Abstract
Low Molecular Weight Tyrosine Phosphatases (LMWTP) are widespread in prokaryotes; however, understanding of the signaling cascades controlled by these enzymes is still emerging. P. gingivalis, an opportunistic oral pathogen, expresses a LMWTP, Ltp1, that is differentially regulated in biofilm communities. Here we characterize the enzymatic activity of Ltp1 and, through the use of mutants that lack Ltp1 or expresses catalytically defective Ltp1, show that tyrosine phosphatase activity constrains both monospecies biofilm development and community development with the antecedent oral biofilm constituent S. gordonii. Exopolysaccharide production is downregulated by Ltp1 through transcriptional regulation of multiple genes involved in biosynthesis and transport. Furthermore, Ltp1 regulates transcriptional activity of luxS and thus impacts AI-2 dependent signaling in biofilm communities. In the absence of Ltp1 transcription across the hmu hemin uptake locus is reduced, and consequently uptake of hemin is impaired in the Ltp1 mutant. The gingipain proteinases Kgp and RgpA/B remain phosphorylated in the Ltp1 mutant. Phosphorylated Rgps are poorly secreted, whereas cell surface activity of phosphorylated Kgp is enhanced. By controlling the activity of several virulence-associated properties, Ltp1 may restrain the pathogenic potential of P. gingivalis and maintain a commensal interaction with the host.
Keywords: phosphatase, biofilm, exopolysaccharide, luxS, hemin
Introduction
Periodontal diseases are among the most common chronic infections of humans (Albandar, 2005). A group of gram-negative anaerobes is associated with the initiation and progression of severe manifestations of the disease, and foremost among these is Porphyromonas gingivalis (Haffajee and Socransky, 1994; Holt et al., 1988; Holt and Ebersole, 2005). P. gingivalis produces numerous adhesins, invasins, proteinases, and hemin uptake systems along with communication signals such as AI-2, that contribute to colonization, persistence and pathogenic potential (Holt et al., 1999; James et al., 2006; Lamont and Jenkinson, 1998). On the hard non-shedding tooth surfaces in the oral cavity bacteria exist in biofilm communities, and P. gingivalis can form both homotypic monospecies biofilms and heterotypic mixed species biofilms with other oral bacteria including antecedent colonizers such as S. gordonii (Kuramitsu et al., 2007; Simionato et al., 2006). Disease can then ensue from the interaction of P. gingivalis biofilms, detached biofilm organisms, and secreted products such as proteolytic enzymes, with the host cells and tissues of the periodontium (Haffajee and Socransky, 1994; Lamont and Jenkinson, 1998). Despite this pathogenic potential, P. gingivalis is frequently present in healthy individuals (Ximenez-Fyvie et al., 2000) and can be considered a host-adapted pathogen, causing disease only when the ecological balance between host and microbe is disrupted (Marsh, 2003). In order to maintain a balanced interaction with the host, many bacteria that encounter mucosal surfaces have devised mechanisms to restrict pathogenic outcomes and to limit the increase in bacterial biomass (Galan and Zhou, 2000; Tierrez and Garcia-del Portillo, 2005). In the oral cavity P. gingivalis may also regulate the extent of biofilm development, and/or orchestrate and control its pathophysiology in order to minimize damage to the host.
Signal transduction based on tightly regulated protein phosphorylation and dephosphorylation is a ubiquitous mechanism responsible for adaptation to environmental conditions in both prokaryotes and eukaryotes. In prokaryotes, the predominant phosphotransfer signaling devices are the two component systems that are based on phosphorylation of histidine and aspartate residues in sensor and response regulator proteins. However, serine/threonine and tyrosine kinases and phosphatases have also been described in bacteria, and are emerging as important in the control of a number physiological processes, including the expression of virulence properties (Cozzone et al., 2004; Cozzone, 2005; Grangeasse et al., 2007). In S. pneumoniae, for example, the tyrosine kinase CpsD and the histidinol phosphatase phosphoesterase CpsB function together in controlling polymerization and export of capsular polysaccharide (Morona et al., 2002). Autophosphorylation of the tyrosine kinase CpsD attenuates its activity and negatively regulates capsule production (Morona et al., 2000), while CpsB increases encapsulation through dephosphorylation and activation of CpsD (Morona et al., 2006). Invasive gram-negative pathogens can deliver tyrosine phosphatases directly into host cells through type III secretion systems. For example, YopH of Yersinia and StpP of Salmonella are tyrosine phosphatases that can uncouple host cell signaling pathways that control actin cytoskeleton stability (Black et al., 2000; Lin et al., 2003; Murli et al., 2001).
