Abstract
Prostaglandin E2 (PGE2) plays an important role in maintaining body fluid homeostasis by activating its receptors on the renal collecting duct (CD) to stimulate renal Na+ and water excretion. The PG carrier prostaglandin transporter (PGT) is expressed on the CD apical membrane, where it mediates PG reuptake as part of the termination of autocrine PG signaling. Here we tested the hypothesis that dietary salt loading regulates PGT gene transcription in renal CDs. We placed green fluorescence protein (GFP) under control of 3.3 kb of the mouse PGT promoter and injected this construct into the pronuclei of fertilized FVB mouse eggs. Four of thirty-eight offspring were GFP positive by genotyping. We extensively characterized one (no. 29) PGT-GFP transgenic mouse line. On microscopic examination, GFP was expressed in CDs as determined by their expression of aquaporin-2. We fed mice a low (0.03% NaCl)-, normal (0.3% NaCl)-, or high-salt (3% NaCl) diet for 2 wk and quantified CD GFP expression. The average number of GFP-positive CD cells per microscopic section varied directly with dietary salt intake. Compared with mice on the control (0.3% sodium) diet, mice on a low-sodium (0.03%) diet had reduced numbers of GFP-positive cells (71% of control, P < 0.001), whereas mice on a high-sodium (3%) diet had increased numbers of GFP-positive cells (139% of control, P < 0.001). This increase in apparent CD PGT transcription resulted in a 51–55% increase (P < 0.001) in whole kidney PGT mRNA levels as determined by real-time PCR. The regulation of PG signal termination via reuptake represents a new pathway for controlling renal Na+ balance.
Keywords: organic anion transport, natriuresis, dietary sodium, transgenic mice
the kidney controls body fluid volume, osmolality, and homeostasis by controlling NaCl and water reabsorption. Although most salt and water reabsorption takes place in the proximal tubule and thick ascending limb of Henle's loop, the collecting duct (CD) is the final regulator of sodium and water balance (20, 37). Na+ and water reabsorption are tightly regulated by nonhormonal and hormonal factors, including the renal-angiotensin-aldosterone system and prostaglandins.
Prostaglandins have potent effects on renal Na+ and water absorption (19, 21–23, 29, 45, 46). Prostaglandin E2 (PGE2), the major prostaglandin produced in the kidney (13, 42), inhibits net Na+ and water reabsorption by the CD (19, 21–23, 46), thus inducing natriuresis and diuresis (21, 29, 45). Indeed, dietary salt loading induces expression of cyclooxygenase (COX)-2 in renal medullary interstitial cells (10, 18, 36, 50, 51). Since these cells also express PGE synthase (10), the resulting PGE2 enters the medullary interstitium, binds to EP1 and/or EP3 receptors on the basolateral membrane of the CD, and inhibits Na+ absorption (19, 22).
Curiously, urinary PGE2 excretion fails to rise at an equal pace with this increase in medullary PGE2 synthesis during dietary salt loading. Although this may be due simply to the inability of medullary interstitial PGE2 to cross CD tight junctions (15), this observation is particularly remarkable since CDs themselves synthesize and release PGE2 into the urinary space (8, 10, 24, 42, 49–51). In fact, urinary PGE2 actually varies inversely with NaCl intake (1–4, 11, 39, 43, 44, 47), implying a mechanism that clears the urinary space of PGE2 during salt loading. Such a clearance mechanism would be especially important since PGE2 residing in the tubule lumen would inhibit the desired natriuresis by stimulating apical EP4 receptors (5, 38, 48).
The prostaglandin transporter (PGT) is constitutively expressed at the CD apical or subapical membrane and mediates PGE2 uptake (6, 7, 28, 34). In a polarized CD cell culture model system, we have shown that PGT extracts PGE2 from the apical (urinary) solution, transports it across the CD epithelium, and delivers it, essentially intact, to the basolateral solution (12). Thus in situ PGT in the CD would reduce luminal PGE2 and minimize the stimulation of apical (antinatriuretic) EP4 receptors while simultaneously increasing basolateral PGE2 and stimulating basolateral (natriuretic) EP1 and/or EP3 receptors.
Because of these predicted effects of PGT to enhance or sustain natriuresis, we asked whether salt loading might increase expression of PGT along the renal CD. To test this hypothesis, we constructed mice transgenic for a PGT promoter- green fluorescence protein (GFP) reporter. Here we show that PGT is transcriptionally expressed along the CD nephron segment and that CD PGT transcriptional levels are significantly increased by high salt intake.
MATERIALS AND METHODS
Materials.
Designed primers were purchased from Invitrogen. Restriction enzymes were from either Promega or New England Biolabs. The expanded high-fidelity PCR system was purchased from Roche. Qiagen kits were used for purification of PCR products. Superfect transfection reagent was obtained from Qiagen. The DNA extraction kit was purchased from Lambda Biotech. The vector pEGFP-1 was purchased from BD Biosciences/Clontech. FVB/N mice were obtained from Charles River Laboratories. Antibody against aquaporin-2 (AQP2) was from Alpha Diagnostic International. The secondary antibodies, Alexa Fluor 350 goat anti-rabbit IgG and Alexa Fluor 568 goat anti-rabbit IgG, were purchased from Molecular Probes.
Plasmid construction.
