Abstract
The torque-velocity relationship is known to be affected by ageing, decreasing its protective role in the prevention of falls. Interindividual variability in this torque-velocity relationship is partly determined by genetic factors (h2: 44–67%). As a first attempt, this genome-wide linkage study aimed to identify chromosomal regions linked to the torque-velocity relationship of the knee flexors and extensors. A selection of 283 informative male siblings (17–36 yr), belonging to 105 families, was used to conduct a genome-wide SNP-based (Illumina Linkage IVb panel) multipoint linkage analysis for the torque-velocity relationship of the knee flexors and extensors. The strongest evidence for linkage was found at 15q23 for the torque-velocity slope of the knee extensors (TVSE). Other interesting linkage regions with LOD scores >2 were found at 7p12.3 [logarithm of the odds ratio (LOD) = 2.03, P = 0.0011] for the torque-velocity ratio of the knee flexors (TVRF), at 2q14.3 (LOD = 2.25, P = 0.0006) for TVSE, and at 4p14 and 18q23 for the torque-velocity ratio of the knee extensors TVRE (LOD = 2.23 and 2.08; P = 0.0007 and 0.001, respectively). We conclude that many small contributing genes are involved in causing variation in the torque-velocity relationship of the knee flexor and extensor muscles. Several earlier reported candidate genes for muscle strength and muscle mass and new candidates are harbored within or in close vicinity of the linkage regions reported in the present study.
Keywords: human muscle strength, torque-velocity relationship, linkage, whole genome
the force generated by a muscle not only depends on the length of the muscle, and thus the sarcomere length or contractile filaments overlap, but it also varies with the velocity at which it shortens. With increasing shortening velocity, the force sustained by the muscle rapidly decreases, finally leading to a velocity at which force can no longer be sustained; this is the maximum velocity of shortening (Vmax). Force at zero velocity of shortening is the isometric force (F0). It is possible to compare muscles of different sizes by expressing the force at a particular velocity as a fraction of F0. The speed of shortening of a contracting muscle is dependent on the number of sarcomeres in series.
The torque-velocity relationship is known to be affected by ageing (6, 34, 44), and concentric torques have been found to be depressed in older individuals (15, 24). The deficit in concentric peak torque between young and elderly individuals has also been found to increase with contraction velocity (6). Explosive muscle power (the product of force and the speed at which force is produced during the first seconds of a movement) is important in correcting a displacement or movement error. To prevent a fall, an individual must have sufficient lower limb muscle power in the stabilizing leg to counteract the kinetic energy of the imbalance (31). The discovery of genomic factors (e.g., DNA sequence variants) causing individual variation in the torque-velocity relationship can elucidate the physiological pathways involved in this trait. Furthermore, final discovery of velocity-dependent gene variants might ultimately be used for screening individuals at risk for low muscle power and be used in individualized exercise prescription.
Several studies suggest that isometric (Fisom) and concentric (Fconc) muscle strength are under moderate to high genetic control with heritabilities ranging between 44 and 78% for Fisom and 31 and 61% for Fconc (23, 35–37, 39, 41, 42). However, muscle strength is a complex multifactorial trait, and high heritabilities do not guarantee the presence of quantitative trait loci (QTLs) with large effect size. Heritability estimates for variation in eccentric and concentric arm flexion torques at different velocities, indicative for torque-velocity specificity, have been reported by Thomis et al. (39). The contribution of genetic factors was larger in eccentric torques at different velocities (66–78% at 110° and 62–82% at 140°) than in concentric torques at different velocities (29–51% at 110° and 52–65% at 140°). The importance of genetic factors gradually decreased with increasing velocity contraction in concentric torques (from 65% at 30°/s to 53% at 120°/s in an elbow angle of 140°).
The “Human gene map for performance and health-related fitness phenotypes” (26) indicates that only a few studies have reported significant association of allelic variants in putative QTLs with muscle strength characteristics, and even fewer associations have consistently been replicated. To our knowledge, only a limited number of studies has been performed to investigate association of candidate genes with the torque-velocity relationship. Vincent et al. (45) demonstrated a genotype-by-velocity interaction with a greater decrease of torque at higher velocities for the ACTN3 XX compared with the RR genotype group.
The linkage studies of Huygens et al. (16, 17) were the first to explore the role of candidate genes in the myostatin pathway as QTLs for knee or trunk muscle strength and estimated muscle cross-sectional area in 329 young male Caucasian siblings from the Leuven Genes for Muscular Strength (LGfMS) study. Results from these single-point linkage explorations with a single microsatellite marker per candidate gene revealed that the chromosomal regions harboring myostatin (GDF8, 2q32.2), p21 (CDKN1A, 6p21.2), MyoD (MYOD1, 11p15.1), and retinoblastoma (RB1, 13q14.2) are potentially interesting regions for further genetic studies [logarithm of the odds ratio (LOD) scores between 1.50 and 2.78, P values between 0.05 and 0.0002]. Strengthened by these preliminary single-point linkage results and the physiological evidence for the entire myostatin pathway, a multipoint linkage analysis was performed on a larger and partially independent sample (18) for these genetic regions, in addition to the regions harboring the muscle regulatory factors Myf5 (MYF5, 12q21.31) and Myf6 (MYF6, 12q21.31), insulin-like growth factor-1 (IGF1, 12q22q23), cyclin-dependent kinase-2 (CDK2, 12q13), and titin (TTN, 2q31.2) (18). Significant or suggestive linkage with knee muscle strength was found for the regions comprising CDK2 (LOD 3.4, P = 0.0004), RB1 (LOD 2.74, P = 0.0002), and IGF1 (LOD 2.6, P = 0.0002).
