Abstract
The rapid modulation of ligand-binding affinity (“activation”) is a central property of the integrin family of cell adhesion receptors. The small GTP-binding protein Ras and its downstream effector kinase Raf-1 suppress integrin activation. In this study we explored the relationship between Ras and the closely related small GTP-binding protein R-Ras in modulating the integrin affinity state. We found that R-Ras does not seem to be a direct activator of integrins in Chinese hamster ovary cells. However, we observed that GTP-bound R-Ras strongly antagonizes the Ras/Raf-initiated integrin suppression pathway. Furthermore, this reversal of the Ras/Raf suppressor pathway does not seem to be via a competition between Ras and R-Ras for common downstream effectors or via an inhibition of Ras/Raf-induced MAP kinase activation. Thus, R-Ras and Ras may act in concert to regulate integrin affinity via the activation of distinct downstream effectors.
INTRODUCTION
Integrins are heterodimeric cell–cell and cell–matrix adhesion receptors that play a key role in cell growth, survival, migration, and tumor metastasis (Hynes, 1992; Schwartz et al., 1995). A characteristic feature of specific integrins is their ability to modulate dynamically their affinity for ligand in response to intracellular signals, a process referred to as “inside-out” signaling or “activation” (Hughes and Pfaff, 1998). Integrin activation is a cell type–specific and energy-dependent process, requiring both the α and β subunit cytoplasmic domains (O’Toole et al., 1994; Hughes et al., 1996; Hughes and Pfaff, 1998).
Currently the cytoplasmic-signaling pathways regulating integrin affinity are incompletely understood. However, a number of recent studies indicate that the Ras family of small GTP-binding proteins and their downstream effectors play a central role in regulating integrin affinity (Shimizu and Hunt, 1996; Z. Zhang et al., 1996; Hughes et al., 1997). The Ras family of proteins functions as molecular switches that are controlled by a GDP/GTP-binding cycle (Bos, 1997). H-Ras and its downstream effector kinase Raf-1 can suppress integrin activation in Chinese hamster ovary (CHO) cells. This suppressive effect is independent of protein synthesis and mRNA transcription and correlates with the activation of the ERK MAP kinase pathway (Hughes et al., 1997). Furthermore R-Ras, a small GTP-binding protein homologous to H-Ras, influences integrin activation. In contrast to H-Ras, activated R-Ras stimulates ligand binding to integrins (Z. Zhang et al., 1996).
R-Ras was originally identified because of its similarity to the H-Ras, K-Ras, and N-Ras oncogenes, being ∼55% identical to each (Lowe et al., 1987). Currently, little is known about R-Ras function and how it compares with that of the Ras proteins. Despite the considerable sequence similarity to the other Ras proteins, several observations suggest that the functions of R-Ras are distinct. For example, activating mutations in H-Ras, K-Ras, or N-Ras will induce the morphological transformation of a variety of fibroblasts and epithelial cell lines (Bos, 1997). In contrast, activated R-Ras causes the transformation of a much more limited spectrum of cell types (Cox et al., 1994; Huff et al., 1997).
R-Ras and the other Ras proteins have highly homologous effector-binding domains; consequently both GTP-bound R-Ras and Ras bind to several common effectors. Like Ras, R-Ras interacts with the p110 catalytic subunit of phosphatidylinositol 3-kinase (PI 3-kinase) in vitro and induces the elevation of the levels of PI 3-kinase lipid products in vivo (Marte et al., 1996). R-Ras also interacts with the Raf serine/threonine kinases and exchange factors for the Ras-related Ral small GTP-binding proteins (Vojtek et al., 1993; Spaargaren and Bischoff, 1994; Spaargaren et al., 1994). However, in contrast to Ras, R-Ras does not activate Raf or Ral guanine nucleotide exchange activity in vivo (Marte et al., 1996; Urano et al., 1996). The GTP-bound state of R-Ras seems to be regulated differently from that of Ras. Both Ras and R-Ras interact with the GTPase-activating proteins p120 Ras GAP, neurofibromin, and p98 R-Ras GAP (Garrett et al., 1989; Rey et al., 1994; Yamamoto et al., 1995). However, in vivo it is thought that p120 GAP and neurofibromin primarily serve as GAPs for Ras and that p98 R-Ras GAP primarily serves as a R-Ras GAP. The physiological stimuli that activate R-Ras in vivo are not known. R-Ras is not activated by SOS or GRF1 and GRF2, guanine nucleotide exchange factors for Ras, and currently no R-Ras–specific guanine nucleotide factors have been identified (Shou et al., 1995; Fam et al., 1997; Huff et al., 1997).
In this study we explored the relationship between Ras and R-Ras in integrin affinity modulation. We found that R-Ras does not seem to be a direct activator of integrins in CHO cells. However, we observed that GTP-bound R-Ras strongly antagonizes the Ras/Raf-initiated integrin suppression pathway. Furthermore, this reversal of the Ras/Raf suppressor pathway does not seem to be via a competition between Ras and R-Ras for common downstream effectors or via an inhibition of Ras/Raf-induced MAP kinase activation. Thus, these observations suggest that R-Ras and Ras could act in concert to regulate integrin affinity via the activation of distinct and novel effectors.
MATERIALS AND METHODS
Antibodies and Reagents
The isolation and characterization of the anti-αIIbβ3 monoclonal antibodies anti-LIBS6 and D57 have been described previously (O’Toole et al., 1994). The activation-dependent anti-αIIbβ3 monoclonal antibody PAC1 was a generous gift of Dr. S. Shattil (Scripps Research Institute, La Jolla, CA) (Shattil et al., 1985). The anti-Tac antibody 7G7B6 was obtained from the American Type Culture Collection (Rockville, MD). Antibodies 7G7B6 and D57 were biotinylated with biotin-N-hydroxysuccinimide (Sigma, St. Louis, MO) according to the manufacturer’s directions. The αIIbβ3-specific peptidomimetic inhibitor Ro43-5054 was a generous gift of Dr. Beat Steiner (Hoffmann La Roche, Basel, Switzerland).
cDNA Constructs, Transfection, and Cell Lines
The CDM8 expression constructs encoding the αIIb chimera αIIbα6A, composed of the αIIb extracellular and transmembrane domain fused to the cytoplasmic domains of α6A, and the chimera β3β1, composed of the β3 extracellular and transmembrane domain fused to the cytoplasmic domain of β1, were constructed as described (O’Toole et al., 1994). The plasmid pDCR-H-Ras(G12V) was a generous gift of Dr. M. H. Wigler (Cold Spring Harbor laboratory, Cold Spring Harbor, NY). pcDNA3-R-Ras(G38V) was a gift of Dr. K. Vuori and Dr. E. Ruoslahti (The Burnham Institute, La Jolla, CA). The plasmids pMT2-HA-Rlf-CAAX, pMT2-HA-RalA, pMT2-HA-RalA(T28N), and pMT2-HA-RalA(G23V) were generous gifts of Dr. Rob Wolthuis (Utrecht University, Utrecht, Netherlands) (Wolthuis et al., 1997). The expression vectors encoding Tac-α5, Raf-BXB, hemagglutinin (HA)-tagged ERK2, HA-tagged Akt, and Myc-tagged R-Ras have been described previously (Marte et al., 1996; Hughes et al., 1997).