A PCR product containing the genomic sequence of PGT from 5′ nucleotides −3,423 to −113 (referenced to the ATG start codon) was amplified at an annealing temperature of 55°C using primers containing SalI and SacII restriction sites (5′ACGCGTCGACCAGTCCAAGCTTGTATCGCTG3′ and 5′TCC CCGCGGGCGAGACGTGGAGACTGAGC3′). This PCR product was digested with SalI and SacII sequentially. The 3.3-kb amplification product, provisionally considered as the PGT promoter, was gel purified and subcloned into the pEGFP-1 vector at the SalI and SacII sites. The sequence of the resulting PGT promoter-GFP reporter was checked by two methods, 1) digestion with restriction enzymes including SalI, SacII, AatII, BamHI, and BbSI and 2) sequencing with primers 5′CGTTATCCCCTGATTC3′, 5′AGGAGCAGCCATCTTGTGAC3′, 5′TTTTGTCCCAACCCCCAGCACTTC3′, 5′AGGCGACAGCGGCTGGG3′, and 5′CGTCGCCGTCCAGCTCGACCA3′.
Transient transfection.
3T3 cells were seeded at 25–30% confluency. Twenty-four hours later, they were transfected with PGT promoter-GFP reporter. Pictures of transfected 3T3 cells were taken 24–48 h after transfection.
Generation and breeding of transgenic mice.
The circular transgene was excised from the vector by digestion with BamHI and AflII and purified by electrophoresis. The linear transgene of 4,361 base pairs containing the PGT promoter-GFP reporter was isolated and purified. Transgenic founder mice were created by the Transgenic Mouse Facility at Albert Einstein College of Medicine. The transgene was injected into the male pronucleus of single-cell embryos derived from FVB/N mice, and the embryos were implanted into pseudopregnant females. Candidate transgenic mice were weaned, and toe and tail clips were obtained before 21 days of age. After genotyping, the founder mice (F0) were mated with wild-type FVB/N mice to generate transgene positive offspring F1, F2, F3, and F4.
Genotyping of transgenic mice.
Genotyping of transgenic mice was conducted by conventional PCR. Mouse genomic DNA was extracted from tails. Transgenic mice were prescreened by PCR for GFP positivity. The primers used to amplify a fragment of 342 base pairs within GFP reporter were 5′GCACCATCTTCTTCAAGGACGAC3′ for forward and 5′TCTTTGCTCAGGGCGGACTG3′ for backward. The annealing temperature was 59°C, and the cycle number was 30.
Real-time quantitative PCR.
To check the stability of the transgene within the first five generations, real-time quantitative PCR was conducted. The primer set in the PGT region consisting of 5′CCTCCTCCGAACTGCCTCAAG3′ for the forward direction and 5′CCTCTCCTGGACTGGGACCTTAG for the backward was used to determine the relative copy numbers of the transgene compared with the native copy number of PGT, which is 2. The primer set in the GFP region consisting of 5′GAACGGCATCAAGGTGAACTTC3′ for the forward direction and 5′TGGGTGCTCAGGTAGTGGTTGTC3′ for backward was used to confirm the relative copy numbers. IL-2β is a housekeeping gene and was used as internal control for the quantity of genomic DNA. A sample (10 ng) of genomic DNA from tails of mice was mixed with SYBR green. The PCR was conducted using the following parameters: 1 cycle of 95°C for 10 min; and then 40 cycles of 95°C for 10 s, 60°C for 35 s, and 72°C for 30 s. The Ct values of the samples were determined as the average of triplicates, where Ct is the cycle threshold. The relative copy numbers were calculated by the following equation (16, 17, 30): relative DNA copy number = 2−ΔΔCt, where −ΔΔCt = Ct(sample) − Ct(wt).
Visual analysis of transgene expression.
F1-F4 transgenic mice of lines 13 and 29 were analyzed for transgene expression. Transgenic mice were euthanized by exsanguination after methoxyflurane or halothane anesthesia, and the kidneys or other organs were dissected for the various studies described below. The expression of GFP was examined by fluorescence microscopy. Slices of fresh kidney were fixed with 4% paraformaldehyde for 0.3 h and sectioned at 50 to 200 μm by microtome. Thin sections were prepared from tissues by perfusion-fixation and/or immersion for 1 h in PBS containing 4% paraformaldehyde. Specimens were cryoprotected overnight in 30% sucrose, embedded in optimal cutting temperature compound, frozen in isopentane, and sectioned at 5 to 10 μm using a Leica Frigocut 2800N cryostat (14, 31, 33). Sections were mounted with aqueous medium (Vectashield) and visualized under fluorescent microscope.
Immunofluorescent staining.
Cryosections (5–10 μm thick) were prepared from formaldehyde-fixed tissues, mounted, and incubated with primary antibody (for PGT or AQP2) in 5% BSA and 2% gold serum overnight at 4°C. Sections were then washed with PBS three or four times, 8–10 min each time. They were then incubated with a 1:1,000 dilution of Alexa Fluor 350-coupled or Alexa Fluor 568-coupled goat anti-rabbit IgG in PBS for 1 h at room temperature and washed. Stained sections were mounted with Vectashield and examined under a fluorescence microscope.
Quantification of urinary sodium and water excretion and of renal GFP expression, as a function of dietary sodium intake.
From one generation of GFP transgenic mice, three sex-matched littermates with similar baseline GFP expression levels (as determined by conventional PCR and copy numbers of the transgene) were selected from each of three separate litters (n = 9). Each littermate was randomized to the low-, normal-, or high-salt diet. This process was repeated with the transgenic offspring of a subsequent generation; thus this generation of offspring also contributed three mice to each of three salt intake groups.
