytoplasmic extracts were produced by use of a standard protocol (10). Unstimulated extracts were used to avoid the presence of endogenous TTP. Cytoplasmic extract (7 μl) was incubated with 10 μg of reporter RNA at 30°C for up to 30 min in 50-μl reactions containing 10 mM HEPES, pH 7.9, 50 mM KCl, 1 mM MgCl2, 1 mM DTT, 15 mM creatine phosphate, and 0.6 mM ATP. Remaining RNA was recovered with the RNeasy Mini kit (Qiagen) and eluted in 30 μl of RNase-free water for reverse transcription and PCR amplification. Reticulocyte lysates (Promega) were programmed with 3 μg of our reporter RNA per the manufacturer's protocol and incubated for 90 min at 30°C, and 20 μl were removed for firefly luciferase assay in a 96-well plate (Dual Luciferase Assay, Promega).
Reverse-transcriptase PCR amplification.
To measure poly(A) tail length, the G-tailing method of Kusov et al. (27) was used, which produces a PCR product between a fixed 5′ PCR primer and a 3′ primer annealing to the junction between the poly(A) tail and an enzymatically added homopolymer guanosine tract. The Qiagen RNEasy purified postlysate RNA was poly(G) tailed as described by using yeast poly(A) polymerase and GTP as the sole source of nucleotide (27), then reverse transcribed with the Superscript First Strand Synthesis kit (Invitrogen) substituting the primer CCC CCC CCC TTT TTT. PCR amplification was performed with the CCC CCC CCC TTT TTT primer and GCC GTG ACT GTA ATT GCC, which anneals 75 bases from the end of the poly(A) tail. Thus the PCR product size in the G-tailing reaction represents the length of the poly(A) tail plus 75 bases. Standard random priming and oligo (dT) priming were performed with commercially available kits (Superscript II First-Strand cDNA kit, Invitrogen). PCR amplification from RNA products of pSEAP-based plasmids was performed using primers GAC CCA CGC AGG CGA GGA C and GGC CAT GAC GTG CGC TAT GAA and amplification of pGL3 luciferase-based products was performed using the primers ACT GCC TGC GTG AGA T and TCA GTG AGC CCA TAT CCT with HiFi Taq Polymerase (Invitrogen). Ethidium bromide-stained DNA TBE PAGE gels were used for evaluating the PCR reactions vs. the 1-kb ladder (Invitrogen), and SYBRgreen quantitative real-time PCR was performed as previously described (42).
RESULTS
Immunoprecipitation of PABP1 with TTP by mass spectrometry sequencing.
In a previous report, we mapped the MK2 and p38 MAP kinase phosphorylation sites on TTP by immunoprecipitation of TTP for mass spectrometry sequencing (10). Although our experiments were not specifically intended to isolate coprecipitating proteins, a significant number of peptides from multiple regions of specific proteins were consistently recovered in a TTP-dependent fashion and in high abundance. Most of these were related to regulation of mRNA stability or splicing (Fig. 1A) and these were not found in the untransfected negative control. The most abundant protein represented after TTP itself was PABP, with PABP peptides present in all TTP-transfected analyses with significant PABP protein coverage (Fig. 1B), including diagnostic peptides for PABP1 (Fig. 1C) and PABP4 (Fig. 1D). This is a striking finding since PABP and TTP have opposing effects on the integrity of the poly(A) tail of mRNAs targeted by TTP, and TTP RNA binding is known to lead to mRNA destruction. No PABP was recovered in the non-TTP-transfected controls. With these findings, on the basis of experiments from two different cell types and the known biology of TTP, we concluded that the detection of PABP only in the TTP-transfected immunoprecipitations was specific.
Fig. 1.
Immunoprecipitation of tristetraprolin (TTP) and identification of poly(A) binding protein (PABP) fragments by mass spectrometry. Epitope-tagged TTP was expressed in transfected BHK and NIH3T3 cells and immunoprecipitated for phosphorylation site analysis (as described in Ref. 10). Peptides from a number of proteins involved in mRNA metabolism were reproducibly recovered in significant quantity only in the TTP-expressed samples (A). A significant percentage of the sequenced peptides were found to be PABP, surprising since the role of TTP is to promote deadenylation and therefore destroy the PABP binding site. Polyadenylate-binding protein 1 (PABP1) protein sequence and peptide coverage (bold and underlined; B) routinely detected from analyses of tryptic digests by online nHPLC-μESI mass spectrometry. Many of these are shared by more than 1 PABP isoform. Peptides that are unique to PABP1 and PABP4 are shown in C and D, respectively.
Development of a TTP-promoted in vitro assay of mRNA degradation.
PABP1 is ubiquitously expressed in eukaryotic cells and is required for cell viability, making inhibitory methods such as small interfering RNA directed at PABP impractical and potentially unreliable (47). PABP1 has been recently shown not to exchange between mRNAs in a complex cellular mRNA pool, resulting in a very stable protein-mRNA complex once formed (39). To test the impact of an interaction of PABP1 and TTP, a TTP-primed mRNA degradation assay was necessary. An existing assay of TTP function in HEK 293 cell lysates requires mixing of immunoprecipitates of both epitope-tagged TTP and PARN to deadenylate an ARE RNA (7, 31), adding a third protein (all at unknown concentrations) would make this assay difficult to interpret. We initially tested the TTP-PABP association in RAW264.7 macrophage cells using overexpression of TTP with and without PABP1 using a TNF 3′ reporter gene, but found that the TTP-transfected cells lost their ability to remain attached to the tissue culture plate (not shown), similar to NIH3T3 cell studies where TTP overexpression led to cell death (21).