In the annotated genome of P. gingivalis there is one predicted tyrosine phosphatase and one a putative tyrosine kinase, that are distant from each other on the chromosome. Interestingly in a microarray based gene identification study, the tyrosine phosphatase gene (PG1641) was found to be upregulated in heterotypic P. gingivalis-S. gordonii biofilms. Moreover, deletion of this gene resulted in more luxuriant biofilm formation (Simionato et al., 2006), indicating a possible role for tyrosine phosphorylation/dephosphorylation in P. gingivalis community organization. PG1641 is a predicted cytoplasmic eukaryotic-type low molecular weight tyrosine phosphatase (LMWTP) and is annotated as Ltp1 in the P. gingivalis database. While the physiological functions of LMWTPs in bacteria are poorly understood; these enzymes are known to participate in secondary metabolite production in Streptomyces coelicolor, in the control of biosynthesis and/or transport of exopolysaccharides in E. coli and Klebsiella pneumoniae (Grangeasse et al., 2007; Preneta et al., 2002), and in stress resistance in Bacillus subtilis (Musumeci et al., 2005). The putative LMWTP of P. gingivalis therefore, is a potential candidate for a component of a regulatory network that controls pathogenic status. In this report, we define the function and role of P. gingivalis Ltp1 in the control of critical physiological and pathogenic processes.
Results
Substrate Specificity of P. gingivalis Ltp1
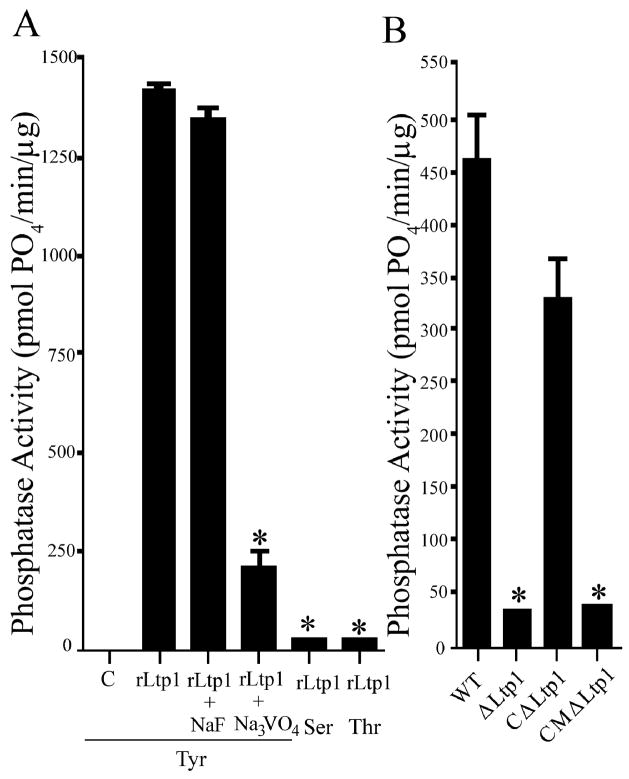
To verify enzyme activity and define the substrate specificity of P. gingivalis Ltp1, purified His-tagged recombinant protein was tested against tyrosine- serine- or threonine- phosphorylated peptides in a malachite green phosphatase colorimetric assay (Tribble et al., 2006). The enzyme was active against a phosphotyrosine peptide with a Km = 71.1 μM, Kcat = 36.9 min−1 and Kcat/Km = 0.52 min−1μM−1. Activity against phosphoserine and phosphothreonine peptides was not significantly above background levels (Fig. 1A). rLtp1 was inhibited by orthovanadate, a tyrosine phosphatase inhibitor, but not by sodium fluoride an inhibitor of phosphoserine phosphatases (Fig. 1A), providing further confirmation that the protein is a phosphotyrosine phosphatase.