The first genome-wide multipoint linkage analysis on different muscle strength characteristics revealed one region on chromosome 14 with significant evidence for linkage with torque-length relationship of the knee flexors (14q24.3) and several other regions with suggestive evidence for linkage (muscle and bone cross-sectional area: 14q32.2; isometric knee peak torque at 30° flexion: 2p24.2; torque-length relationship of the knee extensors: 1q21.3, 2p23.3, and 18q11.2; muscle-cross sectional area adjusted knee extension torque: 18p11.31) (12).
In this paper we report the results of a genome-wide linkage scan using 6,008 single nucleotide polymorphism (SNP) markers, aimed at identifying genomic regions harboring candidate genes that cause variation in the torque-velocity relationship of the knee flexor and extensor muscles.
METHODS
Subjects
A selection of 283 male siblings aged 17–36 yr in 105 families from the total sample of the LGfMS study was made and was composed of 13 quads, 47 trios, and 45 pairs of brothers, resulting in 309 pairwise comparisons. As the absence of parental genotypes reduces the accuracy of identity by descent (IBD) estimation within families and thus the power to detect linkage, emphasis was placed on the inclusion of trios and quads, as well as the selection of the most informative sib pairs based on their level of discordance for different strength phenotypes as Risch and Zhang (29) showed that extreme discordant sib pairs provide substantial power to detect linkage for a QTL. The recruitment procedures and subject characteristics have been described in detail previously (16, 18). Prior to participation, the purpose and procedures of the study were explained in detail, and the subjects gave their written informed consent. The procedures used in this study were approved by the medical and ethical committee of the Katholieke Universiteit Leuven.
Measurements
A detailed overview of the anthropometrical and muscle strength measurements in the LGfMS project can be found in Huygens et al. (16, 18).
Muscle strength.
The Cybex NORM isokinetic dynamometer (Lumex, Ronkonkoma, NY) was used to assess the maximal isometric and concentric knee strength characteristics. After a 5–10 min warm-up period on an ergometer cycle and light stretching exercises, subjects were positioned on the dynamometer, following the instructions of the manufacturer. Anatomical zero was set at full extension of the knee, and the rotation axis of the joint was aligned with the mechanical axis of the dynamometer. Two isometric and four concentric submaximal trials preceded the actual tests for familiarization with the testing procedure (angle, range of motion, or velocity). Maximal isometric knee strength was measured at two angles (30° and 60°). At each angle, highest torque values (Nm) during a 6 s isometric contraction of three maximal flexion and extension contractions were retained for further analysis. Thirty seconds of rest between each contraction were given. Peak torque over the complete range of motion (0°-90°) of concentric knee extension and flexion was measured at 60°/s (3 repetitions), at 120°/s (25 repetitions), and at 240°/s (5 repetitions). During these contractions, torque at specific angles was also recorded: following the force-length relationship of a muscle, optimal strength is generated at longer muscle length, i.e., at an angle of 60° for knee extension (quadriceps) and 30° for knee flexion (hamstrings). Subjects were verbally encouraged to perform at their maximum effort, and visual feedback of their performance was presented after each test.
Torque-velocity calculations.
Dynamic torques at different velocities (0°/s, 60°/s, 120°/s, and 240°/s) for the optimal angles were first expressed as a percentage of maximal isometric torque (relative torque per velocity). A natural logarithmic transformation (Ln) of velocity was performed to linearize the torque-velocity curve with the equation y = a + bx [whereby a = intercept, b = slope, x = Ln(velocity); R2 between 0.96 and 0.99]. The torque-velocity slope of each subjects linear torque-velocity curve for knee flexors (TVSF) and extensors (TVSE) was calculated and retained for further analyses. A second measure of torque-velocity was the ratio of torque at 240°/s and torque at 60°/s, multiplied by 100. This torque-velocity ratio was calculated for knee flexors (TVRF) and extensors (TVRE).
Baecke Sport Index.
The Baecke physical activity questionnaire (5) was used to assess the level of daily physical activity and more specifically the subjects' sports participation in the Sport Index.
DNA Collection
DNA was extracted using the Chemagic DNA Blood Kit on an automated Chemagic Magnetic Separation Module I (Chemagen, Baesweiler, Germany) and a Multiprobe I (PerkinElmer, Waltham, MA) robotic station. An Oragene saliva kit (Genotek, Ottawa, Ontario, Canada) was provided to siblings who were not able to deliver a blood sample. DNA from these saliva kits was extracted following the guidelines of the manufacturer.
Genotyping
The Illumina SNP-based Linkage Panel IVb was selected for genotyping. The panel includes 6,008 biallelic SNP markers distributed evenly across the genome. The average and median intervals between markers are 482 kb (0.64 cM) and 298 kb (0.35 cM), respectively. The largest interval between successfully genotyped markers is 5.02 cM on chromosome 8, and linkage disequilibrium between markers is minimal. The panel is designed to reach an average minor allele frequency and heterozygosity of SNP markers of 37 and 44%, respectively (Illumina technical report). The Illumina markers were typed with the Illumina Beadstation 500GX, in accordance with the manufacturer's standard recommendations. The genotype success rate was 99.6% (33 SNPs were excluded from analysis due to low signal or cluster overlap). Only autosomal SNPs were included for further analyses.
Statistical Analysis
Descriptive statistics.
Descriptive statistics and phenotypic correlations between the different strength characteristics and covariates were calculated using SAS version 9.1 (SAS Institute, Cary, NC). To explore the contribution of velocity-dependent torque declines on the measure of the torque-velocity slope, linear stepwise regression analyses were performed. The declines in torque from 0°/s to 60°/s and 60°/s to 240°/s were expressed as ratios, and the proportion of variance explained by each of these torque-velocity curve ratios was then estimated through regression analyses.
Heritability estimations.