CHO-K1 cells were obtained from the American Type Culture Collection. The αβ-py cells were generated as described (Baker et al., 1997). All cell lines were grown in DMEM (BioWhittaker, Walkersville, MD) containing 10% fetal bovine serum, 1% nonessential amino acids, 2 mM glutamine (Sigma), 100 U/ml penicillin, and 100 μg/ml streptomycin. Transient transfections were undertaken using lipofection (lipofectamine; Life Technologies, Gaithersburg, MD) as described previously (Hughes et al., 1996).
Flow Cytometry
For single-color FAC analysis, 5 × 105 cells were incubated on ice for 30 min with the primary antibody, washed, and then incubated on ice for a further 30 min with an FITC-conjugated goat anti-mouse IgG (Tago, Burlingame, CA) secondary antibody. Cells were pelleted, resuspended, and analyzed on a FACScan (Becton Dickinson, Mountain View, CA). PAC1 binding was analyzed by two-color flow cytometry. Cell staining was performed in DMEM and 1 mg/ml BSA (Sigma). Single-cell suspensions were obtained by incubating cells for 5 min in trypsin and EDTA (Worthington, Freehold, NJ) and diluting with an equal volume of DMEM containing 10% FCS. After washing, 5 × 105 cells were incubated in a final volume of 50 μl containing 0.1% PAC1 ascites in the presence or absence of the competitive inhibitor Ro43-5054 at 1 μM. After a 30-min incubation at room temperature, cells were washed with cold DMEM solution and then incubated on ice with DMEM containing either the biotinylated anti-Tac antibody 7G7B6 or biotinylated D57. After 30 min on ice, the cells were washed and incubated with 10% FITC-conjugated goat anti-mouse IgM (Tago) and 4% phycoerythrin–streptavidin (Molecular Probes, Eugene, OR). Thirty minutes later, cells were washed with 0.5 ml of cold PBS and resuspended in 0.5 ml of cold PBS. The cells were then analyzed on a FACScan (Becton Dickinson) flow cytometer as described (Hughes et al., 1996).
In transiently transfected αβ-py cells, PAC1 binding (FITC staining) was analyzed only on a gated subset of cells positive for Tac-α5 expression (phycoerythrin staining). To define the affinity state, histograms depicting PAC1 staining in the absence or presence of the competitive inhibitor Ro43-5054 were superimposed. Because the peptide mimetic Ro43-5054 is an inhibitor of ligand binding to αIIbβ3, a leftward shift in the histogram in the presence of inhibitor is indicative of the presence of high-affinity αIIbβ3.
To obtain numerical estimates of integrin activation, we calculated an activation index (AI) defined as 100 × (Fo − Fr)/(FoLIBS6 − Fr), where Fo is the median fluorescence intensity of PAC1 binding, Fr is the median fluorescence intensity of PAC1 binding in the presence of competitive inhibitor (Ro43-5054, 1 μM), and FoLIBS6 is the median fluorescence intensity of PAC1 binding in the presence of 2 μM anti-LIBS6. Percent inhibition was calculated by 100(AI0 − AI)/AI0, where AI0 is the activation index in the absence of the cotransfected suppressor and AI is the activation index in its presence.
Measurement of ERK2 and Akt Activity
For ERK2 assays, 2 × 105 cells were transfected using lipofectamine (Life Technologies) with 2 μg of pCMV5 HA-ERK2. The cells were also transfected with 2 μg of the test plasmid [e.g., pDCR-H-Ras(G12V)]. In some experiments, 4–6 μg of a second plasmid [e.g., R-Ras(G38V)] were cotransfected, and the total amount of DNA was standardized at 10 μg, by the addition of pcDNA3, for each transfection. Transfections were done in duplicate to allow parallel analysis of both ERK2 kinase activity and PAC1 binding by flow cytometry, as described above. Forty-eight hours after transfection, cells were harvested and lysed in 0.5% Nonidet P-40 (NP-40) buffer containing phosphatase inhibitors (10 mM sodium pyrophosphate, 10 mM NaF, 3 mM β glycerophosphate, and 1 mM Na3VO4) in addition to protease inhibitors. The HA-ERK2 was immunoprecipitated by the anti-HA antibody 12CA5, and its activity was assessed by an immune-complex kinase assay using myelin basic protein as a substrate. ERK2 expression and recovery were monitored by fractionating 25 μg of whole-cell lysate or one-fifth of the 12CA5 immunoprecipitate on 12.5% SDS-polyacrylamide gels, transferring to Immobilon (Millipore, Bedford, MA) membranes, and immunoblotting with the anti-HA antibody 12CA5 or polyclonal anti-ERK2 (Santa Cruz Biotechnology, Tebu, France).
For Akt kinase assays, 2 × 105 cells were transfected using the lipofectamine method with 2 μg of pSG5-HA-AKT. The cells were also transfected with the appropriate test plasmids, and the total amount of DNA in each transfection was then adjusted to 8 μg by the addition of pcDNA3. Tansfections were done in duplicate to allow parallel analysis of both Akt activity and PAC1 binding. Forty-eight hours after transfection, cells were lysed with 1.0% NP-40 buffer containing phosphatase inhibitors (10 mM sodium pyrophosphate, 10 mM NaF, 3 mM β glycerophosphate, and 1 mM Na3VO4) in addition to protease inhibitors. HA-Akt was immunoprecipitated with the anti-HA antibody 12CA5, as described for the ERK2 kinase assay. The immunoprecipitates were washed two times in cell lysis buffer, followed by two washes in high-salt buffer (0.5 M LiCl, 0.1 M Tris, pH 8.0, and 1 mM EDTA) and a final wash in nonreducing kinase buffer (50 mM Tris, pH 7.5, and 10 mM MgCl2). The immunoprecipitates were resuspended in kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, and 1 mM DTT) and reacted with 2.5 μg of histone 2B, as described (Marte et al., 1996). After incubation at room temperature for 20 min, the reaction was stopped with SDS sample buffer. The samples were then subjected to SDS-PAGE on 16% gels; the gels were dried down and visualized by autoradiography. HA-Akt expression was monitored by fractionating 25 μg of whole-cell lysate on 4–20% SDS-polyacrylamide gels, transferring to Immobilon (Millipore) membranes, and immunoblotting with the anti-HA antibody 12CA5.