Mice were kept in metabolic cages for 1 wk on regular chow and were then placed on low salt (0.03% NaCl), normal salt (0.3% NaCl), or high salt (3% NaCl), respectively, for 2 wk. Urine was collected daily. Urine volumes were recorded; urine Na+ concentration was determined by ion electrodes purchased from Shelfscientific. After 2 wk, the mice were euthanized; one kidney was taken for imaging and the other for mRNA analysis.
For each mouse, cryosections of kidneys were analyzed microscopically to assess the number of GFP-positive cells. First, we identified a low-power (10×) field that contained GFP-positive cells homogeneously distributed across the entire field. GFP-positive cells were then counted at 40× magnification for the field previously demarcated at 10×. For each mouse of each dietary set, the number of GFP-positive cells was taken as the average of three counts of three slides.
For statistical analysis, the three littermates were considered as a paired set. The mean GFP cell count for the littermate on the normal-salt diet was taken as 1.0, and the GFP cell counts of the other two littermates were normalized relative to this count. Taken over the two generations of mice studied, this yielded six cell-count values for the low-salt diet and six for the high-salt diet relative to normal.
Separately, the other kidney from each mouse above was taken for PGT mRNA expression studies as in the next section.
PGT mRNA measurement by real-time quantitative PCR.
Total RNA was extracted from whole kidneys of GFP transgenic mice as above (6 mice in each of 3 dietary groups) and of wild-type mice on the three sodium diets (6 mice in each of 3 dietary groups). cDNA was synthesized from 1 μg of total RNA. Synthesized cDNA was mixed with SYBR green and PGT primers. The PGT primers were 5′ GCAGCCTCACCACTATCGAG 3′ for the forward direction and 5′ TGATGAGGATAGCGTTGCTG 3′ for the backward. β-actin was used as an internal control. The real-time PCR was conducted using the following parameters: 1 cycle of 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 10 s, 55°C for 20 s, 72°C for 30 s; and 1 cycle of 95°C for 15 s, 60°C for 15 s, 95°C for 15 s. For each kidney, PGT mRNA was measured three times, and the average for each mouse was taken from these three measures.
RESULTS
Design and in vitro validation of the PGT promoter-reporter.
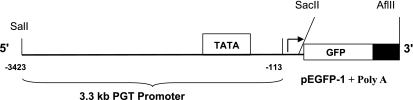
Alignment of GenBank-derived mouse PGT cDNA to mouse genomic DNA sequences generated a sequence that is upstream of the PGT start codon. As shown in Fig. 1, we chose a fragment of 3.3-kb sequence inclusive of the 5′ upstream nucleotides −3,423 to −113 (referenced to the ATG start codon). This fragment contained the TATA-box and the predicted downstream transcription initiation site but did not contain the translation initial site for the PGT gene. A transgene was designed to utilize PGT transcription via the PGT promoter and efficient translation via the enhanced GFP (EGFP) cassette.
Fig. 1.
Structure of prostaglandin transporter (PGT)-enhanced green fluorescence protein (EGFP) transgene. PGT promoter region was fused to the EGFP coding region. The 3.3-kb fragment is a DNA sequence that is upstream of start codon of PGT. The amino acid sequences, −3,423 to −113, are referenced to the ATG start codon.
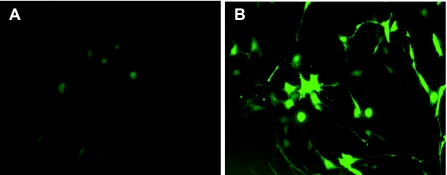
We tested the constructs by transiently transfecting mouse Swiss 3T3 cells. As shown in Fig. 2, transfection with the PGT promoter-GFP reporter construct generated GFP-positive cells constituting more than 30% of the total cell population (Fig. 2B), whereas transfection with the promoter-free GFP vector did not produce any GFP-positive cells (Fig. 2A). Thus the 3.3-kb fragment functions as a promoter to drive the expression of GFP.
Fig. 2.
The PGT promoter drives GFP expression in 3T3 cells. 3T3 cells were transiently transfected with promoter-free EGFP vector (A), as a control, or the transgene consisting of PGT promoter-GFP reporter (B).
Creation, breeding, and genotyping of transgenic mice: a 4.36-kb BamHI.
AflII fragment was used to create transgenic mice by standard pronuclear microinjections. Founders were identified by PCR analysis of tail DNA using oligonucleotide primers specific for EGFP. Out of 36 mice, we obtained four positive founders, 13, 26, 29, and 30 (the F0 generation). When crossed with wild-type FVB mice, all founders transmitted the gene to about 50% of their offspring (F1). F2 and the following generations were created in the same manner. We intensively characterized line 29. The following results are of the first four generations of line 29. We also characterized line 13, the results of which were similar to that of line 29.
Copy number of the transgene.
The stability of transgene passage was assessed by measuring the relative copy number over several generations. We designed two sets of primers to measure the relative copy numbers, one in the PGT region and the other in the GFP region. These two sets of primers generated consistent copy numbers in the range of 4–20 for generations F0 to F4. The copy numbers of the transgene in one subline (29-11-22) slightly changed but consistently decreased in another subline (29-11-21). Overall, there was decline to different degrees in the copy numbers as the transgene was passed down the generations.