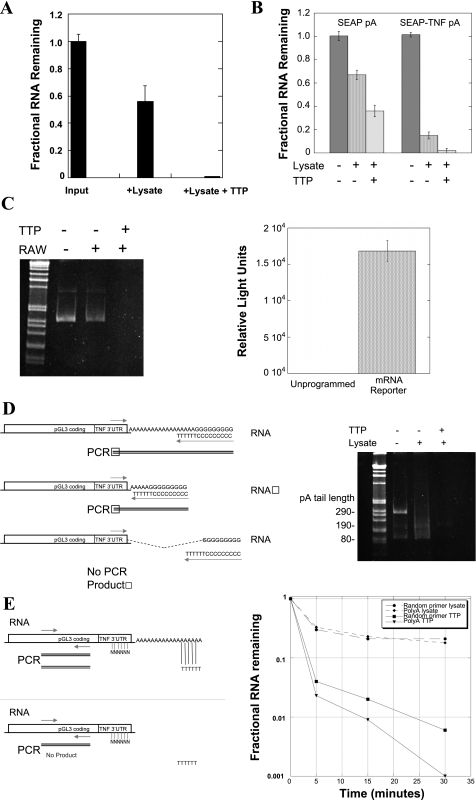
To directly assess whether PABP affects RNA decay promoted by TTP, we developed an assay of in vitro degradation using TNF-based mRNAs, unstimulated RAW264.7 cell cytoplasmic extracts which are known not to express TTP (Ref. 56 and unpublished results) and recombinant TTP protein (53). The initial substrate for this assay was a total cellular mRNA pool containing TNF mRNA, made by treatment of RAW264.7 cells with E. coli LPS, which causes a burst of TNF expression (19, 42). This native total RNA pool was purified to provide a naked RNA substrate for TTP. The substrate mRNA was added back to the RAW264.7 cell cytoplasmic extracts at a time point without endogenous TTP expression, and reactions were initiated with and without recombinant TTP. After incubation for 5 min at ambient temperature, the RNA was recovered and reverse transcribed by oligo(dT) priming, followed by TNF TaqMan real-time PCR. The results, normalized to the amount of input mRNA by SYBRgreen 18S real-time PCR (Fig. 2A), show accelerated degradation of native TNF mRNA with addition of recombinant TTP.
Fig. 2.
Properties of TTP-promoted deadenylation. RAW264.7 (RAW) cytoplasmic extracts and recombinant TTP were used to create an in vitro system for the study of RNA degradation. Total RNA from RAW264.7 cells stimulated for 1 h with LPS was added with carrier RNA to unstimulated RAW264.7 cell cytoplasmic extracts with or without recombinant TTP for 5 min. The remaining TNF mRNA in each condition was quantified by use of oligo(dT)-primed TaqMan real-time RT-PCR and normalized to the input RNA, shown in A. The role of the TNF 3′-untranslated region (UTR) in this process was confirmed using synthetically polyadenylated pSEAP synthetic RNAs that either lack or include the Balb/c TNF 3′-UTR (SYBRgreen real-time PCR, B). Similar results by use of standard RT-PCR are shown on an agarose gel (C, left) by using a synthetic capped luciferase-TNF 3′-UTR-poly(A)-tailed mRNA and luciferase coding region primers. This capped, polyadenylated mRNA is also transcriptionally active in reticulocyte lysates, but not in our macrophage extracts, confirming that translation is not required for TTP-mediated degradation (C, right). Efforts to measure the rate of deadenylation were unsatisfying due to the rapidity of the TTP-dependent process. One of these, the poly(G) tailing method of Kusov et al. (27), is shown schematically in D, left, which primes reverse transcription and PCR amplification from the extreme end of the poly(A) tail by creating a 3′-primer site at the junction of the 6 terminal A residues and an enzymatically added guanosine tract prior to PCR. Our choice of 5′-primer in the TNF 3′-UTR creates a PCR product whose size is equal to the remaining poly(A) tail length plus 75 bases. The reduction in of poly(A) tail length in RAW cell cytoplasmic extracts (D, center lane on gel) is completely lost when recombinant TTP is added (D, right lane on gel). For subsequent studies, the PCR primers in the luciferase coding region as in C (above) were used so that the efficiency of amplification would be the same in all conditions. The 3′ dependence of the TTP effect was addressed using the luciferase-TNF reporter RNA, but by priming the reverse transcription from either end of the poly(A) tail vs. by random priming from the cRNA body, shown schematically in E, left. Both random oligo-primed and poly(A)-tail primed SYBRgreen real-time RT-PCR were evaluated by use of luciferase primers and the results with and without TTP graphed in E, right, expressed as a fraction of the input RNA at time 0. The same data expressed as the ratio of poly(A)-primed to random-primed RNA from 0–30 min are seen in Table 1, with the ratio between them only changing when recombinant TTP is added. Neomycin, a known inhibitor of several deadenylases such as poly(A) ribonuclease and Ccr4 (36, 40), inhibits the effect of TTP on the reporter RNA (F). All of these experiments were repeated at least 3 times or more, with different TTP, lysate, and RNA preparations.
The TNF 3′-UTR dependence of TTP-mediated deadenylation was demonstrated by using two synthetic pSEAP coding region RNAs, one of which contained the Balb/c TNF 3′-UTR after the stop codon. The TNF 3′-UTR contains multiple copies of the TTP binding site, UUAUUUAUU (53). The in vitro transcribed SEAP mRNA targets were enzymatically polyadenylated before incubation in RAW lysates before incubation with recombinant TTP to determine the necessity for the TNF 3′-UTR in this process. The remaining target RNA was isolated and quantified by oligo(dT)-primed real-time PCR (Fig. 2B). Addition of TTP enhanced the degradation of the RNA and was accelerated by the presence of the TNF 3′-UTR.