Upper-limit heritability of the traits (h2) were estimated using the variance-components analysis procedure (VC) in quantitative transmission disequilibrium test (QTDT) software (2). This estimate includes common environmental variation in addition to the additive genetic component, because these factors cannot be separated using sib pairs only. Hence, it is called the upper-limit heritability.
Linkage analysis.
Nonparametric multipoint linkage analyses were performed on 22 autosomes using the revised Haseman-Elston regression algorithm (MERLIN Regress) outlined by Sham et al. (30), implemented in MERLIN software v. 1.1.1 (3). The revised H-E algorithm combines the simplicity and robustness of regression-based methods and the generality and power of variance components methods (30). This method requires specification of trait heritability, population means, and variances, which were calculated using the Pedstats (46) and QTDT programs (2). Average minor allele frequencies and heterozygosities of markers in the present linkage panel within our sample were calculated using MERLIN (3). Before statistical analyses, all genotypes were checked for Mendelian inheritance, using the error detection protocol in MERLIN (3). Deviations from Hardy-Weinberg equilibrium were assessed using the PEDSTATS program (46). Polymorphisms with Hardy-Weinberg P values < 0.001 were removed from further analyses.
Under the assumption of no susceptibility loci, simulated data for 100 genome scans were generated using MERLIN (3), to estimate the significant and suggestive threshold for linkage (10, 21, 43). In each simulation, we retained the original pedigree structure and generated a new data set with the same allele frequencies, marker spacing, phenotypes, and any missing genotypes or data pattern. Any evidence for linkage in these simulated data is due to chance. The cut-off for suggestive linkage (LOD = 2.37) was calculated as the mean of the genome-wide maximum LOD score from each genome scan, which determines the maximum peak size expected once per genome scan by chance alone. The significant linkage threshold (LOD = 3.64) was defined as the maximum LOD score occurring with probability 0.05 in a genome scan (i.e., 5 peaks of equal or greater size observed in the 100 simulations).
It is nowadays a widely accepted practice to obtain empirical P values for reported LOD scores (1, 4, 32). We calculated empirical P values through the use of 1,000; 10,000; or 100,000 gene-dropping simulations (related to the original P values of SNPs with LOD >1.5, see Table 3 under P value) using MERLIN (3), under the assumption of no susceptibility loci. The empirical significance level of an LOD peak was then determined by counting the proportion of simulated (unlinked) LOD scores higher or equal to the original LOD score.
Table 3.
LOD scores >1.5 and P values of multipoint linkage analysis for torque-velocity relationship of the knee flexors and extensors
| Trait | SNP | Chromosomal Region | cM | LOD | P Value | Empirical P Value | P Value Likelihood-Ratio Test |
|---|---|---|---|---|---|---|---|
| Slope flexion | rs717651 | 13q12.3 | 26.87 | 1.70 | 0.003 | 0.002 | 0.95 |
| Ratio flexion | rs1403970 | 2q36.3 | 231.87 | 1.55 | 0.004 | 0.004 | 0.36 |
| rs1384542 | 3q25.32 | 164.47 | 1.53 | 0.004 | 0.002 | 0.92 | |
| rs2998367 | 6q14.1 | 88.25 | 1.93 | 0.0014 | 0.0007 | 0.18 | |
| rs921630 | 7p12.3 | 70.13 | 2.03 | 0.0011 | 0.0006 | 0.32 | |
| Slope extension | rs477449 | 2q14.3 | 139.88 | 2.25 | 0.0006 | <0.0001 | 0.96 |
| rs4751909 | 10q25.3 | 133.38 | 1.85 | 0.002 | <0.001 | 0.095 | |
| rs918044 | 12q24.32 | 149.17 | 1.85 | 0.002 | 0.001 | 0.149 | |
| rs768826 | 13q21.33 | 64.23 | 1.82 | 0.002 | <0.001 | 0.73 | |
| rs1348318 | 15q23 | 76.79 | 2.91 | 0.00013 | <0.00001 | 0.62 | |
| Ratio extension | rs938326 | 2p25.3 | 3.58 | 1.85 | 0.002 | 0.001 | 0.033 |
| rs1039559 | 4p14 | 58.04 | 2.23 | 0.0007 | 0.0001 | 0.70 | |
| rs1866338 | 18q23 | 118.79 | 2.08 | 0.001 | <0.001 | 0.73 |
LOD, logarithm of the odds ratio; SNP, single nucleotide polymorphism.
Combined linkage and association analysis.
Tests for joint linkage and association in regions with suggestive and significant evidence for linkage were carried out using QTDT software (2). SNPs in a −1 LOD support region around SNPs showing suggestive or significant evidence for linkage in the MERLIN-regress linkage analysis were investigated. Fulker et al. (14) developed a method for simultaneous modeling of association and linkage for quantitative traits using sib pair data that also controls for population stratification. Combined linkage and association analysis is a powerful tool for pinpointing functional associated loci responsible for a linkage signal. Assuming there is suggestive/significant linkage before modeling association, the extent to which evidence for linkage diminishes in the joint test of linkage and association reflects the proximity of the marker to the functional QTL (9, 14). If the linkage signal of a certain locus is entirely explained by modeling association (i.e., the linkage signal is no longer significant), that particular locus is either the functional locus or in tight linkage disequilibrium (LD) with the functional locus.
Association analysis.
SNPs in a −1 LOD support region around the SNP showing significant evidence for linkage were tested for association with the MERLIN likelihood-ratio test for the different strength phenotypes (3).
RESULTS
Descriptive Statistics
Average minor allele frequencies and heterozygosities for the autosomes within this sample were 50 and 45%, respectively. The percentage of SNPs with minor allele frequency <5% was 0.65%.
