Abstract
The Izh2p protein from Saccharomyces cerevisiae is a receptor for the plant antifungal protein, osmotin. Since Izh2p is conserved in fungi, understanding its biochemical function could inspire novel strategies for the prevention of fungal growth. However, it has been difficult to determine the exact role of Izh2p because it has pleiotropic effects on cellular biochemistry. Herein, we demonstrate that Izh2p negatively regulates functionally divergent genes through a CCCTC promoter motif. Moreover, we show that Izh2p-dependent promoters containing this motif are regulated by the Nrg1p/Nrg2p and Msn2p/Msn4p transcription factors. The fact that Izh2p can regulate gene expression through this widely dispersed element presents a reasonable explanation of its pleiotropy. The involvement of Nrg1p/Nrgp2 in Izh2p-dependent gene regulation also suggests a role for this receptor in regulating fungal differentiation in response to stimuli produced by plants.
Keywords: Osmotin, Izh2p, iron, zinc, yeast
Introduction
The Izh2p protein from Saccharomyces cerevisiae was found to function as a receptor for a plant protein called osmotin.[1] This discovery is intriguing because osmotin belongs to the PR-5 family of defensins that possesses broad-spectrum antifungal activity and a better understanding of how plant PR-5 defensins affect fungi can be used affect to develop novel antifungal pharmaceuticals. Not surprisingly, there is considerable interest in characterizing the biochemical role of Izh2p and mapping the pathway through which it affects yeast physiology.
However, Izh2p has been implicated in a variety of biochemical processes ranging from iron and zinc homeostasis [2] to the metabolism of lipids and phosphate [3] to programmed cell death.[1] To complicate the issue even further, Izh2p overexpression affects both a general stress responsive transcriptional reporter and the expression of a gene involved in filamentous growth.[4] Thus, IZH2 is a pleiotropic gene that cannot yet be functionally assigned to any one particular biochemical pathway. The purpose of this study is to shed light on the origin of this pleiotropy by developing a clearer picture of how Izh2p regulates specific genes.
As a starting point, we used the fact that Izh2p overexpression represses the expression of two specific genes. This effect can be attributed to basal signaling capability of the receptor, which, when present at elevated levels, can activate its downstream signal transduction pathway.[5] Herein we present an analysis of the promoter regions of these two Izh2p-regulated genes that indicates a CCCTC motif is responsible for their response to Izh2p. This motif has been shown to function as a binding site for the Nrg1p and Nrg2p transcriptional repressors [6] and may competitively bind the Msn2p and Msn4p transcriptional activators.[7] This short motif can be found in the promoters of hundreds of functionally divergent yeast genes, a fact that may help explain the pleiotropy of Izh2p and lead to a better understanding of how PR-5 defensins, through their interaction with Izh2p, affect fungal physiology.
Materials and Methods
Yeast strains and growth conditions
MCY5326 wild type, MCY5338 (msn2Δmsn4Δ), MCY5378 (nrg1Δnrg2Δ) and MCY5385 (msn2Δmsn4Δnrg1Δnrg2) have been described previously.[7] All other yeast strains used in this study were purchased from Euroscarf (Frankfurt, Germany) and are in the BY4742 background. Strains were grown in either chelexed synthetic medium (CSM), Low Iron Medium (LIM) or Low Zinc Medium (LZM) the compositions of which have been previously described.[2; 4; 8] For CSM, which is a nutrient drop-out medium, metal-repletion is achieved by adding 10 μM of the respective metal. For LIM and LZM, which contain EDTA, the addition of 1 μM of each metal is considered metal-deficient, while 1 mM is considered metal-replete. For all media, 2% galactose is used as the carbon source to induce the expression of genes driven by the GAL1 promoter.