For subsequent experiments, a synthetic luciferase/TNF 3′-UTR reporter mRNA was used, since some 3′-exonucleases have been shown to be dependent on translation and the pSEAP protein product was not well suited for this purpose, since it is intended to be secreted. The dependence of RNA degradation on the addition of TTP was again verified by using luciferase primers (Fig. 2C). No luciferase production was seen in the quiescent RAW264.7 cell lysates, although the same RNA produced near maximal relative light units when placed into reticulocyte lysate vs. unprogrammed reticulocyte lysate control (Fig. 2C). This in vitro translated and enzymatically polyadenylated RNA construct was used for subsequent experiments.
To prove that our recombinant TTP led to degradation from 3′-exonuclease activity, we initially used the poly(G) tailing method of Kusov et al. (27) to measure poly(A) tail length by reverse priming from the extreme end of the poly(A) tail. This is shown schematically in Fig. 2D. Using a 5′-primer 75 bases from the start of the poly(A) tail, we found that our in vitro poly(A) tailing kit consistently put on a tail of at least 200 residues, but the great activity of the TTP-primed 3′-exonuclease reaction yielded a very faint smear on ethidium bromide gels that was not readily quantifiable (Fig. 2D). This was not optimal for determining factors that change the rate of deadenylation.
In macrophages, deadenylation and decay of the mRNA body are independently regulated steps (12). This is similar to Xenopus oocytes, in which deadenylated maternal mRNAs can be polyadenylated and therefore recruited for translation to control morphogenesis (44). We compared oligo(dT) and random primed RT-PCR products of each RNA sample and measured the relative amount of the two species by using luciferase coding region primers [poly(A)/total RNA]. A decrease in the ratio of poly(A)-primed to random-primed RNA indicates a loss of poly(A)-tailed mRNA at a more rapid rate than the total mRNA. In this setting, the efficiency of PCR amplification is the same since the products are equivalent (Fig. 2E). The SYBRgreen real-time PCR results using this approach are shown in Fig. 2E and in Table 1, comparing the degradation of the luciferase reporter RNA without and with recombinant TTP. Again, TTP dramatically accelerates RNA degradation (Fig. 2E). In Table 1, this is expressed as a ratio of poly(A)/total mRNA at each time point: Without TTP, the ratio of degradation of poly(A)+ to total RNA by the lysate is essentially ∼1 at all time points, consistent with the degradation as being non-3′-directed. With the addition of TTP, the ratio diminishes over time, consistent with increased deadenylation and 3′-directed mRNA decay.
Table 1.
Changing ratio of polyA-primed to random-primed RNA in RAW264.7 lysates with the addition of recombinant TTP
|
Time |
||||
|---|---|---|---|---|
| 0 min | 5 min | 15 min | 30 min | |
| Lysate alone | 1.0 | 1.1 | 1.1 | 0.9 |
| Lysate + TTP | 1.0 | 0.58 | 0.45 | 0.17 |
The Sybr-Green real-time PCR results in Fig. 2E were normalized to the input RNA by the cycle threshold method, then the ratio between the polyA-primed vs. random-primed RNA calculated at each time point. The poly(A) dependence of the tristetraprolin (TTP) effect is seen as the ratio changes with increasing time.
TTP has been experimentally linked to the 3′-exonucleases CCR4 and PARN (31, 33). Our purified recombinant TTP does not have intrinsic deadenylase activity in vitro, consistent with the requirement for the protein to recruit the exonuclease to the poly(A) tail (33). Both CCR4 (36) and PARN (40) 3′-exonucleases can be inhibited by the antibiotic neomycin B, with ∼80% of PARN activity lost at a 10 μg/ml concentration (40). We used neomycin to assess whether the RNA degradation was consistent with the known biology of these 3′-exonucleases: TTP-promoted deadenylation was inhibited by neomycin at 10 μM concentration (40), with oligo(dT)-primed RT-PCR amplification products shown on an ethidium bromide-stained agarose gel in Fig. 2F.
TTP and PABP interact on the TNF ARE.
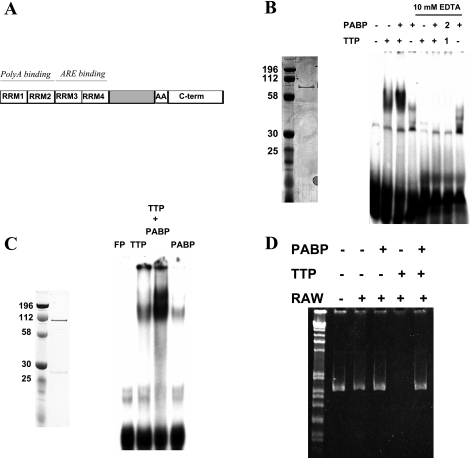
PABP1 is reported to have ARE-binding activity in RNA recognition motif (RRM) domains (3 and 4) distinct from those used for poly(A) binding (RRMs 1 and 2), as shown in Fig. 3A (45). Whether TTP and PABP1 form a complex on a radioactive murine TNF ARE probe was tested, with sequence gggCACUUUAUUUAUUUAUUUGCUUG. (The ARE core is underlined). GST-TTP/His-PABP RNA EMSA (Fig. 3B) led to a striking increase in the amount of shifted probe over TTP or PABP1 alone, with evidence of a supershift. EDTA-coincubations were included in Fig. 3B to determine whether the integrity of the two TTP Cys3His repeats (the protein's known ARE binding domain) were required for this enhanced interaction and whether PABP1 could protect TTP from zinc enucleation. EDTA completely disrupted the interaction of TTP with probe and/or PABP1 without regard to the order of protein or probe addition, suggesting that this interaction requires intact TTP RNA binding domains. To prove that this effect was not due to the polyhistidine tag on His-PABP interacting with the coordinated TTP zinc atoms, this process was confirmed by use of GST-TTP and GST-PABP (Fig. 3C).