Table 1 shows the main somatic and muscle strength statistics of 283 siblings, with corresponding upper-limit heritabilities (h2) estimated for both the total LGfMS sample (n = 748) and the subsample (n = 283). Subjects had normal weight and height; the body mass index (BMI: 22.9 ± 2.9 kg/m2) indicates that they were, on average, rather lean.
Table 1.
Somatic characteristics and muscle strength statistics
| Trait | Mean ± SD | h2, %* | h2, %† |
|---|---|---|---|
| Age | 25.1±4.5 | ||
| Weight, kg | 74.5±10.4 | 85 | 89 |
| Stature, cm | 180.2±6.5 | 92 | 99 |
| Fat-free mass, kg | 63.4±6.5 | 90 | 94 |
| Body mass index, kg/m2 | 22.9±2.9 | 81 | 87 |
| MBA midthigh, cm2 | 171.8±22.7 | 85 | 89 |
| Slope flexion, % | −23.1±5.0 | 71 | 44 |
| Slope extension, % | −27.9±3.6 | 44 | 55 |
| Ratio flexion, % | 62.0±11.1 | 68 | 67 |
| Ratio extension, % | 54.8±8.3 | 59 | 52 |
| Sport Index | 3.0±0.7 | 50 | 63 |
h2 estimated on the total Leuven Genes for Muscular Strength (GfMS) sample (n = 748);
h2 estimated on the subsample (n = 283); ratios were calculated by dividing peak torque at 240°/s by peak torque at 60°/s, multiplied by 100. Slopes were estimated using peak torque at 0°/s, 60°/s, 120°/s, and 240°/s (see materials and methods). Flexion and extension torques were measured at a knee angle of 30° and 60°, respectively; MBA, muscle and bone area.
As expected, the upper-limit heritability estimate of stature was high (99%), and genetic determination of body mass and BMI was somewhat lower (89 and 87%, respectively). Fat-free mass and muscle and bone cross-sectional area (MBA) of the midthigh showed h2 estimates of 94 and 89%, respectively. In this sample, moderate to high upper-limit heritabilities were found for the torque-velocity slopes and ratios of the knee flexors and extensors (44–67%). Variability in the Sport Index could be accounted for by genetic factors for at most 63%.
Correlations between the different torque-velocity phenotypes ranged from 0.20 to 0.73 (Table 2). Moderate to high correlations were found between TVSF and TVRF (0.73), and between TVSE and TVRE (0.62). Low to moderate cross-trait/-estimate correlations were found (r between 0.20 and 0.46), indicating the specificity of both torque-velocity estimates and the torque-velocity characteristics of knee extensor muscles and flexor muscles. Correlations between Sport Index and torque-velocity phenotypes were not significant.
Table 2.
Pearson correlations (P value) between torque-velocity characteristics and scores on the Baecke Sport Index
| Sport Index | Slope Flexion | Slope Extension | Ratio Flexion | |
|---|---|---|---|---|
| Slope flexion | 0.07 (0.24) | |||
| Slope extension | −0.05 (0.40) | 0.32 (<0.0001) | ||
| Ratio flexion | 0.07 (0.25) | 0.73 (<0.0001) | 0.20 (0.0013) | |
| Ratio extension | −0.05 (0.40) | 0.40 (<0.0001) | 0.62 (<0.0001) | 0.46 (<0.0001) |
Correlations are calculated on the total GfMS sample (n = 748).
Regression analyses demonstrated that the ratios for 0°/s to 60°/s and 60°/s to 240°/s for the knee flexors and extensors account for 41 and 55%, and 4 and 44%, respectively, of the total variance in the torque velocity slope from 0°/s to 240°/s.
Linkage Analyses
Figure 1 shows LOD score curves over the autosomal genome for each of the phenotypes. Empirical P values and LOD scores for regions with LOD of 1.5 and higher are shown in Table 3. The highest multipoint LOD score (2.91) was found on chromosome 15q23 for TVSE, with an empirical P value < 10−5. Other interesting linkage regions with LOD scores >2 were found for TVRF (LOD = 2.03 on 7p12.3), TVSE (LOD = 2.25 on 2q14.3), and TVRE (LOD = 2.23 and 2.08 on 4p14 and 18q23, respectively).
Fig. 1.
Logarithm of the odds ratio (LOD) scores for autosomal linkage analysis for ratio and slope characteristics of torque-velocity for the knee flexors and extensors. Cut-off LOD scores for suggestive and significant linkage are 2.37 and 3.64, respectively. TVRF, torque-velocity ratio for knee flexors; TVSF, torque-velocity slope for knee flexors; TVRE, torque-velocity ratio for knee extensors; TVSE, torque-velocity slope for knee extensors.
Combined Linkage and Association Analyses
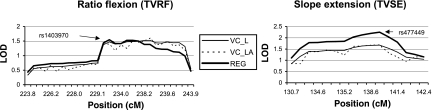
Figure 2 shows two examples of LOD score curves for the effects of combined linkage and association analysis for SNPs in a LOD −1 support region around SNP rs1403970 (2q36.3) for TVRF, and around SNP rs477449 (2q14.3) for TVSE. No significant drop in LOD scores was observed after modeling a joint test for linkage and association, indicative for rather limited effects of allelic association between the investigated SNPs and the studied traits. Similar results were found for all other SNPs in regions with evidence for linkage. However, when applying the MERLIN likelihood-ratio association test, we found significant associations (P < 0.01) (Table 4) between SNPs within these LOD −1 support regions and several phenotypes. Association tests for the SNPs with peak LOD scores are included in Table 3; however, these were not significant. Classical AN(C)OVA-type association analysis confirmed the above MERLIN likelihood-ratio association findings, except for SNPs rs1479371 at chr3 (TVRF) and SNP rs1321834 at chr6 (TVSF) (Table 4). Peak LOD scores for the associated SNPs in Table 4 are also included.
Fig. 2.