Ral Activation Assay
The GTP-bound form of Ral was specifically pulled down from clarified cell lysates by incubation with the GST-tagged form of the Ral-binding domain (RalBD) of RLIP76, as described (Wolthuis et al., 1998). Cells (2 × 105) were transfected using the lipofectamine method with the indicated HA-Ral and HA-Rlf-CAAX constructs and with the total amount of DNA in each transfection adjusted to 8 μg by the addition of pcDNA3. The cells were then washed after transfection, and after 24 h the cells were maintained in media containing 0.5% FCS. The cells were then washed twice with cold PBS and lysed on ice in Ral-binding buffer (15% glycerol, 1% NP-40, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM MgCl2) containing protease inhibitors. Lysates were then clarified by centrifugation, and the supernatants of each sample were incubated with 15 μg of GST-RalBD precoupled to glutathione beads. Samples were then incubated for 1 h on a tumbler at 4°C followed by four washes in Ral-binding buffer. The beads were boiled in Laemmli sample buffer and subjected to SDS-PAGE and Western blotting with the anti-HA antibody 12CA5 to assay the recovery of HA-Ral. In parallel HA-Ral and HA-Rlf-CAAX expression was monitored by fractionating 20 μg of whole-cell lysate on a 4–20% SDS-polyacrylamide gel, followed by transfer to Immobilon (Millipore) membranes and immunoblotting with the anti-HA antibody 12CA5.
RESULTS
R-Ras Does Not Directly Activate the Integrin αIIbβ3 in CHO Cells
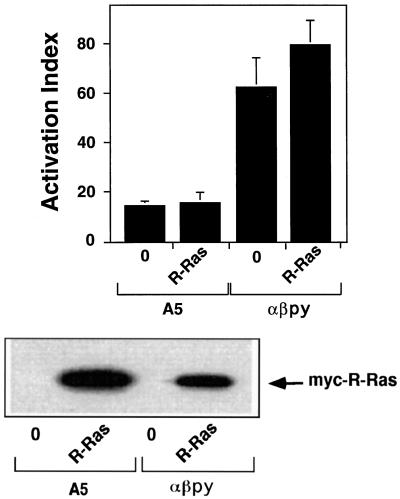
To gain further insight into the role of R-Ras in integrin affinity modulation, we tested the effect of transfecting activated R-Ras on the affinity state of resting and active integrins expressed in CHO cells. When stably expressed in CHO cells (A5 cells), the platelet-specific integrin αIIbβ3 fails to bind activation-specific ligands with high affinity (O’Toole et al., 1990). To determine whether the expression of an activated variant of R-Ras [R-Ras(G38V)] could activate the αIIbβ3 in these cells, an expression vector encoding R-Ras(G38V) was transiently transfected into A5 cells. We assessed activation by the binding of PAC1, an antibody specific for the active conformation of αIIbβ3 (Shattil et al., 1985). Expression of activated R-Ras in A5 cells did not induce the activation of αIIbβ3 as determined by PAC1 binding (Figure 1). However, PAC1 binding to these A5 cells could be induced by the addition of an activating monoclonal antibody, anti-LIBS6 (our unpublished observations). We also tested the effect of transfecting R-Ras(G38V) into CHO cells stably expressing the active chimeric integrin αIIbα6Aβ3β1 (αβ-py cells). We found that transfection of activated R-Ras did not lead to a significant increase in PAC1 binding to αβ-py cells (Figure 1). Western blot analysis of lysates from both the transfected A5 and αβ-py cells revealed that R-Ras(G38V) was well expressed in both cell types (Figure 1).
Figure 1.
R-Ras(G38V) does not directly stimulate integrin αIIbβ3 activation in CHO cells. Both αβ-py and A5 cells were transiently transfected with 3 μg of an expression vector encoding R-Ras(G38V) and or an equivalent amount of an empty control vector. After 48 h, PAC1 binding was analyzed by flow cytometry as described in MATERIALS AND METHODS. Top, The mean activation indices ± SD of three independent experiments. Bottom, Immunoblot analysis of cell lysates illustrating the expression of myc-tagged R-Ras(G38V). Twenty micrograms of cell lysate from each transfection were separated on a 4–20% gradient gel and immunoblotted with the anti-Myc antibody 9E10.
The A5 and αβ-py cells are clonal cell lines. To ensure these results were not caused by artifacts of clonal selection, we examined PAC1 binding to parental CHO cells transiently transfected with expression vectors encoding wild-type αIIbβ3 and αIIbα6Aβ3β1 in the presence or absence of activated R-Ras. In agreement with the observations made in stably transfected cells, activated R-Ras had no significant effect on PAC1 binding (our unpublished observations). Thus, from these data we conclude that activated R-Ras does not directly influence the ligand-binding affinity of αIIbβ3 in CHO cells.
R-Ras Can Reverse the Suppressive Effect of Activated H-Ras and Raf-1
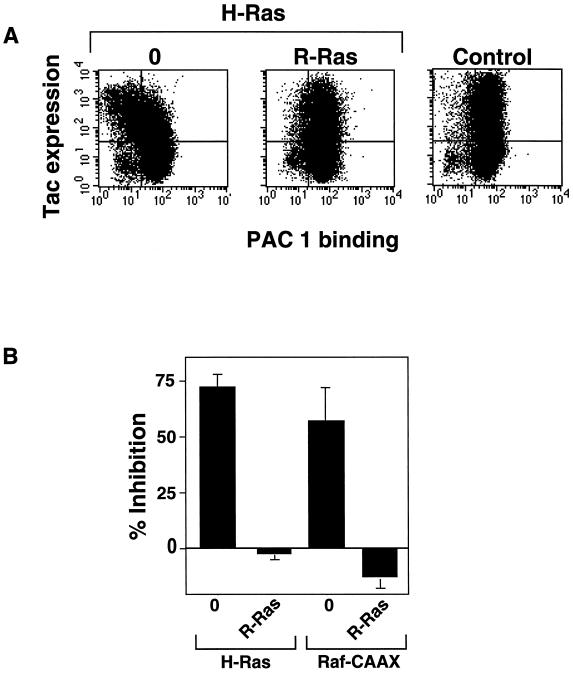
GTP-bound R-Ras did not activate the integrins tested in CHO cells. However, its activating effects might be attributable to it antagonizing the suppressive effect of Ras. To test this hypothesis, we examined the effect of R-Ras(G38V) on suppression caused by activated H-Ras. αβ-py cells were transiently transfected with H-Ras(G12V) in the presence or absence of R-Ras(G38V). H-Ras(G12V) alone caused marked inhibition of PAC1 binding (Figure 2, A and B); however, cotransfection with an expression vector encoding activated R-Ras completely reversed this suppression (Figure 2A). The average median fluorescence intensity (MFI) of PAC1 binding from five independent experiments in the control transfected αβ-PY cells was 37.8 ± 7.44; after transfection with H-Ras(G12V), this was reduced to 15.6 ± 3.92. The cotransfection of activated R-Ras restored the mean MFI to 41.4 ± 8.72. These MFIs are representative of those seen in subsequent experiments.
Figure 2.