Plasmids
All plasmids have been previously described as indicated by the references. pFET3-398 and pFET3-297 are episomal reporter plasmids in which lacZ is driven by different truncations of the FET3 promoter (−398 to +3 and −297 to +3, respectively).[4] Both constructs are induced by iron-deficiency via the Aft1p-transcription factor due to the presence of an iron-response element between −252 and −245. (See Supplemental Figure 1A) pZRT1-521, pZRT1-361, pZRT1-305 and pZRT1-201 are integrating reporter plasmids in which lacZ is driven by different truncations of the ZRT1 promoter (−521 to +3, −361 to +3, −305 to +3 and −201 to +3, respectively). The ZRT1 promoter contains at least three functional zinc response elements (ZREs: ZRE3, −445 to −434; ZRE1, −318 to −309; ZRE2, −202 to −191) to which the Zap1p transcriptional activator binds during zinc deficiency.[9] (See Supplemental Figure 1B) pZRT1-ZRT1ET is a centromeric plasmid containing an HA-epitope tagged ZRT1 open reading frame driven by approximately 600 base pairs of the native ZRT1 promoter.[10] pZPS1-lacZ, pZRC1-lacZ, pOLE1-lacZ and pFET4-lacZ contain ~1000 base pairs of the ZPS1, ZRC1, OLE1 and FET4 promoters fused to lacZ.[2; 8] Plasmids pCYC1(ZRT1ZRE1)-lacZ and pCYC1(IZH1ZRE)-lacZ consist of the lacZ gene driven by the minimal CYC1 promoter into which fragments of the ZRT1 or IZH1 promoters containing functional zinc response elements (ZREs) have been inserted.[8] (See Supplemental Figure 1C) pGAL1-IZH2 contains the IZH2 gene driven by the GAL1 galactose-inducible promoter and is derived from the pRS316 expression vector.[2] pGAL1-NRG2 contains a GAL1-driven TAP-tagged (Tandem Affinity Purification) NRG2 construct and was purchased from OpenBiosystems.
Biochemical Assays
β-galactosidase assays [2] and total membrane protein preparations [4] were performed as previously described. Western blots on SDS-PAGE gels loaded with equal amounts of total membrane protein lysate were performed using standard chemilluminescence protocol with rabbit polyclonal anti-HA primary and goat anti-rabbit IgG-HRP conjugate secondary antibodies (Santa Cruz Biotechnology). Pattern searching for motifs in yeast promoters was performed using RSA tools (http://rsat.ulb.ac.be/rsat/) by defining a promoter as 800 base pairs upstream of ATG excluding overlap with upstream genes.
Results and Discussion
Izh2p-negatively regulates the ZRT1 promoter
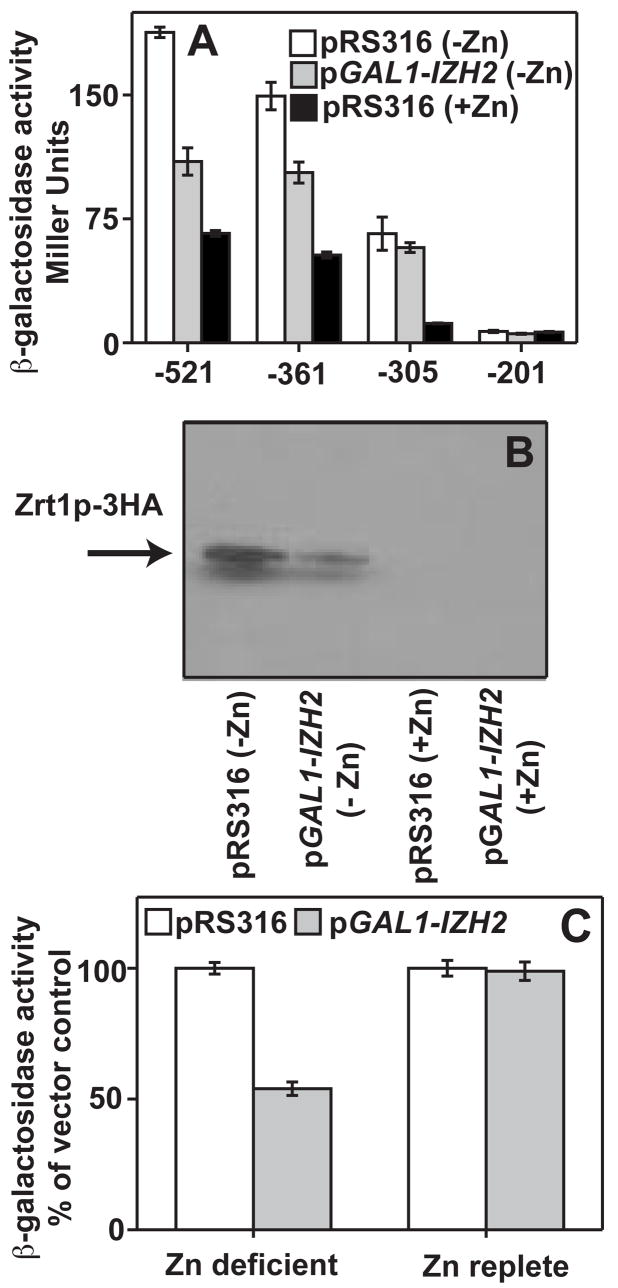
Because of its suspected involvement in zinc metabolism, we analyzed the effect of Izh2p overexpression on ZRT1, the gene that encodes the high-affinity zinc-uptake transporter. Izh2p overexpression repressed the ability of Zap1p to induce the expression of pZRT1-521 and pZRT1-361 during zinc-deficiency (Figure 1A) and resulted in decreased accumulation of HA-tagged Zrt1p protein driven by ~600 base pairs of ZRT1 promoter (Figure 1B). These results indicate that Izh2p negatively regulates the ZRT1 gene. Izh2p overexpression had no effect on basal expression of pZRT1-521 (Figure 1C) or inducible expression of pZRT1-305 (Figure 1A), indicating that the effect of Izh2p requires induction by Zap1p but does not globally affect the ability of Zap1p to activate genes.