Fig. 3.
PABP1 enhances TTP binding to the TNF RNA AU-rich element (ARE). A: structure of PABP1, with RRM 1 and 2 [poly(A) binding] and RRM 3 and 4 (ARE binding). The COOH terminus contains an alanine-rich region and a COOH-terminal oligomerization domain. GST-TTP (from Ref. 53) and His-PABP1 (Coomassie-stained gel on left) were evaluated individually and together by RNA EMSA using a TNF ARE probe (B, lanes 1–4). The requirement for intact Cys3His zinc binding domains and whether PABP1 could protect from zinc enucleation was evaluated in lanes 5–8, with the protein-RNA complex allowed to form before the addition of EDTA to 10 mM. In lane 7, TTP (1) was added to the RNA before PABP1 addition (2) to determine whether PABP binding could protect TTP from zinc enucleation. To confirm that this interaction was not an artifact of the polyhistidine epitope tag on PABP1, a TNF EMSA was performed using GST-PABP1 (Coomassie, C, left) and GST-TTP in TNF ARE EMSA (C, right). FP, free probe (no protein) lane. D: the ability of PABP1 to interfere with TTP-promoted deadenylation was assessed by use of the synthetic luciferase TNF-3′-UTR reporter RNA in unstimulated RAW264.7 cytoplasmic extracts by RT-PCR using luciferase coding region primers. Recombinant GST-TTP and GST-PABP were used. This has been repeated >3 times with equivalent results.
Creation of this TTP-initiated in vitro method of RNA degradation allowed us to probe the impact of PABP on this process, used in the same stoichiometry as TTP. Oligo(dT)-primed RT-PCR was performed on the recovered mRNA, shown in Fig. 3D. The PABP1-TTP complex prevented RNA degradation, suggesting that PABP1 is sufficient to suppress the degradative effects of TTP.
Inhibition of TTP-mediated deadenylation involves non-poly(A) binding portions of PABP1.
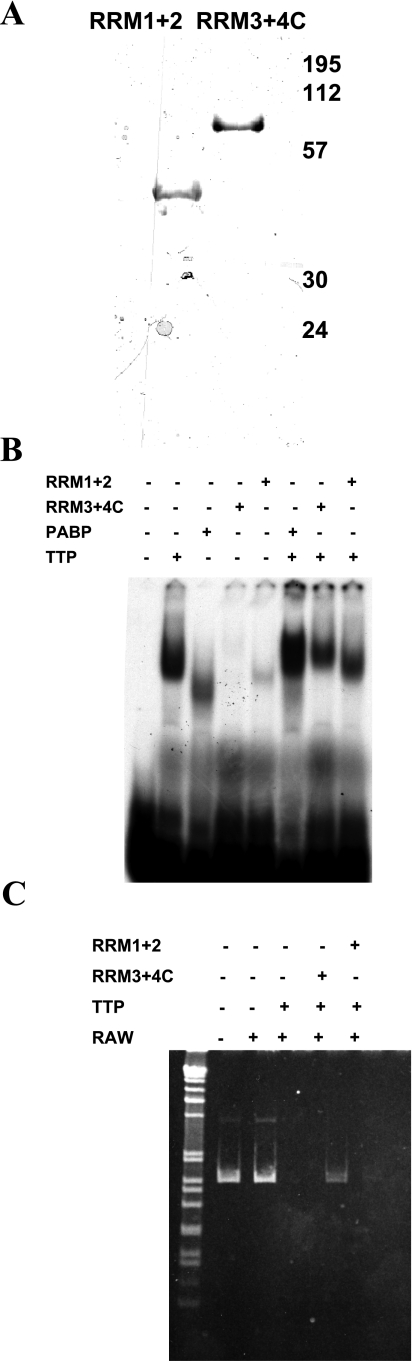
As was shown above, the PABP1 protein has four RRM RNA binding domains, as well as a COOH-terminal oligomerization domain. RRMs 1 and 2 are used to bind the poly(A) scaffold, whereas RRMs 3 and 4 of PABP1 are reported to bind the ARE (43, 45). A reasonable assumption was that the inhibition of TTP-mediated deadenylation was due to poly(A) tail binding by the RRM1+2 portions of PABP, blocking access of a TTP-recruited deadenylase to the poly(A) tail. To test this directly, we purified truncated PABP1 protein fragments that include RRM1+2 and the RRM3+4/COOH terminus, respectively (Fig. 4A) as GST fusions.
Fig. 4.
The non-poly(A) binding portion of PABP1 is sufficient to block TTP-induced deadenylation. A: poly(A) and non-poly(A) binding portions were cloned and expressed in Escherichia coli separately, shown as a wet-gel scan of Coomassie-stained GST-PABP1-RRM1+2 and GST-PABP1-RRM3+4-COOH terminus PABP truncations. B: truncation proteins enhance TTP-binding to a radioactive TNF probe, although less than the full-length PABP1 protein. C: TTP-directed deadenylation is inhibited by the truncation mutants RRM3+4C, which contains the ARE-binding region, but not the poly(A) binding region. These experiments have been performed >3 times with equivalent results.
The separated PABP1 poly(A)-binding (RRM1+2) and AU-rich binding activities (RRM3+4 and COOH terminus) were analyzed with TTP by TNF ARE EMSA, shown in Fig. 4B. The fragments showed weak binding of the TNF ARE probe without TTP, and in combination with TTP incubation caused either no change or a slight decrease in probe binding from TTP alone. It is possible that the fragment containing RRM3+4 and the COOH terminus still interacts with TTP, since the peak of the radioactive band is shifted toward the origin; however, enhanced binding to the TNF ARE probe (in this case 50% more than TTP alone by PhosphorImager quantification) is only observed with full-length PABP.