Fig. 1.
Phosphatase activity of recombinant (r)Ltp1 and P. gingivalis strains. (A) rLtp1 phosphatase activity on a tyrosine phosphopeptide (Tyr) with and without inhibitors (orthovanadate or sodium fluoride), a serine phosphopeptide (Ser) or a threonine phosphopeptide (Thr). Phosphate release was measured with Malachite Green. C represents background spontaneous phosphate release from the Tyr peptide in the absence of Ltp1. * denotes p < 0.01 (t-test) compared to rLtp1 with Tyr peptide. (B) Tyrosine phosphatase activity of cell lysates of P. gingivalis strains 33277 (WT),ΔLtp1 (Ltp1 mutant), CΔLtp1 (complemented mutant) and CMΔLtp1 (mutant complement with catalytically defective C10S Ltp1). * denotes p < 0.01 (t-test) compared to WT. Error bars represent standard deviation (n = 3).
Ltp1 tyrosine phosphatase activity regulates heterotypic community development
To investigate the functionality of Ltp1 tyrosine phosphatase activity, site specific mutagenesis was performed in the conserved catalytic motif 8VCLGNICRS18. Conversion of the nucleophile cysteine 10 within this phosphotransfer loop to a serine abolished phosphatase activity of the recombinant protein. The C10S mutated ltp1 allele was cloned into the E. coli-Bacteroides shuttle vector pT-COW (Gardner et al., 1996) and introduced into the ltp1 strain of P. gingivalis (Simionato et al., 2006) to create P. gingivalis CMΔltp1. In addition, the wild type ltp1 allele was similarly introduced into Δltp1 to create P. gingivalis CΔltp1. Cell lysates of these strains were tested for phosphatase activity (Fig. 1B). Both the wild type and the CΔltp1 strains were able to dephosphorylate a tyrosine phosphorylated peptide, while Δltp1 and CMΔltp1 lost this activity. Hence these strains allow us to evaluate the function and role of Ltp1, and more precisely tyrosine phosphatase catalytic activity, in P. gingivalis.
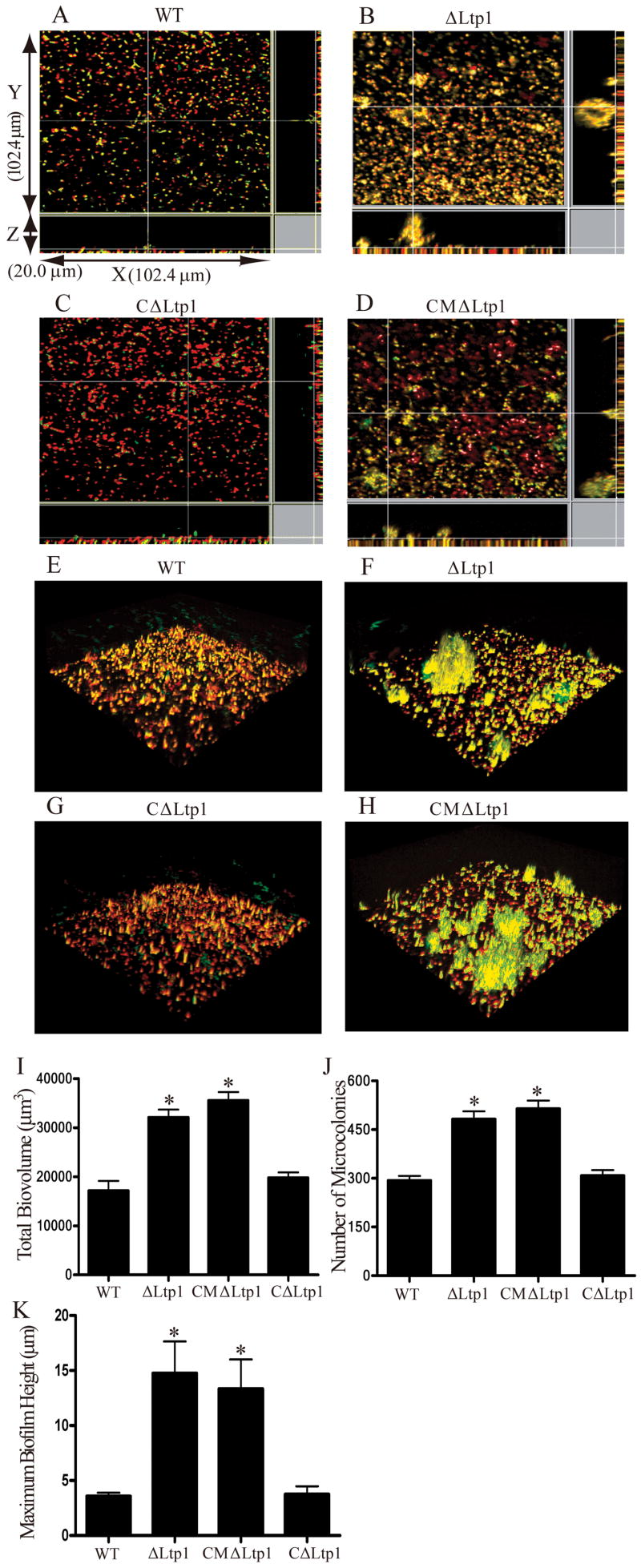
Accumulation of P. gingivalis into heterotypic communities with S. gordonii proceeds through a series of interactive events (Kuboniwa et al., 2006). P. gingivalis cells are first recruited from the fluid to the solid phase where accretion into rudimentary biofilm microcolonies occurs. This developmental process is enhanced in the absence of Ltp1 (Simionato et al., 2006), suggesting that Ltp1 restricts community development. To establish the role of Ltp1 tyrosine