Activated R-Ras rescues the suppression of integrin activation by H-Ras(G12V) and Raf-CAAX. (A) αβ-py cells were transiently transfected with 2 μg of an expression vector encoding Tac-α5 alone and Tac-α5 plus H-Ras(G12V). In a separate transfection Tac-α5 plus H-Ras(G12V) was cotransfected with 3 μg of a plasmid encoding R-Ras(G38V). After 48 h, cells were harvested and stained for Tac expression (y-axis) and PAC1 binding (x-axis). Left, In the H-Ras(G12V)-transfected cells, there is a leftward shift of the dot plot in the upper quadrants representing a reduction in PAC1 binding. Middle, This shift is reversed by the cotransfection with activated R-Ras(G38V). Right, In the empty vector control transfection, there was no suppression of PAC1 binding in the Tac-α5–expressing cells. (B) αβ-py cells were cotransfected with 4 μg of an expression vector encoding Raf-CAAX and H-Ras(G12V). In separate transfections Raf-CAAX or H-Ras(G12V) expression vectors were simultaneously cotransfected with 3 μg of a plasmid encoding R-Ras(G38V). After 48 h, integrin activation was determined by PAC1 binding. Depicted is the mean percent inhibition of integrin activation relative to that of the empty vector ± SE of three independent determinations.
We also tested the ability of R-Ras to reverse suppression mediated by an activated membrane-targeted variant of Raf, Raf-CAAX, and results similar to those observed with H-Ras(G12V) were seen (Figure 2B). Thus R-Ras(G38V) reversed the suppressive effect of activated H-Ras or Raf-1 on integrin affinity, suggesting that R-Ras can regulate the integrin activation state by modulating the activity of the Ras/Raf-1–dependent suppression pathway.
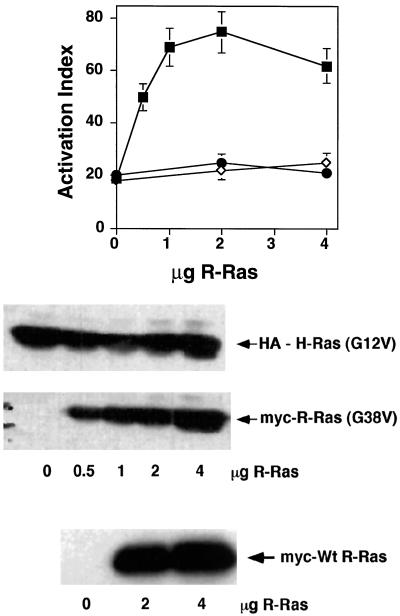
Small GTP-binding proteins must be in the GTP-bound conformation to bind and activate their downstream effectors (Bos, 1997). To determine whether R-Ras needs to be activated to reverse H-Ras suppression, we examined the effects of wild-type and putative dominant-negative R-Ras(S43N) on H-Ras suppression. Transient transfection of R-Ras(S43N) or wild-type R-Ras had no effect on H-Ras suppression of integrin affinity in αβ-py cells (Figure 3). In contrast, transfection of activated R-Ras(G38V) caused a concentration-dependent reversal of H-Ras(G12V) suppression. The cotransfection of R-Ras(G38V) (0.5–4 μg), at ratios of plasmid DNA as low as 1 to 8 with respect to H-Ras(G12V), still antagonized H-Ras suppression (Figure 3). Furthermore, Western blot analysis showed that increasing amounts of transfected R-Ras(G38V) cDNA had no effect on H-Ras expression (Figure 3), eliminating the possibility that the R-Ras “rescue” is caused by a reduction in the expression of H-Ras(G12V). Thus, these data suggest that R-Ras may influence integrin activation via a competition for a common downstream effector.
Figure 3.
Activated but not wild-type or dominant-negative R-Ras can reverse the suppressive effect of activated H-Ras(G12V). Top, αβ-py cells were cotransfected with an expression vector encoding H-Ras(G12V). They were simultaneously cotransfected with 2 and 4 μg of a plasmid encoding wild-type R-Ras (●) or R-Ras(S43N) (⋄) and 0.5, 1, 2, and 4 μg of a plasmid encoding R-Ras(G38V) (▪). After 48 h, integrin activation was determined by PAC1 binding. Depicted is the activation index in the presence of H-Ras(G12V) ± SE of three independent determinations. Bottom, The immunoblot analysis of cell lysates illustrates the expression of wild-type (Wt) R-Ras and R-Ras(G38V) in the presence of H-Ras(G12V). Twenty micrograms of cell lysate from each transfection were separated on a 4–20% gradient gel and immunoblotted with either an anti-HA antibody, 12CA5, or the anti-Myc antibody 9E10. The R-Ras(S43N) was also well expressed, as determined by immunoblot analysis of lysates from R-Ras(S43N)–transfected cells (our unpublished observations).
R-Ras Reversal of H-Ras Suppression Is Not Caused by Simple Competition between H-Ras and R-Ras for the Common Effector Raf-1
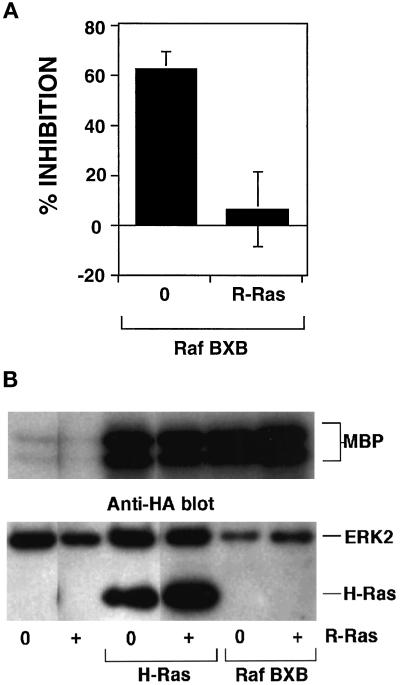
Suppression of integrin activation by H-Ras and its downstream effector kinase Raf-1 correlates with the activation of the ERK MAP kinase pathway. Furthermore, R-Ras binds to Raf in a GTP-dependent manner but fails to stimulate Raf kinase activity. Consequently, reversal of H-Ras suppression by R-Ras could be the result of R-Ras competition with H-Ras for Raf-1. To test this idea, we assessed the capacity of R-Ras(G38V) to reverse the suppressive effect of Raf-BXB, an activated variant of Raf-1 that, in contrast to Raf-CAAX, lacks a Ras-binding domain. As expected, transient transfection of αβ-py cells with Raf-BXB caused a marked suppression of integrin activation. However, the cotransfection of R-Ras(G38V) completely reversed suppression by Raf-BXB (Figure 4A). These data suggest that the R-Ras reversal of H-Ras–induced suppression is not caused by simple competition between the two G-proteins for the common effector Raf-1.
Figure 4.