These truncated products were used in the in vitro deadenylation assay to determine whether TTP-mediated deadenylation could be inhibited with fragments reported to only bind either to poly(A) or ARE RNA. The results, shown in Fig. 4C, reveal that poly(A) tail binding activity of PABP is not required to inhibit deadenylation by TTP. This is consistent with previous PABP tethering experiments that suggest that as long as RRM3 and 4+COOH terminus of PABP are bound to the mRNA, even without association to the poly(A) tail or poly(A) binding activity, the mRNA is protected from degradation (11).
PABP stabilizes TTP protein.
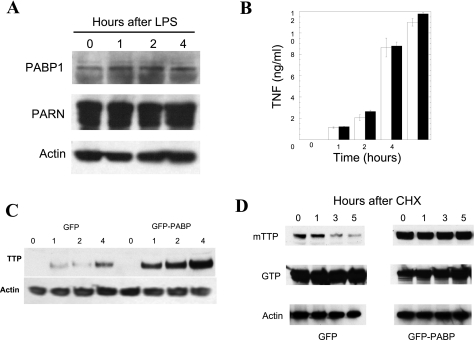
Although identification of the precise deadenylase(s) interacting with TTP in the RAW264.7 lysates was beyond the scope of these experiments, we did find that PARN is abundantly expressed in RAW264.7 cells by Western blotting and, like PABP1, does not significantly change concentration in RAW264.7 cells in response to LPS stimulation (Fig. 5A). To assess the role of PABP1 on macrophage TNF production, we stably transfected RAW264.7 cells with GFP-PABP1, a tagged PABP1 that has previously been validated in functional studies of PABP nucleocytoplasmic shuttling (1) and that has been used to show that PABP trafficked into the stress granule (23). GFP alone was used as a control. G418-selected nonclonal cells were used to avoid possible effects from site-specific integration that could confound analysis. More than 95% of the cells were fluorescent and remained attached after up to 24 h of LPS stimulation (not shown). After stimulation with LPS, TNF production was assayed by ELISA and no significant differences were observed (Fig. 5B).
Fig. 5.
PABP1 alters TTP kinetics in cells. PABP1 or PARN protein levels are not appreciably altered by LPS stimulation in RAW264.7 macrophage cells by Western blotting (A). The interaction between TTP and PABP1 was therefore explored in RAW264.7 cells that were stably transfected with either GFP-PABP or GFP. Pools of G418-selected cells were used to avoid site-integration and selection effects with prolonged propagation. E. coli LPS stimulation of these cells was performed from 0 to 6 h and the supernatants were assayed for TNF ELISA (B). A TTP Western blot of LPS-stimulated GFP- and GFP-PABP expressing cells, with equal protein loading verified by actin immunoblotting, is shown in C. To determine whether PABP expression affects TTP protein levels, the kinetics of TTP in the presence of PABP expression was explored in 293T cells cotransfected with myc epitope-tagged TTP (mTTP) and either GFP or GFP-PABP, with protein synthesis subsequently inhibited with cycloheximide (CHX) for 0 to 5 h to inhibit further protein synthesis (D). Loading was confirmed with an anti-actin antibody, TTP expression with the 9E10 monoclonal anti-myc antibody, and the transfection of GFP was verified by use of an anti-GFP antibody (shown at ∼32 kDa for GFP and ∼100 kDa for GFP-PABP). This experiment has been performed >3 times with equivalent results.
Given the reciprocal functions of PABP1 and TTP, one possibility to explain the lack of significant TNF production was that a counterregulatory increase in TTP expression occurs with PABP1 overexpression. To determine whether PABP1 led to reciprocal changes in TTP expression after LPS stimulation, the GFP and GFP-PABP1 RAW264.7 cell lysates were evaluated for TTP expression by Western blotting. The Western blot in Fig. 5C reveals that indeed there is an upregulation of TTP expression in response to increased PABP1 that is only observed with endotoxin treatment of these macrophages.
One possibility for the increased TTP expression levels in RAW264.7 cells is that PABP1 not only protects the poly(A) tail but also protects TTP from cellular degradation. Since PABP blocks the effect of TTP deadenylation and PABP binds to the poly(A) to protect an mRNA from degradation, the association of TTP with PABP on the same mRNA could protect the mRNA from degradation unless PABP was displaced from the poly(A) tail. Since our experiment in RAW264.7 cells involved LPS stimulation, which increases endogenous TTP production, we looked at TTP protein kinetics in a nonimmune cell line that allows isolated TTP overexpression, the 293T cell. 293T cells were transiently transfected with myc-TTP and either GFP-PABP1 or GFP, with protein synthesis inhibited by cycloheximide. Protein abundance was determined by Western blotting with antibodies to the mTTP myc epitope tag, GFP, and actin (loading control). Overexpression of PABP1 creates similar increases in TTP protein production, by decreasing the rate of TTP protein degradation (Fig. 5D).
p38 MAP kinase phosphorylation of PABP and TTP stabilization.