Representation of combined linkage and association analysis for ratio flexion and slope extension at LOD −1 support region around single nucleotide polymorphisms rs1403970 and rs477449, respectively. LOD scores for variance components analyses in QTDT: Linkage (VC_L) and combined linkage and association (VC_LA); LOD scores for Merlin-regress analyses (REG).
Table 4.
Association results for SNPs with P < 0.01
| SNP | Chromosomal Region | Genetic Location, cM | Torque-Velocity Phenotype | LOD |
Likelihood-Ratio Test |
Analysis of Variance
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Allele Effect Size | P Value | Carriers, n | Noncarriers, n | P Value | |||||
| rs1479371 | 3q12.3 | 113.38 | ratio flexion | 0.692 | G | −4.896 | 0.00031 | 61.7±0.9 (271) | 64.2±6.1 (10) | 0.701 |
| rs765695 | 3q24 | 151.04 | ratio extension | 0.127 | G | −2.339 | 0.0041 | 54.4±0.8 (234) | 58.2±1.6 (47) | 0.042 |
| rs1321834 | 6q13 | 86.07 | slope flexion | 0.379 | G | −1.712 | 0.0041 | −23.2±0.5 (270) | −21.2±1.4 (11) | 0.182 |
| rs1321834 | 6q13 | 86.07 | ratio extension | 1.011 | G | −3.380 | 0.00071 | 54.8±0.7 (270) | 63.5±2.4 (11) | 0.012 |
| rs1545963 | 13q21.31 | 59.33 | slope flexion | 0.008 | G | +1.337 | 0.0038 | −22.8±0.5 (241) | −25.6±0.7 (40) | 0.00022 |
Values are means ± SE.
G carriers vs. CC;
G carriers vs. AA; C carriers vs. AA; A carriers vs. CC. A carriers vs. GG. Genotype frequencies are in Hardy-Weinberg equilibrium (P > 0.01).
DISCUSSION
We performed a genome-wide multipoint linkage analysis to identify quantitative trait loci for the torque-velocity relationship of the knee flexors and extensors, including two measures: 1) the slope of the linearized torque-velocity relationship and 2) the ratio of torque at 240°/s and torque at 60°/s. The most promising evidence for linkage was observed for the torque-velocity slope of the knee extensors with SNP rs1348318 on chromosome 15q23. It is possible that the identified susceptibility locus is muscle specific (flexors vs. extensors) and does not account for variability in knee flexion. As muscle strength and its derived phenotypes are regarded as a complex genetic trait, identification of a large number of markers with LOD scores that meet the criteria of suggestive linkage should not be expected. Another interesting chromosomal region is region 7p12.3, which harbors a candidate gene (IGFBP-1) for TVRF, although this region was not reported in earlier linkage studies investigating strength-related phenotypes.
Our group previously performed a multipoint linkage analysis for muscle strength and muscle mass in a similar cohort of young, male siblings, using microsatellite markers in chromosomal regions harboring myostatin pathway genes (18). Region 12q24.32 was identified in the present study and is in the vicinity of the earlier reported region 12q22–23 (18). Compared with our previous analyses (18) with microsatellites markers (average marker distance 4.81 cM for chromosome 12), the current study used the Illumina predesigned SNP-based Linkage IVb panel with an average marker distance of 0.56 cM for chromosome 12. Although this SNP panel induces higher genetic informativeness than microsatellite markers (13, 47), differences between present (n = 283, Illumina SNP panel) and previous findings (n = 367, microsatellite markers; Applied Biosystems) (18) are probably related to sample size differences or region-specific informativeness of closely chosen microsatellite markers vs. SNP markers (mean heterozygosity of SNP markers was 44.2% on chr12). Of note, also several other overlapping/neighboring linkage regions could be detected between the present study and our genome-wide scan for maximum and length-dependent knee muscle strength (12): region 2p25.3 (TVRE) with 2p24.2 for isometric torque at 30° flexion; region 10q25.3 (TVSE) with 10q26.13 for torque-length flexion; region 2q14.3 (TVSE) with 2q14.2 for muscle-bone area of the midthigh; region 12q24.32 (TVSE) with 12q24.32 and 12q24.31 for torque-length flexion and extension, respectively; and 15q23 (TVSE) with 15q23 for isometric torque at 30° flexion. These regions might harbor candidate genes for underlying shared characteristics like muscle fiber type distribution that cause variability in both torque-length and torque-velocity characteristics.
Several multivariate genetic studies examined genetic and environmental contributions to individual differences in various muscle strength characteristics (11, 36, 37, 39, 41, 42). These studies suggest a shared pleiotropic gene action for the different phenotypic characteristics (the same genes causing variability in different types of contractions, speeds, angles), referring to a common underlying genetic cause of strength generality. This would imply that different examined phenotypes share the same chromosomal region with suggestive or significant evidence for linkage. However, no clear overlap for chromosomal regions between the different phenotypes was found within this study. This is somewhat surprising, given the correlations up to 0.73, which strongly suggest a shared genetic source of variance in muscle strength, although additional shared environmental sources of covariance might induce this correlation. Common linkage signals were also absent between highly intercorrelated VO2max and maximal power output (r = 0.94) in the HERITAGE Family study (27). An explanation might be the different information provided by each measure: the slope measure encompasses the torque-velocity curve in a range of 0°/s to 240°/s, whereas the ratio measure focuses on the faster part of the curve (60°/s to 240°/c). Therefore, the torque-velocity ratio does not pick up linkage signals for slower contraction velocities (0°/s to 60/°s), whereas the torque-velocity slope does.