Figure 1. Regulation of ZRT1 by Izh2p.
In all panels, cells were grown in zinc-replete (+Zn) or zinc-deficient (−Zn) LZM. Cells either carry pRS316 or pGAL1-IZH2. (A) The effect of Izh2p-overexpression on various pZRT1-lacZ fusion constructs. Numbers indicate the amount of upstream sequence contained in each construct. (B) Effect of Izh2p overexpression on the accumulation of HA-tagged Zrt1p expressed from the pZRT1-ZRT1ET plasmid. (C) The effect of Izh2p overexpression on pZRT1-521.
Defining the Izh2p-response element
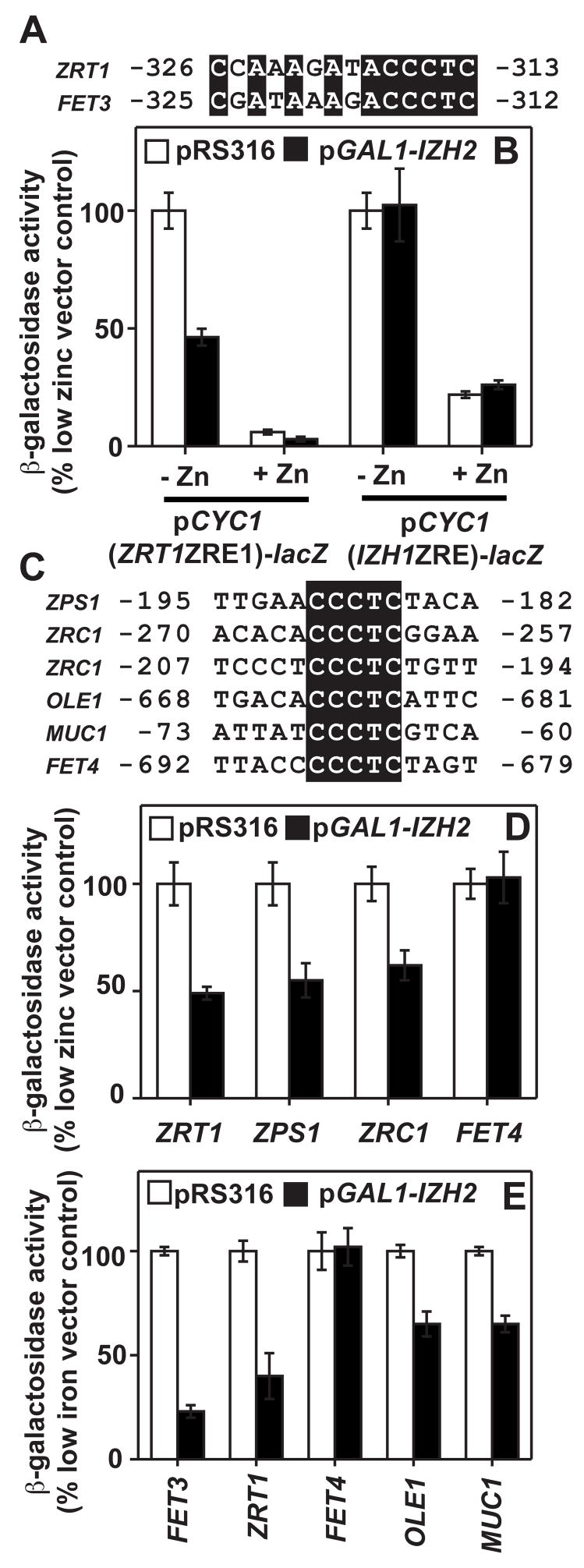
pZRT1-305 is not repressed by Izh2p overexpression, indicating that Izh2p exerts its effects on a regulatory element between −361 and −305 of the ZRT1 promoter. We previously found that Izh2p similarly represses the Aft1p-inducible expression of the FET3 gene by affecting a regulatory element between −398 and −297 of the FET3 promoter.[4] We scanned these regions of the FET3 and ZRT1 promoters for similar sequences and found a conserved ACCCTC motif t (Figure 2A)
Figure 2. Identification of the Izh2p-response element.