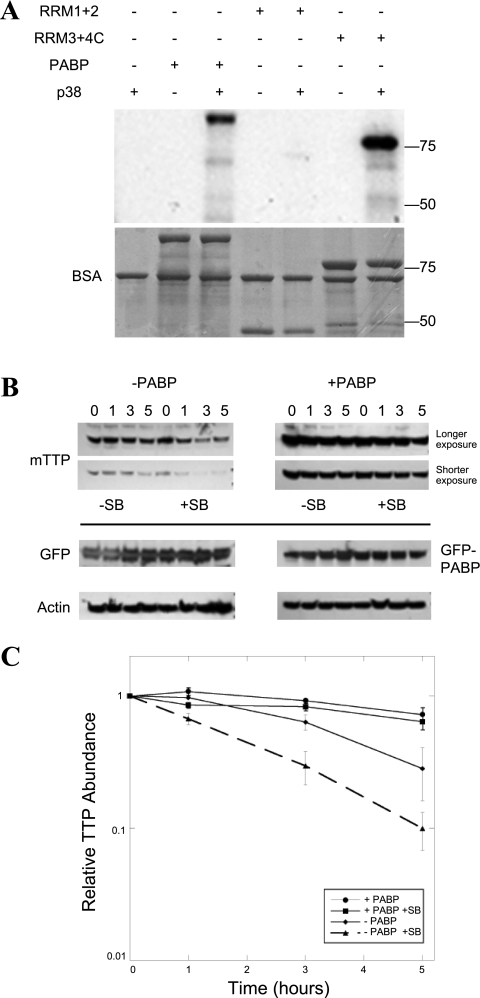
Our initial studies that identified PABP as a potential regulator of TTP function involved the p38 MAP kinase and the downstream MAPKAP kinase (MK2). Using mass spectrometry, in addition to phosphorylation by the MK2 and identification of coprecipitating PABP1 and PABP4, we observed direct phosphorylation of TTP by p38 (10). The p38 pathway prevents ARE-directed deadenylation (13), at least in part by promoting rapid TTP degradation by the proteasome (6). Although a previous report suggested that PABP was a direct target for MK2 (5), using recombinant PABP, a system for producing active MK2 in E. coli (10), and the p38-inhibitor SB 203580, we were unable to show phosphorylation of PABP that was not attributable to trace contaminating p38 activity. Since both PABP and p38 appear to exert an anti-TTP effect, we investigated whether PABP1 is a direct target for p38 MAP kinase. When this was directly addressed by using purified recombinant p38, we observed that p38 phosphorylates PABP, an effect that maps to the non-poly(A) portion of the molecule (Fig. 6A), the same region that can inhibit mRNA decay (Fig. 4C).
Fig. 6.
Loss of p38 MAP kinase activity destabilizes TTP protein and is inhibited by PABP coexpression, another p38 target. The p38 pathway prevents ARE-directed deadenylation (13), at least in part by promoting rapid TTP degradation by the proteasome (6). We previously mapped p38 phosphorylation sites on TTP (10) and predicted that p38 phosphorylation of PABP might also be possible, given primary sequence of the protein. A: recombinant p38 MAP kinase was used to phosphorylate full-length PABP and the truncated PABP fragments from Fig. 4A in an in vitro kinase assay, then separated on a protein gel that was both autoradiographed on a PhosphoImager to measure phosphorylation (top) and Coomassie stained (bottom). Kinase assays were performed at 30°C in 25-μl reactions containing (final concentrations) 25 mM HEPES, pH 7.4, 10 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 0.1 mg/ml substrate protein, 10 μg/ml p38 MAP kinase, 10 mM MgCl2, 50 μM [γ-32P]ATP (∼4,000 cpm/pmol). The effect of p38 inhibition on the stabilization of TTP by PABP coexpression was assayed as in Fig. 5E. The kinetics of TTP in the presence of PABP expression was explored in 293T cells cotransfected with myc epitope-tagged TTP (mTTP) and either GFP or GFP-PABP. Protein synthesis was inhibited with CHX for 0–5 h (B), except that parallel wells were also incubated with the p38 MAP kinase inhibitor, SB 203580 (SB). Western blotting was performed as above, with each Western for a given antibody developed on the same chemiluminescent film. Two exposures of mTTP are shown, so that the relative rates of mTTP elimination could be compared between images of comparable intensity at time 0. C: this experiment has been performed >3 times with equivalent results. The densitometry of the TTP films is expressed as means ± SE.
The identification of PABP as a p38 kinase target suggested that that phosphorylation might influence the kinetics of TTP disappearance. Activation of p38 inhibits deadenylation (13), with deadenylation accelerated in the presence of the p38 MAP kinase inhibitor SB 203580. The same experiment was performed in 293T cells, except that parallel wells were incubated with SB 203580 (Fig. 6B) vs. mock-treated cells. Two exposures of the same TTP Western allow relative comparison of the rate of disappearance of TTP: each row represents exposures from the same autoradiogram film. The densitometry of TTP abundance from three separate experiments normalized to time 0 are shown in Fig. 6C. In the absence of PABP overexpression, p38 inhibition accelerates the loss of TTP. In the presence of PABP, there is minimal change in TTP abundance.
DISCUSSION
Our studies of tristetraprolin add another function to the versatile PABP1, that of inhibiting TTP-mediated deadenylation in a riboprotein complex, thereby protecting the mRNA from decay. This is not a simple protein-protein interaction but requires the presence of mRNA, since we were unable to establish a TTP/PABP complex by pull-down assays with recombinant purified protein. A supershifted complex is captured by RNA EMSA using both full-length PABP1 and the fragment of PABP1 that lacks RRM1+2, the poly(A) binding domains suggesting that this interaction is weak. These same proteins are capable of inhibiting TTP-promoted deadenylation of a TNF 3′-reporter mRNA in RAW264.7 cell extracts, a complex mixture of cytoplasmic proteins.