Tissue expression of PGT.
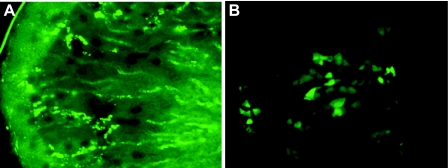
Using the PCR genotype-positive mouse 29–2, we made fresh sections of one kidney and cryosections of another. Both kinds of sections revealed multiple bright, GFP-positive cells (Fig. 3). A representative image of a fresh section at lower magnification (Fig. 3A) shows that GFP is expressed in the cortex, outer medulla, and inner medulla. Expression in the outer medulla is stronger than in the other regions. Close examination of the cryosections (Fig. 3B) suggested that GFP is expressed along the CD. We did not observe definitive PGT-GFP expression in other tissues, suggesting that this promoter regulates PGT expression specifically in the renal CD. In kidney, the relative copy numbers of the transgene varied from one generation to another and from one subline to another. Nonetheless, we obtained a relatively stable subline of transgenic mice (line 29-11-22), which allowed us to localize PGT in the kidney and examine its regulation.
Fig. 3.
PGT expression is distributed in the radial tubules of the cortex and outer and inner medulla. Fluorescence microscopy of kidney sections of a PGT-GFP transgenic mouse. A: fresh section (200 μm) cut by microtome after fixation with 4% paraformaldehyde for 0.5 h, 10× magnification. B: frozen cryosection (6 μm), 60× magnification.
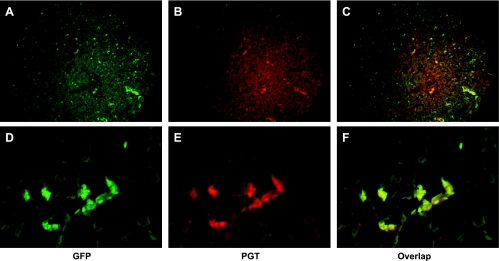
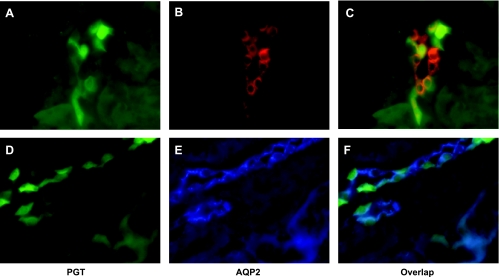
To verify the localization of renal PGT-GFP in the CD, we labeled cryosections of kidney with anti-AQP2 polyclonal antibody and subsequent Alexa Fluor 568 (red in Fig. 4B) or 350 goat anti-rabbit IgG (blue in Fig. 4E). AQP2 is a marker of principal cells (14, 31, 33). As shown in Fig. 4, most of the GFP-positive cells (Fig. 4, A or D) colocalize with AQP2 labeling (Fig. 4, B or E), indicating that PGT is expressed in principal cells (Fig. 3, C or F).
Fig. 4.
PGT expression is localized to the principal cells of the collecting duct. Fluorescence microscopy of kidney cryosections of a PGT-GFP transgenic mouse (green, A and D); immunofluorescence labeling for aquaporin-2 (AQP2) using Alexa Fluor 568 (red, B and E). C and F are the merges of A and B, and D and E, respectively.
GFP-positive cells are PGT-positive cells.
To confirm that the GFP expression is driven by a PGT promoter, i.e., that GFP-positive cells are PGT-positive cells, we labeled cryosections of kidneys from adult mice with anti-PGT antibody and subsequently with Alexa Fluor 568 goat anti-rabbit IgG (red). As shown in Fig. 5, GFP expression and PGT protein expression were highly concordant. Thus GFP expression appears to be driven by the PGT promoter in these transgenic animals.
Fig. 5.
GFP-positive cells are PGT-expressing cells. Fluorescence microscopy of kidney cryosections of a PGT-GFP transgenic mouse (A and D), immunofluorescence labeling for PGT using Alexa Fluor 568 (red, B and E). C and F are the merges of A and B, and D and E, respectively. A, B, and C are by 10× magnification; D, E, and F are 20× magnification.
Salt intake regulates PGT transcriptional expression in kidney CD.
As described in materials and methods, we placed six sets of mice in metabolic cages on diets containing 0.03%, 0.3%, or 3% NaCl. Each set of mice consisted of sex-matched littermates that had similar baseline GFP expression levels as determined by conventional PCR and the copy numbers of the transgene. As shown in Table 1, the daily total Na+ excretion of transgenic mice on the high-salt diet was about 16-fold that of the mice with normal-salt diet. The daily total Na+ excretion of mice on a normal-salt diet was about eightfold that of mice on a low-salt diet.
Table 1.
Daily urine volume, urine Na+ concentration, and total Na+ excretion of mice on different diets
| NaCl Content in Diet |
Vurine, ml/day |
[Na+]urine, mM
|
Total NaCl Excretion, μEq/day
|
|||
|---|---|---|---|---|---|---|
| WT (n = 3) | Transgenic (n = 3) | WT (n = 3) | Transgenic (n = 3) | WT (n = 3) | Transgenic (n = 3) | |
| 0.03% | 0.83±0.20 | 0.71±0.10 | 4.3±1.1 | 6.2±0.4 | 2.9±0.2 | 4.3±0.9 |
| 0.3% | 0.79±0.16 | 0.78±0.12 | 38±9 | 45±2 | 26.1±1.2 | 34.3±3.8 |
| 3% | 2.44±0.59 | 2.67±0.12 | 202±31 | 218±11 | 418±48 | 563±20 |
Values are means ± SE. WT, wild-type; [Na+], Na concentration.