In panels (B), (D) and (E), cells carry either pRS316 or pGAL1-IZH2. (A) Conserved regions of the FET3 and ZRT1 promoters. (B) The effect of Izh2p overexpression on either pCYC1(ZRT1ZRE1)-lacZ or pCYC1(IZH1ZRE)-lacZ. Cells are grown in LZM. (C) Location of putative Izh2p-response elements in various genes. (D+E) Effect of Izh2p overexpression on the ability to induce various promoter-lacZ constructs. Cells were grown in zinc-deficient (D) or iron-deficient (E) CSM.
In the ZRT1 promoter, this motif directly overlaps ZRE1 (−318 to −309). To determine if this motif is sufficient to confer Izh2p-responsiveness onto a promoter, we measured the effect of Izh2p overexpression on pCYC1(ZRT1ZRE1)-lacZ. This construct was both zinc-responsive due to the presence of ZRE1 and Izh2p-repressible due to the presence of the ACCCTC motif. (Figure 2B) pCYC1(IZH1ZRE)-lacZ, another reporter containing a different ZRE, did not respond to Izh2p-overexpression, confirming that Izh2p did not generally repress all reporters containing ZREs. (Figure 2B)
Thus, this ACCCTC motif, which we are calling the Izh2p-Response Element (IzRE) is sufficient to confer Izh2p-responsiveness onto promoters and we found putative IzREs in the promoters of over 600 yeast genes. We already possessed promoter-lacZ fusions constructs for three of these genes - ZRC1, ZPS1 and OLE1.[2; 8] pZPS1-lacZ and pZRC1-lacZ are induced by zinc-deficiency and, as expected, their zinc-dependent induction was repressed by Izh2p overexpression. (Figure 2D) pOLE1-lacZ is inducible by iron-deficiency [2] and its iron-dependent induction was repressible by Izh2p overexpression. (Figure 2E) The IzRE in the OLE1 promoter is in the opposite orientation relative to ATG, suggesting the orientation of the IzRE may not be important. We also previously showed that a fifth promoter construct, pMUC1-lacZ, is repressible by Izh2p overexpression [4], yet this promoter contains a variant TCCCTC motif, suggesting that the functional IzRE can tolerate changes at the first position. However, the pFET4-lacZ construct, which contains CCCCTC, is actually inducible by both zinc-deficiency and iron-deficiency [11], but unresponsive to Izh2p overexpression under either condition. (Figures 2D and 2E) Thus, the consensus IzRE is (A/T)CCCTC at this point, although more work is required to define all functional variations in the motif.