In support of our findings, Lykke-Andersen and Wagner (33) have shown immunoprecipitation of TTP with an anti-PABP antibody, suggesting that they are bound to the same cellular mRNA. Although their focus was not on possible TTP/PABP functional interactions, their findings are compatible with ours. Like their findings, our experiments did not include RNase pretreatment of samples since that was not the original experimental design, but cellular RNases were not inactivated. We were not able to immunoprecipitate recombinant TTP with PABP (or vice versa) using the short mRNAs used for gel shifts. Since the two proteins functionally interact in either RNA EMSAs, where free diffusion is limited by the gel matrix, or in RNA degradation assays, in cytoplasmic lysates, we believe that the interaction is weak and likely facilitated by other factors. Since both proteins are phosphoproteins, phosphorylation may play a role in the regulation of this process. An intriguing possibility is that PABP4 (iPABP), an inducible form of cytoplasmic PABP that is known to be upregulated fivefold in activated T cells, might function similarly to PABP1, since it shares similar structure and functional properties (38, 45, 54).
PABP1 has poly(A), ARE (45), and 5′-cap binding properties (25) and is part of a complex of proteins binding to the GM-CSF ARE (5), one of the labile mRNAs that are stabilized in the TTP knockout mouse (8). We have also previously described a weak poly(A) binding activity in TTP by TNF RNA EMSA (53). We find that, like TTP (10, 34), PABP1 is a target for p38 MAP kinase phosphorylation, but not MK2, since we are unable to show MK2 phosphorylation not attributable to contaminating p38 activity. Since we have previously mapped p38 phosphosites on TTP (10) and since p38 activation inhibits deadenylation, our finding that p38 inhibition by SB 203580 increases TTP degradation is consistent with this kinase as a major GM-CSF and TNF mRNA regulatory factor that promotes poly(A) tail integrity and therefore mRNA stability (13). Since we also find that PABP1 is a p38 MAP kinase target on the fragment that both interacts with TTP and inhibits deadenylation, we speculate that our present efforts to map and mutate the PABP1 and TTP p38 phosphosites will provide additional information regarding these factors in regulating TNF and GM-CSF gene expression. PABP1 overexpression has a stabilizing effect on TTP degradation as well as a TNF 3′-UTR reporter, and the interaction of PABP1 and TTP is dependent on TTP RNA binding.
TTP-bound mRNA can be targeted to translating polysomes (41), a stress granule in which the mRNA remains untranslated but undegraded (3), or a processing body, for ultimate mRNA degradation and gene silencing (18). Localization of TTP to stress granules requires its RNA binding activity (37). TTP is a component of all three subcellular bodies, but only exerts a deadenylating influence at the processing body. PABP1 has only been described in polysomes, where it promotes translation, or in stress granules, which are translationally silent. PABP1 has been shown to shuttle in and out of stress granules by use of a GFP fusion protein similar to those we used for these studies (23). That PABP could block TTP-promoted deadenylation in polysomes or stress granules is reasonable on the basis of our data, but the specifics of this process and the role of p38 phosphorylation of PABP will require additional experiments. p42/ERK kinase is also required for TNF mRNA stabilization (14), which our studies do not address. There is a known ERK phosphosite on TTP at serine 220 (48). Our studies do show that a PABP1-TTP complex at the TNF ARE limits the ability of TTP to promote degradation of the TNF poly(A) tail, but this alone is not able to increase TNF production in the intact cell, suggesting additional levels of regulation. Inhibition of deadenylation does not require the poly(A) binding portions of PABP1 found in RRM1 and RRM2, similar to previous PABP tethering experiments (11). We found that the ability of PABP1 to bind to a TNF ARE probe was weak compared with that for TTP (Fig. 4B).
Our results are consistent with the model of Deleault et al. (14) for MAP kinase regulation of TNF expression by TTP. They studied factors that stabilize TNF after LPS activation in macrophages and found that ERK and p38 MAP kinases are both required to promote maximal TTP inhibition and subsequent TNF mRNA stabilization. Both TTP degradation and TNF mRNA decay were prevented by inhibition of the proteosome, suggesting that the two processes are closely linked. Although the focus of that investigation was stabilization of the burst of TNF mRNA produced in response to LPS activation, part of their model for its regulation by TTP in macrophages is the drop in p38 activity that occurs ∼90 min after activation and is associated with both TTP protein degradation and TNF mRNA decay (14). This time point corresponds to the downregulation of TNF expression (42), with the loss of p38 activity signaling the return to a quiescent macrophage (14). Our findings that PABP prevents TTP degradation, prevents TNF mRNA decay, and mitigates the loss of p38 activity by inhibiting TTP degradation suggest that an important function of PABP is to prevent entry of both TNF mRNA and its associated TTP into terminal decay pathways. This may have implications for the treatment of inflammatory diseases such as rheumatoid arthritis and Crohn's disease, in which the pathology is mediated by aberrant TNF production (4) and in which unmasking the degradative effects of TTP could be beneficial.
GRANTS
This work was made possible by National Institutes of Health (NIH) Grant DK-060720 and a Crohn's and Colitis Foundation Senior Research Award to M. T. Worthington, NIH Grant GM-62890 to T. W. Sturgill, and U.S. Army Medical Research and Materiel Command Grant under DAMD17-03-1-0555 to C. A. Chrestensen.