The examined phenotypes related to the torque-velocity relationship are regulated by numerous genes. In addition, gene-gene and gene-environment interactions and contributions of environmental factors for muscle strength can be assumed to affect the power of linkage studies. The identification of susceptibility loci using a small sample size is therefore difficult. We calculated the successful estimation of IBD status for each pairwise comparison over all 133 SNP markers at chromosome 22 by counting a >0.95 probability level as a successful classification of IBD status (IBD0, IBD1, or IBD2). The information content for duo, trio, and quad families was 60, 78, and 88%, respectively, which is even higher than reported in Ref. 33. The power to detect regions harboring susceptibility loci was estimated using the Genetic Power Calculator (25) based on the heritabilities estimated in QTDT [assumptions: θ = 0; additive QTL variance = 30%; common residual shared variance = 40% (= genetic variance not attributable to the QTL and/or common environmental variance); no dominance; residual nonshared variance = 30%; α level = 0.05]. A weighted noncentrality parameter was derived based on the proportion of quads, trios, and sib pairs. With the current sample, it is estimated that a power of 70% can be reached for the detection of a QTL accounting for 30% of the phenotypic variance, 30% residual shared variance, no dominance, and residual nonshared variance of 40%. Much higher sample sizes are needed when small QTL effect sizes are aimed to be detected.
Heritability of slope flexion in the subsample (n = 283) is low, which might in part explain the single LOD peak >1.5 for this trait. Muscle strength is a complex multifactorial trait, and high heritabilities do not guarantee the presence of a QTL with large effect size, although it increases the probability of finding one.
Different mechanisms underlie the variability in the torque-velocity relationship. First, Rijkelijkhuizen et al. (28) showed in rats that the mean curvature of the torque-velocity relationship for fast oxidative fibers was higher than for fast glycolytic fibers. They concluded that the degree of curvature is fiber-type dependent. This is in line with the single-fiber study of Bottinelli et al. (8), who demonstrated in humans that curvature was higher for slow fibers than for fast fibers; among fast fibers, curvature was higher for fast oxidative (IIa) than for fast glycolytic (IIb) fibers although the difference was not statistically significant. Furthermore, a shortened velocity of knee extensor muscles was found to be significantly correlated with the percentage of fast-twitch fibers in the vastus lateralis muscles (40). Second, previous studies have also suggested the presence of unique genetic causes of variance (genes causing nonshared variability in one specific muscle characteristic) (11, 36, 37, 39, 41, 42). For example, the ACTN3 (11q13.1) gene, which is expressed in Z-lines of fast but not of slow muscle fibers, is believed to be one of the contributing genes in the heritability of fiber type distribution (45) and might be (in)directly involved in the presence of different curvatures of the torque-velocity relationships. ACTN3 R577X genotype associations were reported for relative torques at 300°/s (45), which is at much higher speeds than measured in the present study. We did not find an important linkage signal in the vicinity of ACTN3, which might be attributed to the absence of a causal SNP or lack of LD with the causal SNP in the current Illumina SNP panel. An exploratory association analysis of ACTN3 R577X genotypes at 60°/s, 120°/s, and 240°/s in a subsample of the LGfMS study (n = 362) demonstrated stronger associations for relative dynamic knee extension torques with increasing velocities (38). The lower relative dynamic torques at higher contraction speeds for XX carriers might indicate a downward shift of the force-velocity curve in subjects lacking the ACTN3 protein. Second, the torque-velocity relationship can also be determined by fascicle length (muscle fibers packed in bundles that extend from the proximal to distal tendons) and pennation angle of muscle fibers (angle between the muscle fiber and the axis of the muscle). In a group of 37 male 100-m sprinters (fast and slow group), Kumagai et al. (20) found significant, negative correlations between 100-m personal best performance and fascicle length (absolute and relative to limb length) of the vastus lateralis, and gastrocnemius medius and lateralis muscles. They argued that longer fascicle length might lead to greater maximal velocity of shortening and allows for a greater force output at an identical shortening velocity, which results in greater power production. Furthermore, sprinters in the fast group had a significantly lower pennation angle than did the slow group for the selected muscles. Both muscle architectural characteristics might be the result of genes and/or gene-environment (i.e., training) interactions.
A candidate gene for the torque-velocity relationship is the insulin-like growth factor-binding protein 1 gene (IGFBP-1), which is located on chromosome 7p12p13, within the LOD peak of 2.03 at 7p12.3 (for TVRF). It binds to the insulin-like growth factor 1 protein (IGF-1), which has previously been associated with quadriceps-muscle strength gains in response to strength training (19). Lang et al. (22) demonstrated that an acute in vivo elevation in circulating levels of IGFBP-1 is capable of selectively decreasing protein synthesis in fast-twitch skeletal muscle and concluded that elevations in circulating and tissue levels of IGFBP-1 may be an important mediator for the muscle catabolism observed in various stress conditions. Several candidate genes have been associated with muscle strength phenotypes (7, 26). However, these genes are not located within linkage regions reported in the present study. Growth and differentiation factor 8 (GDF8, 190.6 Mb) is located in relative vicinity to region 2q36.3 (221.8 Mb), myosin light chain kinase (MYLK, 124.9 Mb) to region 3q21 (141.8 Mb), both linked to TVRF, and insulin like growth factor 1 (IGF1, 101.3 Mb) to region 12q24.32 (122.2 Mb). Further fine mapping analysis will be required to assess the potential contribution of each of these candidate genes to explain velocity-dependent muscle strength variation.