phosphatase activity in the development of heterotypic P. gingivalis-S. gordonii communities, the accumulation of P. gingivalis parental and mutant strains on substrata of S. gordonii was examined. Complementation of the Ltp1 deficient strain with the C10S catalytic domain mutant Ltp1 resulted in a biofilm phenotype indistinguishable from that of the Ltp1 deficient host strain (Fig. 2). In contrast, complementation of the Ltp1 deficient strain with the wild type ltp1 allele restored the biofilm phenotype to parental levels. In addition to total biovolume, biofilm parameters of colony number and height were significantly elevated in strain CMΔltp1 compared to parent or CΔltp1 (Fig. 2). These results support the concept that signal transduction through tyrosine phosphorylation/dephosphorylation constrains the development of heterotypic P. gingivalis communities to proportions that are exhibited by wild type strains.
Fig. 2.
Tyrosine phosphatase activity of Ltp1 constrains heterotypic P. gingivalis-S. gordonii community development. (A–D) Analysis of dual species communities by confocal laser scanning microscopy. Substrata of S. gordonii cells (red) were reacted with P. gingivalis 33277 (WT) or mutant strains (green) for 24 h. The resulting heterotypic community was observed on a Yokogawa spinning disc confocal scanning laser microscope with a 60x 1.4 N.A. objective. A series of fluorescent optical 0.2 μm x-y sections were collected to create digitally reconstructed images (x-z section, y-z section and x-y section) of the two species with Imaris software. Colocalized bacteria appear yellow. (E–H) 3D reconstruction of the heterotypic communities present in A-D using Imaris software. (I) P. gingivalis accumulation measured by grain area analysis of fluorescence. Mean with standard deviation of total biovolume for three 102.4 × 102.4 μm x-y sections with a 2 μm spaced z-series for the strains indicated. * denotes p < 0.01 (t-test) compared to WT (33277). (J) Numbers of microcolonies formed by P. gingivalis strains (mean with standard deviation). Microcolonies were defined as clusters of colocalized cells larger than 40 μm3 in 3 random 102.4 × 102.4 μm areas. * denotes p < 0.01 compared WT. (K) Maximum extent (with standard deviation) of P. gingivalis accumulation in the z dimension measured across 3 random x-z sections. * denotes p < 0.01 (t test) compared WT.