Acknowledgments
The authors thank Dr. Michael Wormington for helpful discussions; Drs. Wormington, Christophe Moroni, and Jiahuai Han for supplying reagents; and Dr. Antony Rosen for the use of laboratory resources. Special thanks to Joani Christensen for assistance in preparing figures.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem 273: 13015–13021, 1998. [DOI] [PubMed] [Google Scholar]
- 2.Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry 42: 217–221, 2003. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7: 213–221, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutler B, Bazzoni F. TNF, apoptosis and autoimmunity-a common thread. Blood Cells Mol Dis 24: 216–230, 1998. [DOI] [PubMed] [Google Scholar]
- 5.Bollig F, Winzen R, Gaestel M, Kostka S, Resch K, Holtmann H. Affinity purification of ARE-binding proteins identifies poly(A)-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem Biophys Res Commun 301: 665–670, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol 26: 2408–2418, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo E, Gilkeson GS, Blackshear PJ. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (-/-) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J Clin Invest 100: 986–995, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899, 2000. [PubMed] [Google Scholar]
- 9.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281: 1001–1005, 1998. [DOI] [PubMed] [Google Scholar]
- 10.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem 279: 10176–10184, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev 12: 3226–3235, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford EK, Ensor JE, Kalvakolanu I, Hasday JD. The role of 3′ poly(A) tail metabolism in tumor necrosis factor-alpha regulation. J Biol Chem 272: 21120–21127, 1997. [DOI] [PubMed] [Google Scholar]
- 13.Dean JL, Sarsfield SJ, Tsounakou E, Saklatvala J. p38 Mitogen-activated protein kinase stabilizes mRNAs that contain cyclooxygenase-2 and tumor necrosis factor AU-rich elements by inhibiting deadenylation. J Biol Chem 278: 39470–39476, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Deleault KM, Skinner SJ, Brooks SA. Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45: 13–24, 2008. [DOI] [PubMed] [Google Scholar]
- 15.DuBois RN, Bishop PR, Graves-Deal R, Coffey RJ. Transforming growth factor alpha regulation of two zinc finger-containing immediate early response genes in intestine. Cell Growth Differ 6: 523–529, 1995. [PubMed] [Google Scholar]
- 16.DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem 265: 19185–19191, 1990. [PubMed] [Google Scholar]
- 17.Ehrenfried JA, Townsend CM Jr, Thompson JC, Evers BM. Increases in nup475 and c-jun are early molecular events that precede the adaptive hyperplastic response after small bowel resection. Ann Surg 222: 51–56, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21: 719–735, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med 171: 465–475, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Johnson BA, Geha M, Blackwell TK. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene 19: 1657–1664, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Khanna R, Kiledjian M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J 23: 1968–1976, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol 1: 94–97, 1999. [DOI] [PubMed] [Google Scholar]
- 27.Kusov YY, Shatirishvili G, Dzagurov G, Gauss-Muller V. A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res 29: E57, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24: 1779–1790, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A). Mol Cell Biol 14: 7984–7995, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU- rich element-containing mRNAs. J Biol Chem 277: 9606–9613, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23: 3798–3812, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem 279: 12974–12979, 2004. [DOI] [PubMed] [Google Scholar]
- 33.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol 21: 6461–6469, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem 275: 32268–32276, 2000. [DOI] [PubMed] [Google Scholar]
- 36.Milone J, Wilusz J, Bellofatto V. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10: 448–457, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murata T, Morita N, Hikita K, Kiuchi K, Kiuchi K, Kaneda N. Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain. Exp Cell Res 303: 287–299, 2005. [DOI] [PubMed] [Google Scholar]
- 38.Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10: 151–163, 2005. [DOI] [PubMed] [Google Scholar]
- 39.Penalva LO, Burdick MD, Lin SM, Sutterluety H, Keene JD. RNA-binding proteins to assess gene expression states of co-cultivated cells in response to tumor cells. Mol Cancer 3: 24, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren YG, Martinez J, Kirsebom LA, Virtanen A. Inhibition of Klenow DNA polymerase and poly(A)-specific ribonuclease by aminoglycosides. RNA 8: 1393–1400, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigby WF, Roy K, Collins J, Rigby S, Connolly JE, Bloch DB, Brooks SA. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J Immunol 174: 7883–7893, 2005. [DOI] [PubMed] [Google Scholar]
- 42.Rowlett RM, Chrestensen CA, Nyce M, Harp MG, Pelo JW, Cominelli F, Ernst PB, Pizarro TT, Sturgill TW, Worthington MT. MNK kinases regulate multiple TLR pathways and innate proinflammatory cytokines in macrophages. Am J Physiol Gastrointest Liver Physiol 294: G452–G459, 2008. [DOI] [PubMed] [Google Scholar]
- 43.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 7: 3268–3276, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salles FJ, Darrow AL, O'Connell ML, Strickland S. Isolation of novel murine maternal mRNAs regulated by cytoplasmic polyadenylation. Genes Dev 6: 1202–1212, 1992. [DOI] [PubMed] [Google Scholar]
- 45.Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem 271: 450–457, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Stoecklin G, Ming XF, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol Cell Biol 20: 3753–3763, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tallada VA, Daga RR, Palomeque C, Garzon A, Jimenez J. Genome-wide search of Schizosaccharomyces pombe genes causing overexpression-mediated cell cycle defects. Yeast 19: 1139–1151, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem 270: 13341–13347, 1995. [DOI] [PubMed] [Google Scholar]
- 49.Wakiyama M, Honkura N, Miura KI. Interference with interaction between eukaryotic translation initiation factor 4G and poly(A)-binding protein in Xenopus oocytes leads to inhibition of polyadenylated mRNA translation and oocyte maturation. J Biochem (Tokyo) 130: 737–740, 2001. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J 15: 900–909, 1996. [PMC free article] [PubMed] [Google Scholar]
- 52.Worthington MT, Amann BT, Nathans D, Berg JM. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc Natl Acad Sci USA 93: 13754–13759, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem 277: 48558–48564, 2002. [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Duckett CS, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol 15: 6770–6776, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yonan CR, Duong PT, Chang FN. High-efficiency staining of proteins on different blot membranes. Anal Biochem 338: 159–161, 2005. [DOI] [PubMed] [Google Scholar]
- 56.Zhu W, Brauchle MA, Di Padova F, Gram H, New L, Ono K, Downey JS, Han J. Gene suppression by tristetraprolin and release by the p38 pathway. Am J Physiol Lung Cell Mol Physiol 281: L499–L508, 2001. [DOI] [PubMed] [Google Scholar]
- 57.Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol 15: 2219–2230, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]