We also measured daily Na+ excretion of wild-type mice on low-, normal-, or high-salt diet. Dietary salt had similar effects on the daily total Na+ excretion of wild-type mice compared with transgenic mice (Table 1, P = not significant for wild-type vs. transgenic for all comparisons).
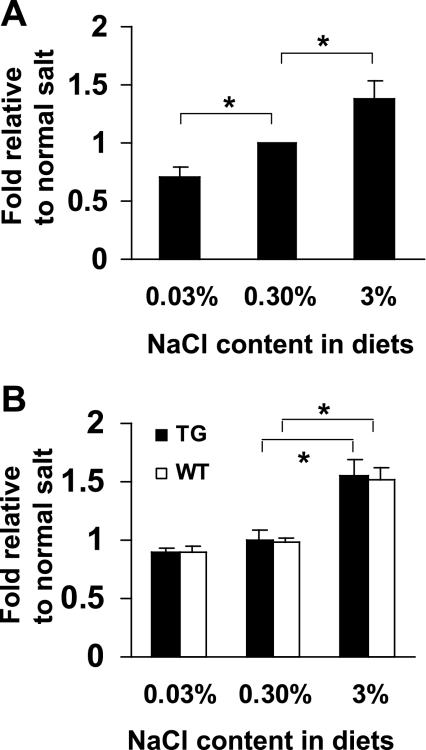
Following measurements in the metabolic cages, we made kidney cryosections in which care was taken to cut at similar positions relative to kidney anatomy. As shown in Fig. 6A, the number of GFP-positive cells increased in a statistically significant way as a function of dietary sodium. Mice on the low-sodium diet had only 71 ± 7.3% (SD) of the GFP-positive cells per low-power field compared with control animals (P < 0.001). In contrast, mice on a high-sodium diet had 139 ± 14% (SD) the control number of GFP-positive cells (P < 0.001). These results indicate that PGT gene transcription is induced in renal CDs by high salt intake.
Fig. 6.
A: effect of dietary salt intake on collecting duct PGT transcription using promoter-GFP reporter mice. Cell counts of mice on low- and high-salt diets were normalized relative to those on a normal-salt diet. See materials and methods for details. Values are the mean ratios of 6 sets of mice. Bars represent standard deviations. *P < 0.001 (unpaired t-test). B: effect of dietary salt intake on whole kidney PGT mRNA expression. The mRNA levels of mice on low- and high-salt diets were normalized relative to those on a normal-salt diet. See materials and methods for details. Values are the mean ratios of 6 sets of mice. Bars represent standard deviations. *P < 0.001 (unpaired t-test). WT, wild-type; TG, transgenic.
To confirm an increase in PGT transcription by salt loading, we measured PGT mRNA in whole kidneys of both wild-type and transgenic mice fed 0.03%, 0.3%, or 3% NaCl. As shown in Fig. 6B, PGT mRNA in kidneys of wild-type mice as a function of salt diet were (means ± SD): 0.90 ± 0.05 (0.03% diet), 0.99 ± 0.03 (0.3% diet), and 1.52 ± 0.10 (0.3% diet). In the promoter-GFP transgenic mice, the corresponding PGT mRNA values were: 0.90 ± 0.02 (0.03% diet), 0.99 ± 0.09 (0.3% diet), and 1.55 ± 0.15 (0.3% diet). In both sets of animals, the increase with high salt was statistically significant compared with the other two groups (P < 0.001).
DISCUSSION
The present studies used a promoter-reporter transgenic mouse as a model system with which to determine whether the PGT is regulated transcriptionally in the kidney in response to dietary salt loading. The transgene was expressed, as is the PGT protein, in renal CDs, especially in principal cells, the site of Na+ reabsorption. Higher levels of dietary NaCl intake were associated with higher degrees of PGT transcription in the CDs, suggesting that high salt intake increases the transport of PGs by the renal CD.
These studies address one component of a larger question, namely, the mechanism by which the cell-surface concentrations of prostanoids are controlled. Much experimental effort has been devoted to understanding the regulation of prostanoid synthesis by the COXs, the tissue-specific isomerases, the various G protein-coupled cell-surface prostanoid receptors, and the downstream signaling events. These studies have revealed that PGs signal a vast array of cellular events. Because many PGs, such as the nearly ubiquitous PGE2, are structurally stable in plasma, unwanted signaling at a distance would occur were it not for highly localized signal termination. The latter consists of two sequential steps: 1) active PG uptake across the plasma membrane, and 2) intracellular oxidation and inactivation (40, 41).
Our laboratory has previously shown, using the renal CD as a model system, that PG synthesis and release, on the one hand, and PG reuptake and intracellular oxidation, on the other, occur in the same cell (34). These results have suggested that PG signaling is analogous to neurotransmitter release/reuptake at the synaptic cleft. In our model, PG (re)uptake after release is mediated by the transporter PGT (6, 12, 28, 31, 35).