The results of the joint test for linkage and association showed no evidence for association with any of the SNPs in regions with suggestive or significant evidence for linkage. However, the association analysis using Merlin software (3) revealed significant association in six different chromosomal regions (Table 4). These SNPs are located in the vicinity of the SNPs with highest LOD scores within their respective chromosomal region. Finding a “true” association between a trait and an SNP requires complete LD between the marker and the functional SNP, or the SNP being the actually functional SNP itself. Marker spacing in the Linkage IVb Panel of Illumina, designed to perform linkage analyses rather than association analyses, is relatively large (average marker spacing = 482 kb or 0.64 cM), with low between-marker LD. Furthermore, the panel is not specifically enriched for functional SNPs (e.g., nonsynonymous variants), as minor allele frequency and heterozygosity are the main criteria for markers to be included. This might explain the observation that the SNPs that identified the highest linkage LOD score did not show up as most significant in the association analysis. A much denser marker panel (e.g., microarray chips) or a functional fine mapping panel of SNPs located in relevant ranked candidate genes (48) will be needed to assess association in regions with suggestive or significant evidence for linkage.
Conclusions
This genome-wide multipoint linkage analysis of different torque-velocity characteristics revealed several promising regions with evidence for linkage with the torque-velocity relationship of the knee flexors and extensors. Several candidate genes that might be relevant from a physiological point of view are located within the identified regions and warrant further fine mapping analyses by additional linkage and/or association studies. Although only the largest families from the LGfMS project were selected for this study, the relatively small sample size limits the possibility to detect chromosomal regions where genes with small effect sizes are harbored. Therefore, fine mapping and replication studies should focus on highly informative samples (e.g., large families) of sufficiently large size and preferably extend to females and other age groups.
GRANTS
G. De Mars is funded by Research Foundation Flanders Grant G.0496.05 [Fonds Wetenschappelijk Onderzoek (FWO)]. A. Windelinckx is supported by the Research Fund of the Katholieke Universiteit Leuven (OT/04/44). W. Huygens was funded by grant OT/98/39. M. W. Peeters is a Postdoctoral Fellow (FWO). The project is funded by OT/98/39 for the phenotyping phase, and OT/04/44, G.0496.05, and mainly FWO G.0567.07 for the genome scan.
Acknowledgments
We thank the siblings for their participation in the study and their enthusiasm in performing at their maximum during the strength testing protocol. The authors thank I. Salden for support during the preparation of DNA samples and R. van't Slot for careful assistance with the genome scan.
Address for reprint requests and other correspondence: M. A. I. Thomis, Dept. of Biomedical Kinesiology, Research Center for Exercise and Health, FABER, Tervuursevest 101, 3001 Leuven, Belgium (e-mail: martine.thomis@faber.kuleuven.be).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Abecasis GR, Burt RA, Hall D, Bochum S, Doheny KF, Lundy SL, Torrington M, Roos JL, Gogos JA, Karayiorgou M. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am J Hum Genet 74: 403–417, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66: 279–292, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KE, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet 74: 482–494, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baecke JA, Burema J, Frijters JE. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am J Clin Nutr 36: 936–942, 1982. [DOI] [PubMed] [Google Scholar]
- 6.Bazzucchi I, Marchetti M, Rosponi A, Fattorini L, Castellano V, Sbriccoli P, Felici F. Differences in the force/endurance relationship between young and older men. Eur J Appl Physiol 93: 390–397, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Beunen G, Thomis M. Gene powered? Where to go from heritability (h2) in muscle strength and power? Exerc Sport Sci Rev 32: 148–154, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Bottinelli R, Canepari M, Pellegrino MA, Reggiani C. Force-velocity properties of human skeletal muscle fibres: myosin heavy chain isoform and temperature dependence. J Physiol 495: 573–586, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardon LR, Abecasis GR. Some properties of a variance components model for fine-mapping quantitative trait loci. Behav Genet 30: 235–243, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Mars G, Thomis MA, Windelinckx A, Van Leemputte M, Maes HH, Blimkie CJ, Claessens AL, Vlietinck R, Beunen G. Covariance of isometric and dynamic arm contractions: multivariate genetic analysis. Twin Res Hum Genet 10: 180–190, 2007. [DOI] [PubMed] [Google Scholar]
- 12.De Mars G, Windelinckx A, Huygens W, Peeters MW, Beunen GP, Aerssens J, Vlietinck R, Thomis MA. Genome-wide linkage scan for maximal and length-dependent knee muscle strength in young men: significant evidence for linkage at chromosome 14q24.3. J Med Genet 45: 275–283, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet 75: 687–692, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet 64: 259–267, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes VA, Frontera WR, Wood M, Evans WJ, Dallal GE, Roubenoff R, Fiatarone Singh MA. Longitudinal muscle strength changes in older adults: influence of muscle mass, physical activity, and health. J Gerontol A Biol Sci Med Sci 56: B209–B217, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck RF, Beunen G. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics 17: 264–270, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen RG, Vlietinck RF, Beunen G. A quantitative trait locus on 13q14.2 for trunk strength. Twin Res 7: 603–606, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Huygens W, Thomis MA, Peeters MW, Aerssens J, Vlietinck R, Beunen GP. Quantitative trait loci for human muscle strength: linkage analysis of myostatin pathway genes. Physiol Genomics 22: 390–397, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Kostek MC, Delmonico MJ, Reichel JB, Roth SM, Douglass L, Ferrell RE, Hurley BF. Muscle strength response to strength training is influenced by insulin-like growth factor 1 genotype in older adults. J Appl Physiol 98: 2147–2154, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Kumagai K, Abe T, Brechue WF, Ryushi T, Takano S, Mizuno M. Sprint performance is related to muscle fascicle length in male 100-m sprinters. J Appl Physiol 88: 811–816, 2000. [DOI] [PubMed] [Google Scholar]