R-Ras reversal of H-Ras– and Raf-1–initiated suppression is not caused by inhibition of Ras/Raf activation of ERK MAP kinase. (A) αβ-py cells were cotransfected with an expression vector encoding either Raf-BXB or a control cDNA. In a separate transfection Raf-BXB cDNA was cotransfected with a plasmid encoding activated R-Ras(G38V). After 48 h, integrin activation was determined by PAC1 binding. Depicted is the mean percent inhibition relative to that of the empty vector control ± SE of three independent determinations. (B) αβ-py cells were cotransfected with an expression vector encoding HA-tagged ERK2. The cells were also cotransfected with a control expression vector or vectors containing inserts encoding either R-Ras(G38V), Raf-BXB, or H-Ras(G12V). In separate transfections expression vectors encoding either Raf-BXB or H-Ras(G12V) were simultaneously cotransfected with a plasmid encoding R-Ras(G38V). The transfected ERK2 kinase was immunoprecipitated with the anti-HA antibody 12CA5. ERK-2 activity was measured by phosphorylation of myelin basic protein (MBP) using an immunocomplex kinase assay. Top, The relative ERK activation is shown. Bottom, Immunoblots with the anti-HA (12CA5) antibody illustrate the comparable expression of HA-tagged ERK2 in all transfections. The H-Ras(G12V) construct bore an HA-tag and was detected as the lower band in lanes transfected with that construct. Note the similar expression of recombinant activated H-Ras(G12V) in both the control and R-Ras(G38V)–cotransfected cells.
As described previously, H-Ras– and Raf-1–mediated suppression correlates with the activation of Raf and the ERK MAP kinase pathway. Consequently, activated R-Ras could rescue suppression by affecting the ability of H-Ras and Raf-1 to activate ERK2. However, the coexpression of R-Ras(G38V) with H-Ras(G12V) or Raf-BXB did not influence the ability of either to activate ERK2 kinase (Figure 4B). In addition, the coexpression of R-Ras(G38V) did not effect ERK2 activation induced by Raf-CAAX (our unpublished observations). As reported previously, transfection of R-Ras(G38V) alone did not activate ERK2 (Marte et al., 1996). These results indicate that the ability of R-Ras(G38V) to rescue the suppressive effects of H-Ras(G12V) and Raf-BXB is not caused by an inactivation of the ERK MAP kinase pathway and further argue against the notion that R-Ras competes with H-Ras for binding to Raf.
The Small GTP-binding Protein Ral and PI 3-Kinase Do Not Play a Role in H-Ras–dependent Suppression or R-Ras–mediated Rescue
The previously described experiments excluded competition for Raf binding as the mechanism for R-Ras reversal of Ras suppression. GTP-bound R-Ras and H-Ras can also bind to the p110 catalytic subunit of PI 3-kinase and Rlf, a guanine nucleotide exchange factor for the Ral family of small GTP-binding proteins. Consequently, we examined the role of these effectors in integrin affinity modulation.
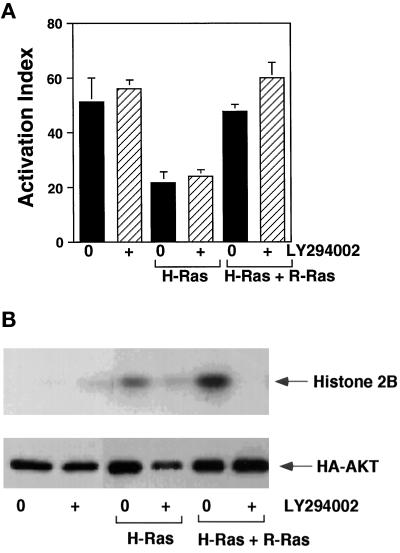
We used a PI 3-kinase inhibitor, LY294002, to test the role of PI 3-kinase in R-Ras’s capacity to oppose H-Ras as a suppressor of integrin activation. Pretreatment of αβ-py cells with 20 μM LY294002 for 24 h had no effect on basal activation of the αIIbβ3 chimera (see Figure 6A). Furthermore 20 μM LY294002 had little effect on the H-Ras(G12V)–induced suppression or the R-Ras rescue of integrin activation in αβ-py cells (Figure 5A). In parallel experiments, PI 3-kinase activity was assessed by measuring the in vitro kinase activity of the PI 3-kinase effector protein kinase B (PKB, Akt). LY294002 inhibited PI 3-kinase activity, and in the absence of detectable PI 3-kinase activation, R-Ras(G38V) was still able to reverse H-Ras(G12V) suppression (Figure 5, A and B). These results demonstrate that basal integrin activation in CHO cells is not affected by inhibition of PI 3-kinase. Furthermore, the modulation of integrin affinity by H-Ras and R-Ras is not via the activation of PI 3-kinase in these cells.
Figure 6.
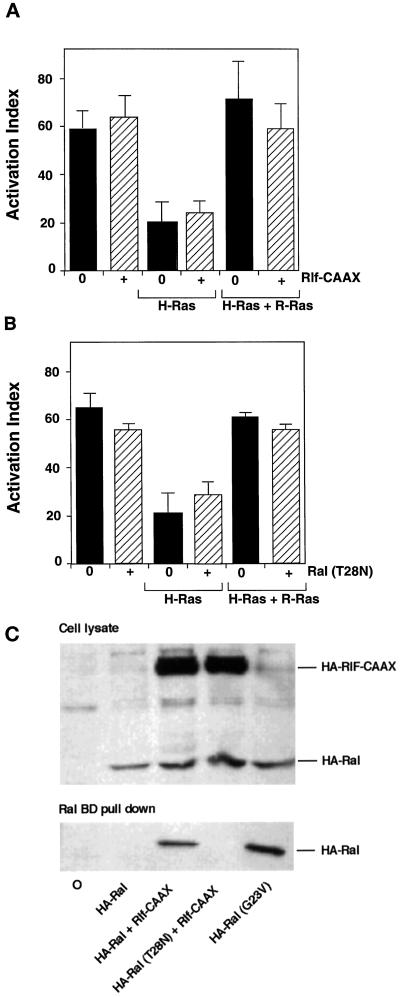
Ral activation does not mediate H-Ras– or R-Ras–dependent regulation of integrin affinity. (A) αβ-py cells were cotransfected with expression vectors encoding H-Ras(G12V),R-Ras(G38V), or Rlf-CAAX in the combinations depicted on the y-axis. After 48 h, integrin activation was determined by PAC1 binding. Depicted are the activation indices ± SE of three independent determinations. Immunoblot analysis of cell lysates demonstrated that HA-tagged Rlf-CAAX was well expressed in all conditions (our unpublished observations). (B) αβ-py cells were cotransfected with expression vectors encoding H-Ras(G12V), R-Ras(G38V), or dominant-negative RalA(T28N) in the combinations depicted on the y-axis. After 48 h, integrin activation was determined by PAC1 binding. Depicted are the activation indices ± SE of three independent determinations. (C) CHO cells were transfected with HA-RalA, HA-RalA(G23V), HA-RalA(T28N), and HA-Rlf-CAAX as indicated and grown in media containing 0.5% FCS before cell lysis. Ral-GTP was precipitated from the clarified cell lysates with glutathione-sepharose–bound GST-RalBD. Precipitated HA-Ral (bottom) and HA-Ral present in the cell lysate (top) were then identified by Western analysis using the anti-HA monoclonal 12CA5.
Figure 5.