Nrg1p and Nrg2p as Izh2p-dependent repressors
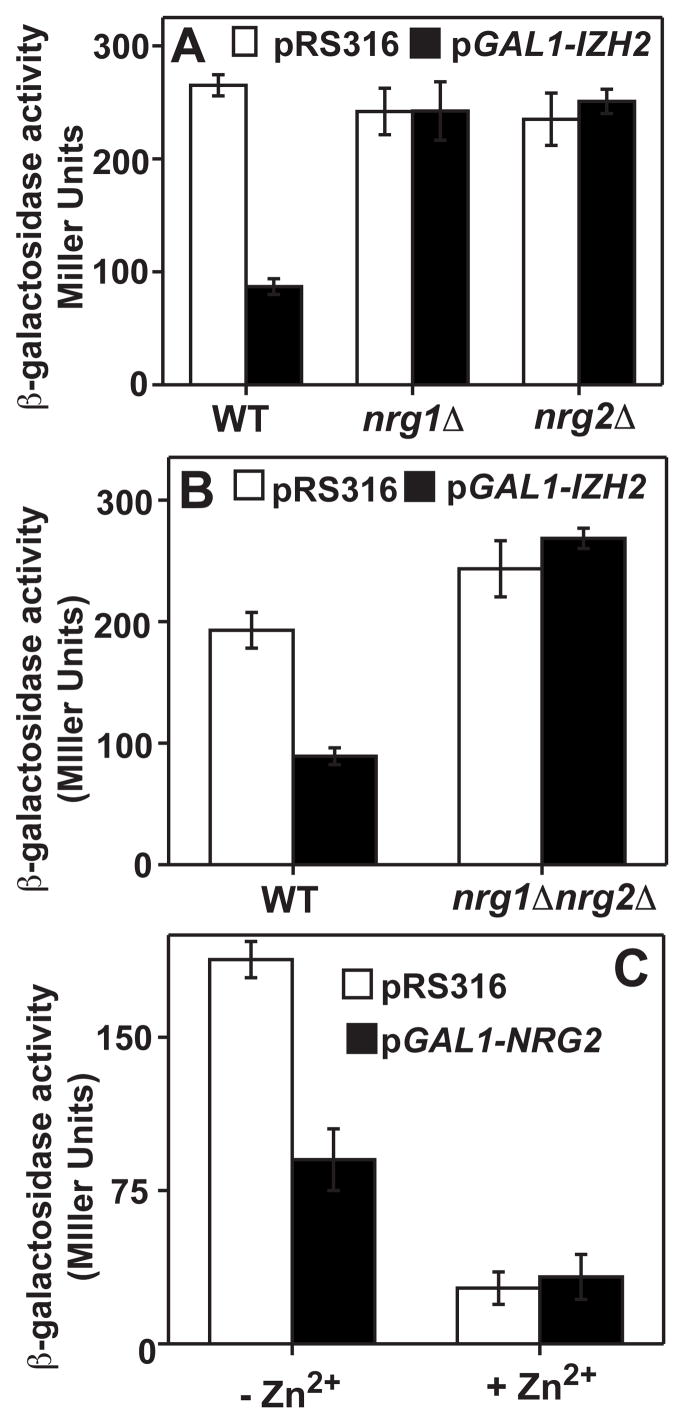
Footprinting analysis revealed that CCCTC is a binding site for the Nrg1p/Nrg2p transcriptional repressors [6], suggesting that the IzhRE is an Nrg1p/Nrg2p binding site. Indeed, Nrg1p has already been identified as a repressor of the Izh2p-regulated ZPS1 and MUC1 genes [7] and we previously showed that Nrg1p/Nrg2p are required for Izh2p-dependent FET3 repression.[4] Figures 3A and 3B show that Nrg1p/Nrg2p were also required for Izh2p-dependent repression of pZRT1-521 and pCYC1(ZRT1ZRE1)-lacZ. Consistent with the involvement of Nrg1p/Nrg2p in Izh2p-dependent repression, we also previously showed that overexpression of Nrg2p had the same effect on FET3 as Izh2p. Herein, we show that overexpression of Nrg2p also repressed the Zap1p-dependent induction of pCYC1(ZRT1ZRE1)-lacZ. (Figure 3C) The fact that the IzRE and ZRE overlap in this construct suggests that Nrg2p competes with Zap1p, and thereby represses inducible expression. The precise mechanism through which Izh2p affects the ability of Nrg1p and Nrg2p to serve as transcriptional repressors is still under investigation.
Figure 3. Izh2p-dependent repression requires Nrg1p and Nrg2p.
In all panels, cells carry either pRS316 or pGAL1-IZH2 and were grown in LZM. (A) The repression of pZRT1-521 by Izh2p overexpression in wild type (WT, BY4742) and isogenic mutant strains lacking either Nrg1p (nrg1Δ) or Nrg2p (nrg2Δ). (B) The repression of pCYC1(ZRT1ZRE1)-lacZ by Izh2p overexpression in wild type (WT, MCY5326) and an isogenic mutant strain lacking both Nrg1p and Nrg2p (nrg1Δnrg2Δ).