- 21.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247, 1995. [DOI] [PubMed] [Google Scholar]
- 22.Lang CH, Vary TC, Frost RA. Acute in vivo elevation of insulin-like growth factor (IGF) binding protein-1 decreases plasma free IGF-I and muscle protein synthesis. Endocrinology 144: 3922–3933, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Loos R, Thomis M, Maes HH, Beunen G, Claessens AL, Derom C, Legius E, Derom R, Vlietinck R. Gender-specific regional changes in genetic structure of muscularity in early adolescence. J Appl Physiol 82: 1802–1810, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Pousson M, Lepers R, Van Hoecke J. Changes in isokinetic torque and muscular activity of elbow flexors muscles with age. Exp Gerontol 36: 1687–1698, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Rankinen T, Bray MS, Hagberg JM, Perusse L, Roth SM, Wolfarth B, Bouchard C. The Human Gene Map for Performance and Health-Related Fitness Phenotypes: The 2005 Update. Med Sci Sports Exerc 38: 1863–1888, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE Family Study. Physiol Genomics 16: 256–260, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Rijkelijkhuizen JM, de Ruiter CJ, Huijing PA, De Haan A. Force/velocity curves of fast oxidative and fast glycolytic parts of rat medial gastrocnemius muscle vary for concentric but not eccentric activity. Pflügers Arch 446: 497–503, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Risch NJ, Zhang H. Mapping quantitative trait loci with extreme discordant sib pairs: sampling considerations. Am J Hum Genet 58: 836–843, 1996. [PMC free article] [PubMed] [Google Scholar]
- 30.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet 71: 238–253, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skelton DA, Kennedy J, Rutherford OM. Explosive power and asymmetry in leg muscle function in frequent fallers and non-fallers aged over 65. Age Ageing 31: 119–125, 2002. [DOI] [PubMed] [Google Scholar]
- 32.Song KK, Weeks DE, Sobel E, Feingold E. Efficient simulation of P values for linkage analysis. Genet Epidemiol 26: 88–96, 2004. [DOI] [PubMed] [Google Scholar]
- 33.The International Multiple Sclerosis Genetics Consortium. Enhancing linkage analysis of complex disorders: an evaluation of high-density genotyping. Hum Mol Genet 13: 1943–1949, 2004. [DOI] [PubMed] [Google Scholar]
- 34.Thom JM, Morse CI, Birch KM, Narici MV. Influence of muscle architecture on the torque and power-velocity characteristics of young and elderly men. Eur J Appl Physiol 100: 613–619, 2007. [DOI] [PubMed] [Google Scholar]
- 35.Thomis M, Claessens AL, Vlietinck R, Marchal G, Beunen G. Accuracy of anthropometric estimation of muscle cross-sectional area of the arm in males. Am J Hum Biol 9: 73–86, 1997. [DOI] [PubMed] [Google Scholar]
- 36.Thomis M, Van Leemputte M, Maes H, Blimkie CJR, Claessens AL, Marchal G, Willems E, Vlietinck R, Beunen G. Multivariate genetic analysis of maximal isometric muscle force at different elbow angles. J Appl Physiol 82: 959–967, 1997. [DOI] [PubMed] [Google Scholar]
- 37.Thomis MA, Beunen GP, Maes HH, Blimkie CJ, Van Leemputte M, Claessens AL, Marchal G, Willems E, Vlietinck RF. Strength training: importance of genetic factors. Med Sci Sports Exerc 30: 724–731, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Thomis MA, De Mars G, Windelinckx A, Vincent B, Huygens W, Peeters M, Vlietinck R, Beunen G. Alpha-actinin-3 R577X genotype and muscle power in young male adults of the Leuven genes for muscular strength study. Med Sci Sports Exerc 38: S365–S366, 2006. [Google Scholar]
- 39.Thomis MAI, Beunen GP, Van Leemputte M, Maes HH, Blimkie CJ, Claessens AL, Marchal G, Willems E, Vlietinck RF. Inheritance of static and dynamic arm strength and some of its determinants. Acta Physiol Scand 163: 59–71, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Thorstensson A, Grimby G, Karlsson J. Force-velocity relations and fiber composition in human knee extensor muscles. J Appl Physiol 40: 12–16, 1976. [DOI] [PubMed] [Google Scholar]
- 41.Tiainen K, Sipila S, Alen M, Heikkinen E, Kaprio J, Koskenvuo M, Tolvanen A, Pajala S, Rantanen T. Shared genetic and environmental effects on strength and power in older female twins. Med Sci Sports Exerc 37: 72–78, 2005. [DOI] [PubMed] [Google Scholar]
- 42.Tiainen K, Sipila S, Alen M, Heikkinen E, Kaprio J, Koskenvuo M, Tolvanen A, Pajala S, Rantanen T. Heritability of maximal isometric muscle strength in older female twins. J Appl Physiol 96: 173–180, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, Carey A, Ewen-White KR, Duffy DL, O'connor DT, Barlow DH, Martin NG, Kennedy SH. Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet 77: 365–376, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valour D, Ochala J, Ballay Y, Pousson M. The influence of ageing on the force-velocity-power characteristics of human elbow flexor muscles. Exp Gerontol 38: 387–395, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Vincent B, De Bock K, Ramaekers M, Van den Eede E, Van Leemputte M, Hespel P, Thomis MA. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol Genomics 32: 58–63, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 21: 3445–3447, 2005. [DOI] [PubMed] [Google Scholar]
- 47.Wilcox MA, Pugh EW, Zhang H, Zhong X, Levinson DF, Kennedy GC, Wijsman EM. Comparison of single-nucleotide polymorphisms and microsatellite markers for linkage analysis in the COGA and simulated data sets for Genetic Analysis Workshop 14: Presentation Groups 1, 2, and 3. Genet Epidemiol 29, Suppl 1: S7–28, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Windelinckx A, Vlietinck R, Aerssens J, Beunen G, Thomis MA. Selection of genes and single nucleotide polymorphisms for fine mapping starting from a broad linkage region. Twin Res Hum Genet 10: 871–885, 2007. [DOI] [PubMed] [Google Scholar]