PI 3-kinase activation does not mediate H-Ras– or R-Ras–dependent regulation of integrin affinity. (A) αβ-py cells were transiently transfected with either a control expression vector or vectors containing inserts encoding either R-Ras(G38V) and H-Ras(G12V) or H-Ras(G12V). Each transfection was performed in duplicate; 24 h after transfection, the PI 3-kinase inhibitor LY294002 was added at a final concentration of 20 μM to one of the duplicates. After 48 h, integrin activation was determined by PAC1 binding. Depicted are the activation indices ± SE of three independent determinations. (B) αβ-py cells were transiently transfected with an expression vector encoding HA-tagged Akt and either a control expression vector or vectors containing inserts encoding either R-Ras(G38V) and H-Ras(G12V) or H-Ras(G12V). Each transfection was performed in duplicate, and 24 h after transfection, the PI 3-kinase inhibitor LY294002 was added at a final concentration of 20 μM to one of the duplicates. Forty-eight hours after transfection, the cells were lysed, and the transfected Akt was immunoprecipitated with the anti-HA antibody 12CA5. Akt activity was then assayed using an immunocomplex kinase assay with histone 2B as a substrate. Top, The relative Akt activation is depicted; note the inhibition of Akt activity by LY294002. Bottom, Immunoblots with the anti-HA (12CA5) antibody illustrate comparable expression of HA-tagged Akt in all transfections.
The transient transfection of Rlf-CAAX [an activated membrane-targeted variant of the Ral guanine nucleotide exchange factor (Ral-GEF [Rlf])] or dominant-negative RalA(T28N) into αβ-py cells had no effect on basal integrin activation, as measured by PAC1 binding (Figure 6, A and B). Moreover, there was no effect of Rlf-CAAX or RalA(T28N) coexpression on the ability of H-Ras(G12V) to suppress activation of the chimeric integrin or of R-Ras(G38V) to rescue suppression (Figure 6, A and B). Furthermore, an H-Ras effector loop mutant, H-Ras(G12V, T35S), which interacts with Raf-1 but not with Ral-GEFs (Rodriguez-Viciana et al., 1997), was a potent suppressor of integrin activation in CHO cells (our unpublished observations), providing further evidence that the suppression of integrin activation by Ras is independent of Ral activation. The overexpression of an activated variant of RalA(G23V) was also tested as an alternative to an activated Ral exchange factor in these experiments, and this construct produced results similar to those observed with Rlf-CAAX (our unpublished observations).
In parallel experiments the activity of Rlf-CAAX and dominant-negative RalA(T28N) was measured by an affinity precipitation assay for GTP-bound Ral using the Ral-binding domain of the RLIP76 (Wolthuis et al., 1998). The cotransfection of Rlf-CAAX with RalA led to a substantial precipitation of GTP-bound RalA, compared with that observed after the transfection of RalA alone, demonstrating that Rlf-CAAX is a potent activator of Ral in CHO cells (Figure 6C). In contrast, in the presence of Rlf-CAAX, the dominant-negative RalA(T28N) was not precipitated by the GST-RalBD (Figure 6C), demonstrating that this variant exists in the GDP-bound state in CHO cells. Thus, activation or inhibition of the Ral arm of the Ras effector pathway does not contribute to modulation of integrin affinity by H-Ras and R-Ras in CHO cells.
DISCUSSION
H-Ras and its downstream effector kinase Raf-1 suppress integrin activation. Here we report that the small GTP-binding protein R-Ras regulates integrin affinity by modulating the activity of the Ras/Raf-initiated suppression pathway. The major findings of this article are as follows. First, activated R-Ras does not seem to be a direct activator of integrins in CHO cells. Second, GTP- but not GDP-bound R-Ras can reverse the suppressive effect of both activated H-Ras and its effector kinase Raf-1. Third, this property of activated R-Ras is not caused by simple competition between H-Ras and R-Ras for Raf-1 or guanine nucleotide exchange factors for the small GTP-binding protein Ral. Fourth, the ability of activated R-Ras to rescue H-Ras–initiated suppression did not correlate with the activation of PI 3-kinase. Furthermore, the inhibition of PI 3-kinase and Ral activity had no effect on basal integrin activation or the ability of activated H-Ras to suppress integrin activation in these cells. Taken together, these data suggest that R-Ras and H-Ras could act in concert to regulate the ligand-binding affinities of integrins via the activation of specific H-Ras and R-Ras effectors.
The expression of an activated variant of the small GTP-binding protein R-Ras [R-Ras(G38V)] did not stimulate high-affinity ligand binding to either wild-type αIIbβ3 or an active αIIbβ3 chimera. These data were obtained by transfecting R-Ras(G38V) into CHO cells stably expressing αIIbβ3 and the chimeric integrin αIIbα6Aβ3β1. When expressed in CHO cells (A5 cells), the platelet-specific integrin αIIbβ3 is the low-affinity conformation, as measured by the binding of activation-specific ligands such as PAC1 and fibrinogen (O’Toole et al., 1991). We found that the transfection of R-Ras(G38V) did not stimulate significant PAC1 binding to A5 cells, even though R-Ras(G38V) was well expressed. In agreement with our observation that R-Ras is not a direct activator of integrins in CHO cells, we also found that activated R-Ras failed to increase further PAC1 binding to CHO cells expressing the active integrin chimera αIIbα6Aβ3β1. We also found that R-Ras failed to stimulate PAC1 binding to parental CHO cells transiently transfected with αIIbβ3. This result is in contrast to that observed by Z. Zhang et al. (1996), who reported that activated R-Ras could stimulate PAC1 binding to CHO cells stably expressing αIIbβ3. It is possible that this apparent difference is caused by clonal variations in the CHO cell lines.
R-Ras(G38V) could reverse the suppressive effects of activated variants of both H-Ras and Raf-1. These results suggest that R-Ras could modulate integrin affinity by antagonizing the H-Ras/Raf-1–dependent suppressor pathway. The Ras GTPases function as molecular switches controlled by a GDP/GTP-binding cycle, binding downstream effectors only in the activated, GTP-bound conformation (Bos, 1997). R-Ras and the other Ras proteins have highly homologous effector-binding domains; consequently both GTP-bound R-Ras and H-Ras bind to several common effectors. Like H-Ras, R-Ras binds the p110 catalytic subunit of PI 3-kinase in vitro and induces an elevation in the levels of PI 3-kinase lipid products in vivo (Marte et al., 1996). R-Ras also binds the Raf serine/threonine kinases and Ral-GDS, an exchange factor for the Ras-related Ral GTP-binding proteins (Spaargaren and Bischoff, 1994; Marte et al., 1996). However, in contrast to Ras, R-Ras does not activate Raf or Ral-GDS in vivo (Marte et al., 1996; Urano et al., 1996). The observation that R-Ras can reverse the suppressive effect of activated H-Ras and its downstream effector kinase Raf-1 suggested that a possible mechanism for this effect of activated R-Ras is via a competition with H-Ras for common effectors. A precedent for such a model is illustrated by the Rap family of small GTP-binding proteins that have been reported to function as suppressors of Ras-mediated downstream signaling (Kitayama et al., 1989; Zhang et al., 1990; Cook et al., 1993). The antagonism between Ras and Rap function seems to be attributable to the ability of Rap and Ras to interact with the same downstream effectors, so that the GTP-bound Rap sequesters Ras effectors in inactive complexes (Bos, 1997). For example, Rap1 can suppress the activation of the ERK MAP kinase via the inactivation of Raf-1, which occurs upon its association with Rap1 (Boussiotis et al., 1997).