Msn2p and Msn4p as Izh2p-dependent activators
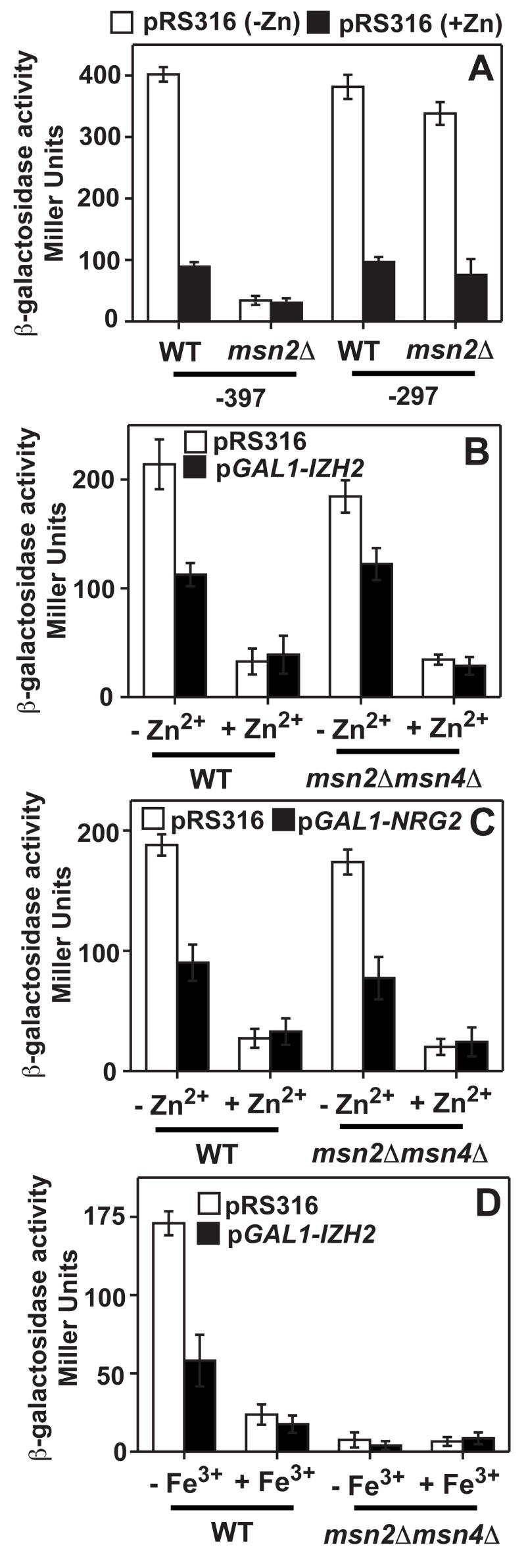
Izh2p affects the ability to induce FET3 and ZRT1, but has no effect on either the Aft1p or Zap1p activators of these genes. This suggests that Izh2p negatively regulates the activity of a co-activator. We previously showed that the Msn2p and Msn4p transcription factors were essential co-activators of FET3 expression.[4] This is demonstrated in Figure 4A, which shows that, while pFET3-398 required Msn2p for iron-dependent induction, pFET3-297 did not. Hence, in addition to the IzRE, there is an Msn2p-dependent upstream co-activating element in the FET3 promoter between −398 and −297. This finding is intriguing because recent findings suggest Msn2p/Msn4p compete with Nrg1p/Nrg2p for similar binding sites, including CCCTC, in a subset of promoters.[7] Thus, Izh2p could function by activating Nrg1p/Nrg2p or by inactivating Msn2p/Msn4p. Clearly, Izh2p does not work solely by inactivating Msn2p/Msn4p because its overexpression still represses the zinc-dependent induction of pZRT1-521 in a strain lacking Msn2p and Msn4p. (Figure 4B) In addition, Nrg2p overexpression still represses pCYC1(ZRT1ZRE1)-lacZ in the msn2Δmsn4Δ strain. (Figure 4C) Thus, activation of Nrg1p/Nrg2p is sufficient for Izh2p-dependent gene repression.
Figure 4. Msn2p and Msn4p are essential co-activators of ZRT1.
In all panels, cells carry either pRS2316 or pGAL1-IZH2 and were grown in iron- or zinc-deficient CSM. (A) The effect of deletion of Msn2p (msn2Δ, BY4742 background) on the iron-responsiveness of two truncations of the pFET3-398 and pFET3-297 reporters. (B, C and D) The effect of deletion of Msn2p and Msn4p (msn2Δmsn4Δ, MCY background) on the zinc- and Izh2p-responsiveness of pZRT1-521 (B), the zinc- and Nrg2p-responsiveness of pCYC1(ZRT1ZRE1)-lacZ (C) and the iron- and Izh2p-responsiveness of pZRT1-521 (D).
