Abstract
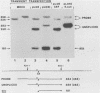
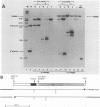
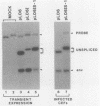
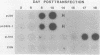
The full-length retroviral RNA transcript serves as (i) mRNA for the gag and pol gene products, (ii) genomic RNA that is assembled into progeny virions, and (iii) a pre-mRNA for spliced subgenomic mRNAs. Therefore, a balance of spliced and unspliced RNA is required to generate the appropriate levels of protein and RNA products for virion production. We have introduced an insertion mutation near the avian sarcoma virus env splice acceptor site that results in a significant increase in splicing to form functional env mRNA. The mutant virus is replication defective, but phenotypic revertant viruses that have acquired second-site mutations near the splice acceptor site can be isolated readily. Detailed analysis of one of these viruses revealed that a single nucleotide change at -20 from the splice acceptor site, within the original mutagenic insert, was sufficient to restore viral growth and significantly decrease splicing efficiency compared with the original mutant and wild-type viruses. Thus, minor sequence alterations near the env splice acceptor site can produce major changes in the balance of spliced and unspliced RNAs. Our results suggest a mechanism of control in which splicing is modulated by cis-acting sequences at the env splice acceptor site. Furthermore, this retroviral system provides a powerful genetic method for selection and analysis of mutations that affect splicing control.
Full text
PDF




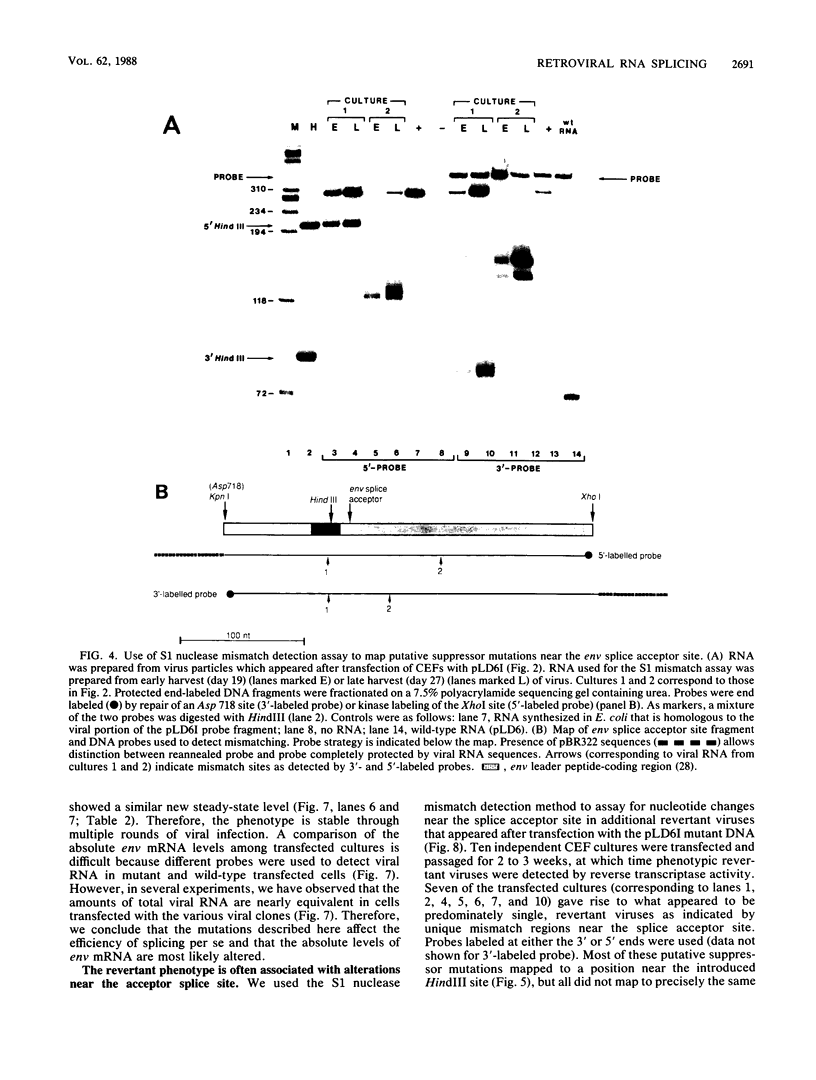
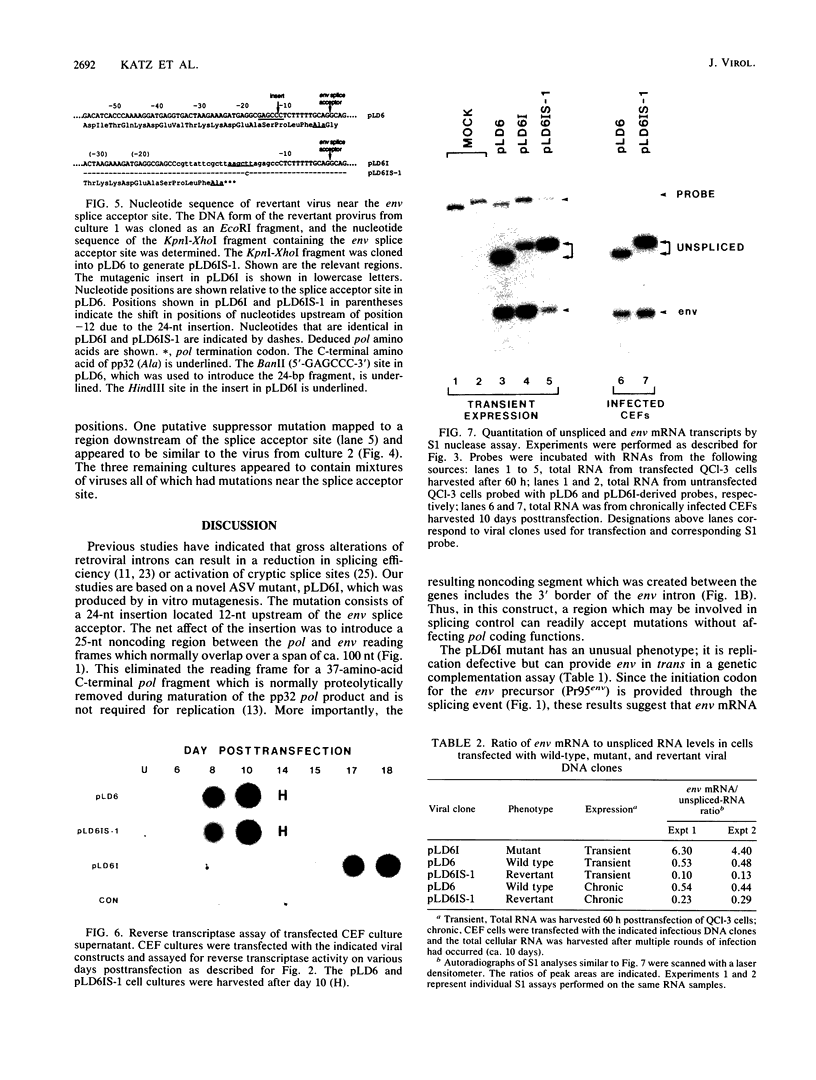



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Raymond K., Ju G. Functional analysis of the transcription control region located within the avian retroviral long terminal repeat. Mol Cell Biol. 1985 Mar;5(3):438–447. doi: 10.1128/mcb.5.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M. B., Jarrett R. F., Aldovini A., Gallo R. C., Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986 Sep 12;46(6):807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- Ficht T. A., Chang L. J., Stoltzfus C. M. Avian sarcoma virus gag and env gene structural protein precursors contain a common amino-terminal sequence. Proc Natl Acad Sci U S A. 1984 Jan;81(2):362–366. doi: 10.1073/pnas.81.