Controlled PG reuptake may be especially important in the renal CD, where PGE2 is by far the major PG synthesized (8, 13) and where the spatial determinants of PGE2 signaling are unusually nuanced. In the renal CD, the predominant PGE2 receptors are EP1 and EP3 (9, 26, 32); these appear to be located on the basolateral membrane, where they inhibit Na+ resorption (5, 38). The EP4 receptor is predominantly expressed in the glomerulus but is also expressed at a low level in the CD (9, 32), where it is likely localized to the luminal/apical membrane and stimulates Na+ resorption (5, 38). In a CD cell model system, PGT is expressed with fidelity at the apical plasma membrane, where it extracts PGE2 from the apical (urinary) solution, transports it across the CD epithelium, and delivers it, essentially intact, to the basolateral solution (12). Thus in situ PGT in the CD would be predicted to reduce luminal PGE2 and minimize the stimulation of apical (antinatriuretic) EP4 receptors, while simultaneously increasing basolateral PGE2 and stimulating basolateral (natriuretic) EP1 and/or EP3 receptors.
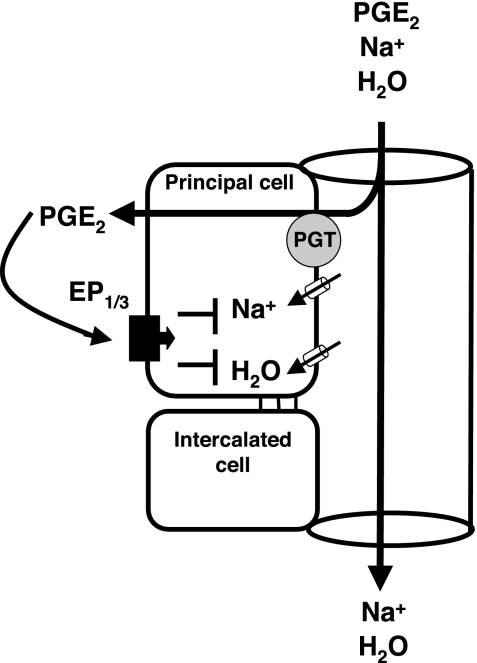
Our present findings are in accord with an emerging picture in which high dietary sodium intake induces expression of medullary COX (24, 51), CD PGE synthase (27), collecting EP3 (but not EP4) (25), and CD PGT (present results). This coordinated response would generate PGE2 and position it at basolateral EP3 receptors in the CD so as to reduce Na+ resorption and thus induce natriuresis (Fig. 7). PGT represents a novel element in this schema, in so far as part of the control of PG signaling appears to occur at the level of PG metabolism.
Fig. 7.
Model of PGT facilitated enhanced prostaglandin E2 (PGE2) stimulation of Na+ and water excretion. Upon sodium loading, cyclooxygenase-2 in kidney is induced resulting in more PGE2. The PGE2 level is increased on both luminal and basolateral sides of the collecting duct. PGT is located on the luminal side of the collecting duct. Upon salt loading, PGT is also increased and takes up more luminal PGE2 into the principal cells. The concentrated PGE2 in the principal cells diffuses into the basolateral side of collecting duct and stimulates Na+ and water excretion by interacting with its basolateral receptors EP1 and EP3.
GRANTS
This work was supported by National Institutes of Health Grants RO1-DK49688 and P50-DK064236.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Agnoli GC, Borgatti R, Cacciari M, Garutti C, Ikonomu E, Lenzi P, Stipo L. Responses of the renal prostanoids to a short-term depletion of sodium or potassium in healthy women. Boll Soc Ital Biol Sper 72: 109–116, 1996. [PubMed] [Google Scholar]
- 2.Agnoli GC, Borgatti R, Cacciari M, Ikonomu E, Lenzi P, Marinelli M. Effective role of the renin-angiotensin system in the control of prostanoid synthesis and renal function in healthy women with moderate salt depletion. Clin Physiol 16: 41–59, 1996. [DOI] [PubMed] [Google Scholar]
- 3.Agnoli GC, Borgatti R, Cacciari M, Lenzi P, Marinelli M, Stipo L. Effects of experimental salt depletion on urinary prostanoid excretions in normal women. Prostaglandins Leukot Essent Fatty Acids 58: 237–242, 1998. [DOI] [PubMed] [Google Scholar]
- 4.Agnoli GC, Borgatti R, Cacciari M, Lenzi P, Marinelli M, Stipo L. Renal prostanoids: physiological relevance in healthy salt-depleted women. Clin Physiol 19: 22–31, 1999. [DOI] [PubMed] [Google Scholar]
- 5.Ando Y, Asano Y. Luminal prostaglandin E2 modulates sodium and water transport in rabbit cortical collecting ducts. Am J Physiol Renal Fluid Electrolyte Physiol 268: F1093–F1101, 1995. [DOI] [PubMed] [Google Scholar]
- 6.Banu SK, Arosh JA, Chapdelaine P, Fortier MA. Molecular cloning and spatio-temporal expression of the prostaglandin transporter: a basis for the action of prostaglandins in the bovine reproductive system. Proc Natl Acad Sci USA 100: 11747–11752, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao Y, Pucci ML, Chan BS, Lu R, Ito S, Schuster VL. Prostaglandin transporter PGT is expressed in cell types that synthesize and release prostanoids. Am J Physiol Renal Physiol 282: F1103–F1110, 2002. [DOI] [PubMed] [Google Scholar]
- 8.Bonvalet JP, Pradelles P, Farman N. Segmental synthesis and actions of prostaglandins along the nephron. Am J Physiol Renal Fluid Electrolyte Physiol 253: F377–F387, 1987. [DOI] [PubMed] [Google Scholar]
- 9.Breyer MD, Davis L, Jacobson HR, Breyer RM. Differential localization of prostaglandin E receptor subtypes in human kidney. Am J Physiol Renal Fluid Electrolyte Physiol 270: F912–F918, 1996. [DOI] [PubMed] [Google Scholar]