Finally, it is intriguing that the Aft1p-dependent induction of FET3 is constitutively repressed in an msn2Δmsn4Δ strain [4], but the zinc-dependent induction of ZRT1 is not. These results suggest that Msn2p/Msn4p plays no role in ZRT1 expression. However, recent genome-wide transcriptional analysis suggested that ZRT1 is induced by iron-deficiency [12] and has a putative FeRE in its promoter between −364 and −358. (See Supplemental Figure 1B) Figure 4D shows that pZRT1-521 was both inducible by low iron and repressible by Izh2p overexpression in a manner that depends on the presence of Msn2p/Msn4p.
Summary
These results show that Nrg1p/Nrg2p are repressors of both FET3 and ZRT1, while Msn2p/Msn4p are co-activators. Moreover, Msn2p/Msn4p likely compete with Nrg1p/Nrg2p for binding to the IzRE. Izh2p affects gene expression by influencing the balance of this competition. Since Nrg1p/Nrg2p and Msn2p/Msn4p regulate hundreds of genes, their involvement in Izh2p-dependent gene regulation provides an explanation for the pleiotropy of this receptor. More importantly, however, their involvement provides a tantalizing clue to the physiological function of this receptor. Nrg1p/Nrg2p are negative regulators of fungal filamentation [7], suggesting that Izh2p, through Nrg1p/Nrg2p, could inhibit the yeast to filament transition. This is supported by a recent study in which IZH2 was identified as a gene that, when overexpressed, repressed filamentous growth.[13] Thus, it is possible that plant PR-5 defensins are designed to influence fungal developmental programs by activating Izh2p. This would represent a new paradigm in plant-fungal interactions.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Narasimhan ML, Coca MA, Jin J, Yamauchi T, Ito Y, Kadowaki T, Kim KK, Pardo JM, Damsz B, Hasegawa PM, Yun DJ, Bressan RA. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell. 2005;17:171–80. doi: 10.1016/j.molcel.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 2.Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ. Metalloregulation of yeast membrane steroid receptor homologs. Proc Natl Acad Sci U S A. 2004;101:5506–11. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karpichev IV, Cornivelli L, Small GM. Multiple regulatory roles of a novel Saccharomyces cerevisiae protein, encoded by YOL002c, in lipid and phosphate metabolism. J Biol Chem. 2002;277:19609–17. doi: 10.1074/jbc.M202045200. [DOI] [PubMed] [Google Scholar]
- 4.Kupchak BR, Garitaonandia I, Villa NY, Mullen MB, Weaver MG, Regalla LM, Kendall EA, Lyons TJ. Probing the mechanism of FET3 repression by Izh2p overexpression. Biochim Biophys Acta. 2007;1773:1124–32. doi: 10.1016/j.bbamcr.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JL, Kupchak BR, Garitaonandia I, Hoang LK, Maina AS, Regalla LM, Lyons TJ. Heterologous expression of human mPRalpha, mPRbeta and mPRgamma in yeast confirms their ability to function as membrane progesterone receptors. Steroids. 2008 doi: 10.1016/j.steroids.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SH, Koh SS, Chun JH, Hwang HJ, Kang HS. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2044–50. doi: 10.1128/mcb.19.3.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vyas VK, Berkey CD, Miyao T, Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1882–91. doi: 10.1128/EC.4.11.1882-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci U S A. 2000;97:7957–62. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem. 1998;273:28713–20. doi: 10.1074/jbc.273.44.28713. [DOI] [PubMed] [Google Scholar]
- 10.Gitan RS, Luo H, Rodgers J, Broderius M, Eide D. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J Biol Chem. 1998;273:28617–24. doi: 10.1074/jbc.273.44.28617. [DOI] [PubMed] [Google Scholar]
- 11.Waters BM, Eide DJ. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem. 2002;277:33749–57. doi: 10.1074/jbc.M206214200. [DOI] [PubMed] [Google Scholar]
- 12.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2005;280:10135–40. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 13.Jin R, Dobry CJ, McCown PJ, Kumar A. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol Biol Cell. 2008;19:284–96. doi: 10.1091/mbc.E07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.