To explore this hypothesis further, we examined whether R-Ras needs to be in the activated, GTP-bound conformation to antagonize suppression mediated by activated H-Ras. We found that activated R-Ras(G38V) but not wild-type or the putative dominant-negative R-Ras(T43N) was able to reverse suppression. R-Ras(T43N) has a higher affinity for GDP than GTP, which indicates that this variant is in the inactive GDP-bound conformation and as such is unable to bind to downstream effectors (Huff et al., 1997). This result is consistent with the hypothesis that activated R-Ras is mediating reversal either via a competition with H-Ras for a common effector or via the activation of a specific downstream effector that stimulates an integrin activation pathway.
The R-Ras rescue of H-Ras–mediated suppression was not caused by competition between H-Ras and R-Ras for the common downstream effector Raf-1. The inhibition of integrin activation by activated H-Ras and Raf-1 correlates with activation of the ERK MAP kinase pathway. We found that whereas R-Ras potently reversed the suppressive effect of H-Ras and Raf, ERK2 activation induced by activated Raf and H-Ras was unaffected by cotransfection of activated R-Ras. This result indicates that R-Ras reversal is not caused by an inhibition of Ras- and Raf-induced MAP kinase activation. This result, combined with the observation that R-Ras can reverse suppression induced by an active Raf variant that lacks a Ras-binding domain, clearly demonstrates that R-Ras reversal is not a result of competition between H-Ras and R-Ras for Raf-1. We have shown previously that the MAP kinase phosphatase-1 can reverse Ras- and Raf-mediated suppression (Hughes et al., 1997). The observation that R-Ras reverses suppression without affecting MAP kinase activation demonstrates that reversal is not the result of R-Ras activating a MAP kinase phosphatase.
R-Ras reversal is not dependent on the activation of PI 3-kinase. GTP-bound R-Ras can bind to the p110 subunit of PI 3-kinase and stimulate the production of PI 3-lipids, demonstrating that PI 3-kinase is a downstream effector of R-Ras (Marte et al., 1996). Studies on integrin function in platelets and leukocytes have identified a role for PI 3-kinase in regulating the activation of β1, β2, and β3 integrins (Shimizu and Hunt, 1996; J. Zhang et al., 1996). These observations combined with the fact that R-Ras can stimulate PI 3-kinase activity provided a possible explanation for the involvement of R-Ras in integrin activation. We used the PI 3-kinase inhibitor LY294002 to examine the role of PI 3-kinase in integrin affinity modulation. The inhibition of PI 3-kinase, as measured by the activation of the downstream effector Akt, had little effect on basal integrin affinity or on H-Ras–induced suppression and R-Ras rescue. Also, the overexpression of an activated variant of PI 3-kinase, p110-CAAX, did not reverse suppression by Raf-CAAX even though it was a potent activator of Akt (our unpublished observations). These data would appear to indicate that PI 3-kinase does not play a role in integrin affinity modulation, at least in CHO cells.
The small GTP-binding protein Ral is not involved in integrin affinity modulation in CHO cells. Both GTP-bound H-Ras and R-Ras can bind to GEFs for the small GTP-binding protein Ral; however, only H-Ras is capable of stimulating Ral-GEF activity in vivo (Urano et al., 1996). This suggested that R-Ras may reverse suppression by competing with H-Ras for Ral-GEFs. To investigate this possibility, we examined the effect on integrin affinity modulation of both blocking and stimulating Ral activity by the coexpression of an activated Ral-GEF and a Ral dominant negative. Our results indicated that Ral does not contribute to the modulation of integrin affinity by H-Ras and R-Ras.
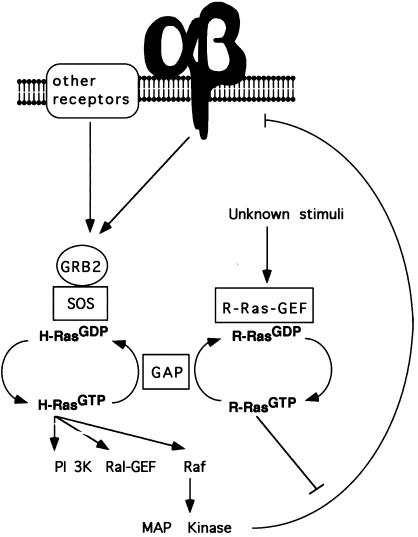
These data suggest that R-Ras and H-Ras mediate their opposing effects on integrin affinity via the activation of a distinct effector. Figure 7 illustrates a model that fits current data, demonstrating how R-Ras and H-Ras could act in concert to regulate integrin affinity. GTP-bound R-Ras could activate an effector that stimulates an undefined signaling pathway that impacts on the integrin suppressor pathway at a point downstream of MAP kinase, inactivating the integrin suppressor pathway. H-Ras can be activated via the dimerization of growth factor receptors and by the ligation and clustering of integrins. In both cases, Ras activation is mediated by the translocation of a complex between the adapter protein GRB2 and the Ras-GEF SOS to the plasma membrane.
Figure 7.
A hypothetical scheme of how the GTP-binding cycles of Ras and R-Ras could influence integrin affinity modulation. In this model, ligation of integrins and other cell surface receptors leads to the formation of the GRB2–SOS complex necessary for Ras activation. Ras can then activate its downstream effectors, including Raf-1, stimulating the MAP kinase–dependent negative feedback loop that can suppress activation of integrin. The activation of R-Ras, by as yet undefined stimuli, could then antagonize the Ras/Raf suppressor pathway downstream of MAP kinase.
In contrast to that of Ras, the stimuli that lead to the activation of R-Ras in vivo have yet to be fully defined. Recently, Ramos et al. (1998) have demonstrated that the overexpression of PEA-15, a small death effector domain–containing protein enriched in astrocytes, is able to reverse the suppressive effect of activated H-Ras. Significantly, the activity of PEA-15 is blocked by dominant-negative R-Ras (Ramos et al., 1998), suggesting that the activation of endogenous R-Ras is capable of reversing H-Ras suppression. This observation suggests that PEA-15 may be a component of a signal transduction pathway that regulates the activity of R-Ras, and in the future it will be of interest to characterize the relationship between PEA-15 and R-Ras. In addition, there is a preliminary report suggesting that thrombin can induce a clear activation of R-Ras in megakaryoblasts (Bos, 1997). Until the stimuli and guanine-nucleotide exchange factors that activate R-Ras in vivo are identified, it will not be possible to test the model outlined in Figure 7 and to define further the physiological role for R-Ras in integrin affinity modulation. It is possible that H-Ras and R-Ras are activated by distinct stimuli that induce either positive or negative effects on integrin affinity. Alternatively, the same stimuli may activate both H-Ras and R-Ras, with integrin affinity reflecting the ratio of the GTP-bound state of these two small G-proteins.