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D., Quinn T., Hippenmeyer P. J., Oroszlan S. Structural characterization of the avian retrovirus reverse transcriptase and endonuclease domains. J Biol Chem. 1985 Jul 15;260(14):8243–8249. [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J., Yoshida M., Seiki M. Transcriptional (p40x) and post-transcriptional (p27x-III) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. A C-terminal domain in the avian sarcoma-leukosis virus pol gene product is not essential for viral replication. J Virol. 1988 Feb;62(2):528–533. doi: 10.1128/jvi.62.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Terry R. W., Skalka A. M. A conserved cis-acting sequence in the 5' leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986 Jul;59(1):163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomedico P. T. Use of recombinant DNA technology to program eukaryotic cells to synthesize rat proinsulin: a rapid expression assay for cloned genes. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5798–5802. doi: 10.1073/pnas.79.19.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. Insertion of several different DNAs in reticuloendotheliosis virus strain T suppresses transformation by reducing the amount of subgenomic mRNA. J Virol. 1986 Apr;58(1):75–80. doi: 10.1128/jvi.58.1.75-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M. A., Brizzard B. L., Wong J. L., Murphy E. C., Jr Murine sarcoma virus ts110 RNA transcripts: origin from a single proviral DNA and sequence of the gag-mos junctions in both the precursor and spliced viral RNAs. J Virol. 1985 Feb;53(2):624–633. doi: 10.1128/jvi.53.2.624-633.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Lorenzen S. K., Berberich S. L. Noncoding region between the env and src genes of Rous sarcoma virus influences splicing efficiency at the src gene 3' splice site. J Virol. 1987 Jan;61(1):177–184. doi: 10.1128/jvi.61.1.177-184.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Hardwick J. M., Shaw K., Hunter E. Alterations in the transport and processing of Rous sarcoma virus envelope glycoproteins mutated in the signal and anchor regions. J Cell Biochem. 1983;23(1-4):81–94. doi: 10.1002/jcb.240230109. [DOI] [PubMed] [Google Scholar]