- 10.Campean V, Theilig F, Paliege A, Breyer M, Bachmann S. Key enzymes for renal prostaglandin synthesis: site-specific expression in rodent kidney (rat, mouse). Am J Physiol Renal Physiol 285: F19–F32, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Davila D, Davila T, Oliw E, Anggard E. The influence of dietary sodium on urinary prostaglandin excretion. Acta Physiol Scand 103: 100–106, 1978. [DOI] [PubMed] [Google Scholar]
- 12.Endo S, Nomura T, Chan BS, Lu R, Pucci ML, Bao Y, Schuster VL. Expression of PGT in MDCK cell monolayers: polarized apical localization and induction of active PG transport. Am J Physiol Renal Physiol 282: F618–F622, 2002. [DOI] [PubMed] [Google Scholar]
- 13.Farman N, Pradelles P, Bonvalet JP. PGE2, PGF2 α, 6-keto-PGF1 α, and TxB2 synthesis along the rabbit nephron. Am J Physiol Renal Fluid Electrolyte Physiol 252: F53–F59, 1987. [DOI] [PubMed] [Google Scholar]
- 14.Fushimi K, Sasaki S, Yamamoto T, Hayashi M, Furukawa T, Uchida S, Kuwahara M, Ishibashi K, Kawasaki M, Kihara I, Marumo F. Functional characterization and cell immunolocalization of AQP-CD water channel in kidney collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 267: F573–F582, 1994. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Perez A, Smith WL. Apical-basolateral membrane asymmetry in canine cortical collecting tubule cells. Bradykinin, arginine vasopressin, prostaglandin E2 interrelationships. J Clin Invest 74: 63–74, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginzinger DG, Godfrey TE, Nigro J, Moore DH 2nd, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res 60: 5405–5409, 2000. [PubMed] [Google Scholar]
- 17.Granfar RM, Day CJ, Kim MS, Morrison NA. Optimised real-time quantitative PCR assays for RANKL regulated genes. Mol Cell Probes 19: 119–126, 2005. [DOI] [PubMed] [Google Scholar]
- 18.Guan Y, Chang M, Cho W, Zhang Y, Redha R, Davis L, Chang S, Du Bois RN, Hao CM, Breyer M. Cloning, expression, and regulation of rabbit cyclooxygenase-2 in renal medullary interstitial cells. Am J Physiol Renal Physiol 273: F18–F26, 1997. [DOI] [PubMed] [Google Scholar]
- 19.Guan Y, Zhang Y, Breyer RM, Fowler B, Davis L, Hebert RL, Breyer MD. Prostaglandin E2 inhibits renal collecting duct Na+ absorption by activating the EP1 receptor. J Clin Invest 102: 194–201, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guyton AC Blood pressure control—special role of the kidneys and body fluids. Science 252: 1813–1816, 1991. [DOI] [PubMed] [Google Scholar]
- 21.Haas JA, Hammond TG, Granger JP, Blaine EH, Knox FG. Mechanism of natriuresis during intrarenal infusion of prostaglandins. Am J Physiol Renal Fluid Electrolyte Physiol 247: F475–F479, 1984. [DOI] [PubMed] [Google Scholar]
- 22.Hebert RL, Jacobson HR, Fredin D, Breyer MD. Evidence that separate PGE2 receptors modulate water and sodium transport in rabbit cortical collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 265: F643–F650, 1993. [DOI] [PubMed] [Google Scholar]
- 23.Jabs K, Zeidel ML, Silva P. Prostaglandin E2 inhibits Na+-K+-ATPase activity in the inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 257: F424–F430, 1989. [DOI] [PubMed] [Google Scholar]
- 24.Jensen BL, Kurtz A. Differential regulation of renal cyclooxygenase mRNA by dietary salt intake. Kidney Int 52: 1242–1249, 1997. [DOI] [PubMed] [Google Scholar]
- 25.Jensen BL, Mann B, Skott O, Kurtz A. Differential regulation of renal prostaglandin receptor mRNAs by dietary salt intake in the rat. Kidney Int 56: 528–537, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Jensen BL, Stubbe J, Hansen PB, Andreasen D, Skott O. Localization of prostaglandin E2 EP2 and EP4 receptors in the rat kidney. Am J Physiol Renal Physiol 280: F1001–F1009, 2001. [DOI] [PubMed] [Google Scholar]
- 27.Jia Z, Zhang A, Zhang H, Dong Z, Yang T. Deletion of microsomal prostaglandin E synthase-1 increases sensitivity to salt loading and angiotensin II infusion. Circ Res 99: 1243–1251, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Kanai N, Lu R, Satriano JA, Bao Y, Wolkoff AW, Schuster VL. Identification and characterization of a prostaglandin transporter. Science 268: 866–869, 1995. [DOI] [PubMed] [Google Scholar]