The deregulation of the MAP kinase pathway is often associated with oncogenic transformation. Unregulated activity of the MAP kinase–dependent integrin suppressor pathway can lead to the loss of the fibronectin matrix assembly and changes in integrin-dependent cell morphology, which may explain some of the integrin-dependent defects associated with the transformed phenotype. Indeed, such defects may account for the high metastatic potential of certain tumors. However, it is unclear whether these defects are primarily caused by the suppression of integrin activation or whether additional factors contribute to these phenotypes. Because R-Ras reverses H-Ras– and Raf-1–mediated suppression of integrin affinity, it will be of interest to determine whether the activation of R-Ras can also reverse these phenotypic defects.
ACKNOWLEDGMENTS
We thank Sandy Shattil and Martin Schwartz for their critical review of the manuscript and Rob Wolthius for his help with the Ral activation assays. T.S. was supported by a Medical Research Council (United Kingdom) traveling fellowship. P.E.H is the recipient of a senior fellowship from the Leukemia Society of America. M.H.G is supported by grants from the National Institutes of Health. J.D is supported by the Imperial Cancer Research Fund.
REFERENCES
- Baker EK, Tozer EC, Pfaff M, Shattil SJ, Loftus JC, Ginsberg MH. A genetic analysis of integrin function: Glanzmann thrombasthenia in vitro. Proc Natl Acad Sci USA. 1997;94:1973–1978. doi: 10.1073/pnas.94.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Ras-like GTPases. Biochim Biophys Acta. 1997;1333:19–31. doi: 10.1016/s0304-419x(97)00015-2. [DOI] [PubMed] [Google Scholar]
- Boussiotis VA, Freeman GJ, Berezovskaya A, Barber DL, Nadler LM. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124–148. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- Cook SJ, Rubinfeld B, Albert I, McCormick F. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Brtva TR, Lowe DG, Der CJ. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- Fam NP, Fan WT, Wang Z, Zhang L, Chen H, Moran MF. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol Cell Biol. 1997;17:1396–1406. doi: 10.1128/mcb.17.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett MD, Self AJ, van Oers C, Hall A. Identification of distinct cytoplasmic targets for ras/R-ras and rho regulatory proteins. J Biol Chem. 1989;264:10–13. [PubMed] [Google Scholar]
- Huff SY, Quilliam LA, Cox AD, Der CJ. R-Ras is regulated by activators and effectors distinct from those that control Ras function. Oncogene. 1997;14:133–143. doi: 10.1038/sj.onc.1200815. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Diaz-Gonzalez F, Leong L, Wu C, McDonald J, Shattil SJ, Ginsberg MH. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J Biol Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- Hughes PE, Renshaw MW, Pfaff M, Forsyth J, Keivens VM, Schwartz MA, Ginsberg MH. Suppression of integrin activation: a novel function of a Ras/Raf-initiated MAP kinase pathway. Cell. 1997;88:521–530. doi: 10.1016/s0092-8674(00)81892-9. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- Lowe DG, Capon DJ, Delwart E, Sakaguchi AY, Naylor SL, Goeddel DV. Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell. 1987;48:137–146. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr Biol. 1996;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- O’Toole TE, Katagiri Y, Faull RJ, Peter K, Tamura R, Quaranta V, Loftus JC, Shattil SJ, Ginsberg MH. Integrin cytoplasmic domains mediate inside-out signal transduction. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole TE, Loftus JC, Du XP, Glass AA, Ruggeri ZM, Shattil SJ, Plow EF, Ginsberg MH. Affinity modulation of the αIIbβ3 integrin (platelet GPIIb-IIIa) is an intrinsic property of the receptor. Cell Regul. 1990;1:883–893. doi: 10.1091/mbc.1.12.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole TE, Mandelman D, Forsyth J, Shattil SJ, Plow EF, Ginsberg MH. Modulation of the affinity of integrin αIIbβ3 (GPIIb-IIIa) by the cytoplasmic domain of αIIb. Science. 1991;254:845–847. doi: 10.1126/science.1948065. [DOI] [PubMed] [Google Scholar]
- Ramos JW, Kojima TK, Hughes PE, Fenczik CA, Ginsberg MH. The death effector domain of PEA-15 is involved in its regulation of integrin activation. J Biol Chem. 1998;273:33897–33900. doi: 10.1074/jbc.273.51.33897. [DOI] [PubMed] [Google Scholar]
- Rey I, Harris-Taylor P, van Earp H, Hall A. R-ras interacts with rasGAP, neurofibromin and c-raf but does not regulate cell growth or differentiation. Oncogene. 1994;9:685–692. [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin DJ, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Shattil SJ, Hoxie JA, Cunningham M, Brass LF. Changes in the platelet membrane glycoprotein IIb.IIIa complex during platelet activation. J Biol Chem. 1985;260:11107–11114. [PubMed] [Google Scholar]
- Shimizu Y, Hunt S. Regulating integrin-mediated adhesion: one more function for PI 3-kinase? Immunol Today. 1996;17:565–573. doi: 10.1016/s0167-5699(96)10061-x. [DOI] [PubMed] [Google Scholar]
- Shou C, Wurmser A, Suen KL, Barbacid M, Feig LA, Ling K. Differential response of the Ras exchange factor Ras-GRF to tyrosine kinase and G protein mediated signals. Oncogene. 1995;10:1887–1893. [PubMed] [Google Scholar]
- Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaargaren M, Martin GA, McCormick FM, Fernandez SM, Bischoff JR. The Ras-related protein R-ras interacts directly with Raf-1 in a GTP-dependent manner. Biochem J. 1994;300:303–307. doi: 10.1042/bj3000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano T, Emkey R, Feig LA. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wolthuis RM, de Ruiter ND, Cool RH, Bos JL. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis RM, Franke B, van Triest M, Bauer B, Cool RH. Activation of the small GTPase Ral in platelets. Mol Cell Biol. 1998;18:2486–2491. doi: 10.1128/mcb.18.5.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Matsui T, Nakafuku M, Iwamatsu A, Kaibuchi K. A novel GTPase-activating protein for R-Ras. J Biol Chem. 1995;270:30557–30561. doi: 10.1074/jbc.270.51.30557. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang J, Shattil SJ, Cunningham MC, Rittenhouse SE. Phosphoinositide 3-kinase γ and p85/phosphoinositide 3-kinase in platelets. Relative activation by thrombin receptor or β-phorbol myristate acetate and roles in promoting the ligand-binding function of αIIbβ3 integrin. J Biol Chem. 1996;271:6265–6272. doi: 10.1074/jbc.271.11.6265. [DOI] [PubMed] [Google Scholar]
- Zhang K, Noda M, Vass WC, Papageorge AG, Lowy DR. Identification of small clusters of divergent amino acids that mediate the opposing effects of ras and Krev-1. Science. 1990;249:162–165. doi: 10.1126/science.2115210. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Vouri K, Wang H, Reed JC, Ruoslahti E. Integrin activation by R-ras. Cell. 1996;85:61–69. doi: 10.1016/s0092-8674(00)81082-x. [DOI] [PubMed] [Google Scholar]