- 29.Katayama S, Attallah AA, Stahl RA, Bloch DL, Lee JB. Mechanism of furosemide-induced natriuresis by direct stimulation of renal prostaglandin E2. Am J Physiol Renal Fluid Electrolyte Physiol 247: F555–F561, 1984. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 31.Lu R, Kanai N, Bao Y, Schuster VL. Cloning, in vitro expression, and tissue distribution of a human prostaglandin transporter cDNA (hPGT). J Clin Invest 98: 1142–1149, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morath R, Klein T, Seyberth HW, Nusing RM. Immunolocalization of the four prostaglandin E2 receptor proteins EP1, EP2, EP3, and EP4 in human kidney. J Am Soc Nephrol 10: 1851–1860, 1999. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen S, DiGiovanni SR, Christensen EI, Knepper MA, Harris HW. Cellular and subcellular immunolocalization of vasopressin-regulated water channel in rat kidney. Proc Natl Acad Sci USA 90: 11663–11667, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura T, Chang HY, Lu R, Hankin J, Murphy RC, Schuster VL. Prostaglandin signaling in the renal collecting duct: release, reuptake, and oxidation in the same cell. J Biol Chem 280: 28424–28429, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Nomura T, Lu R, Pucci ML, Schuster VL. The two-step model of prostaglandin signal termination: in vitro reconstitution with the prostaglandin transporter and prostaglandin 15 dehydrogenase. Mol Pharmacol 65: 973–978, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Pucci ML, Endo S, Nomura T, Lu R, Khine C, Chan BS, Bao Y, Schuster VL. Coordinate control of prostaglandin E2 synthesis and uptake by hyperosmolarity in renal medullary interstitial cells. Am J Physiol Renal Physiol 290: F641–F649, 2006. [DOI] [PubMed] [Google Scholar]
- 37.Rossier BC Negative regulators of sodium transport in the kidney: key factors in understanding salt-sensitive hypertension? J Clin Invest 111: 947–950, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakairi Y, Jacobson HR, Noland TD, Breyer MD. Luminal prostaglandin E receptors regulate salt and water transport in rabbit cortical collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 269: F257–F265, 1995. [DOI] [PubMed] [Google Scholar]
- 39.Scherer B, Siess W, Weber PC. Radioimmunological and biological measurement of prostaglandins in rabbit urine: decrease of PGE2 excretion at high NaCl intake. Prostaglandins 13: 1127–1139, 1977. [DOI] [PubMed] [Google Scholar]
- 40.Schuster VL Molecular mechanisms of prostaglandin transport. Annu Rev Physiol 60: 221–242, 1998. [DOI] [PubMed] [Google Scholar]
- 41.Schuster VL Prostaglandin transport. Prostaglandins Other Lipid Mediat 68–69: 633–647, 2002. [DOI] [PubMed] [Google Scholar]
- 42.Smith WL, Bell TG. Immunohistochemical localization of the prostaglandin-forming cyclooxygenase in renal cortex. Am J Physiol Renal Fluid Electrolyte Physiol 235: F451–F457, 1978. [DOI] [PubMed] [Google Scholar]
- 43.Somova L, Zaharieva S, Ivanova M. Humoral factors involved in the regulation of sodium-fluid balance in normal man. I. Effect of dietary sodium chloride intake on renal prostaglandins, vasopressin and renin-angiotensin-aldosterone system. Acta Physiol Pharmacol Bulg 10: 21–28, 1984. [PubMed] [Google Scholar]
- 44.Stahl RA, Jonassen AJ, Paravicini M, Schollmeyer P. Sodium chloride as regulator of renal prostaglandin E2 production in patients with essential hypertension. Klin Wochenschr 60: 579–581, 1982. [DOI] [PubMed] [Google Scholar]
- 45.Stokes JB Integrated actions of renal medullary prostaglandins in the control of water excretion. Am J Physiol Renal Fluid Electrolyte Physiol 240: F471–F480, 1981. [DOI] [PubMed] [Google Scholar]
- 46.Strange K Volume regulation following Na+ pump inhibition in CCT principal cells: apical K+ loss. Am J Physiol Renal Fluid Electrolyte Physiol 258: F732–F740, 1990. [DOI] [PubMed] [Google Scholar]
- 47.Tan SY, Sandwisch DW, Mulrow PJ. Sodium intake as a determinant of urinary prostaglandin E2 excretion. Prostaglandins Med 4: 53–63, 1980. [DOI] [PubMed] [Google Scholar]
- 48.Wegmann M, Nusing RM. Prostaglandin E2 stimulates sodium reabsorption in MDCK C7 cells, a renal collecting duct principal cell model. Prostaglandins Leukot Essent Fatty Acids 69: 315–322, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Yang T Regulation of cyclooxygenase-2 in renal medulla. Acta Physiol Scand 177: 417–421, 2003. [DOI] [PubMed] [Google Scholar]
- 50.Yang T, Schnermann JB, Briggs JP. Regulation of cyclooxygenase-2 expression in renal medulla by tonicity in vivo and in vitro. Am J Physiol Renal Physiol 277: F1–F9, 1999. [DOI] [PubMed] [Google Scholar]
- 51.Yang T, Singh I, Pham H, Sun D, Smart A, Schnermann JB, Briggs JP. Regulation of cyclooxygenase expression in the kidney by dietary salt intake. Am J Physiol Renal Physiol 274: F481–F489, 1998. [DOI] [PubMed] [Google Scholar]