Homotypic biofilm formation is controlled by Ltp1
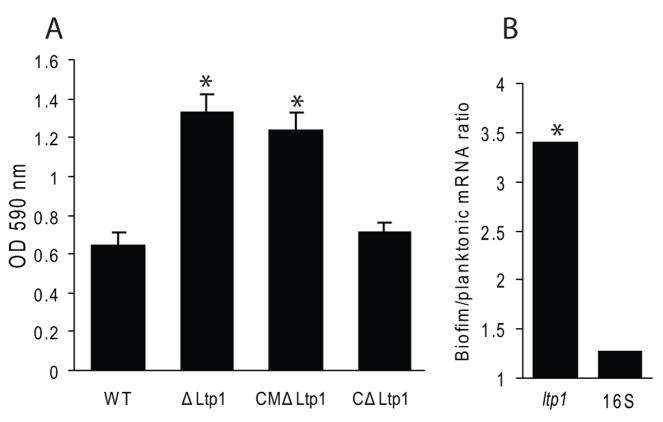
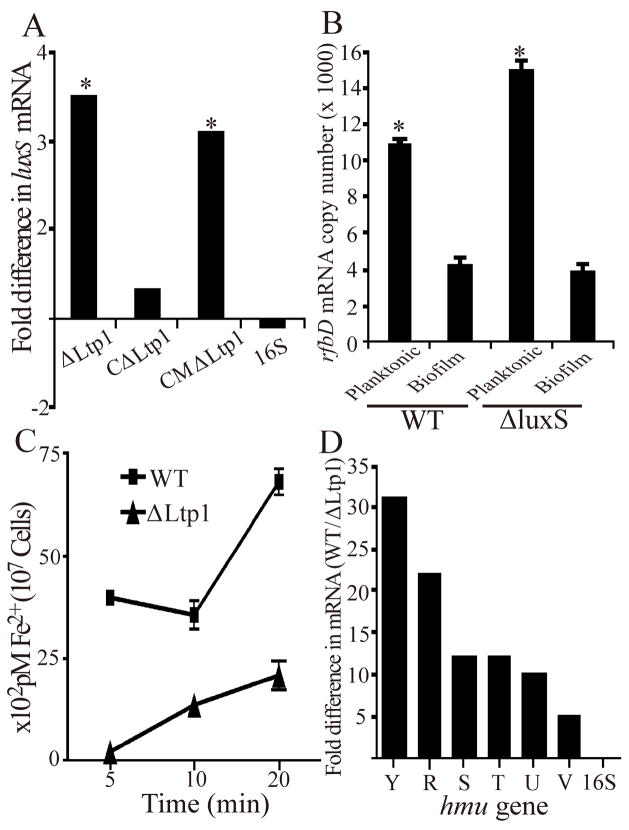
In addition to mixed species consortia, P. gingivalis also forms monospecies biofilms on abiotic surfaces (Capestany et al., 2008; Chen et al., 2002; Davey and Duncan, 2006). Hence we investigated whether Ltp1-mediated control is specific for heterotypic biofilms or also functions to regulate the accumulation of homotypic P. gingivalis biofilms. In a microtiter plate assay (O’Toole and Kolter, 1998), the biomass of the ltp1 and CMΔltp1 strains was enhanced in comparison to the parental and CΔltp1 strains (Fig 3A). Thus, Ltp1-dependent control of P. gingivalis biofilm accumulation is substratum independent. Furthermore, expression of ltp1 mRNA in monospecies biofilms was elevated 3.4 fold as compared to planktonic cells (Fig 3B), similar to the level of induction observed in dual species biofilms (Simionato et al., 2006). These results provided a basis for us to investigate Ltp1 in the context of a simple model P. gingivalis community without the potential confounding influences of S. gordonii.
Fig. 3.
Ltp1 controls homotypic P. gingivalis biofilm growth. (A) Microtiter plate biofilms of wild type (WT) or mutant strains after 24 h were stained with crystal violet. Biofilms were quantified by solubilizing crystal violet in 95% ethanol and measurement of absorbance at 595 nm. * denotes p < 0.01 (t test) compared to WT. (B) Increased expression of ltp1 mRNA in monospecies biofilms by quantitative Real Time RT-PCR. Data are expressed as ratio of transcript levels in biofilm:planktonic conditions. 16S rRNA was used as a control. * denotes p < 0.01 (t test) between biofilm and planktonic cells.
Ltp1 controls exopolysaccharide production
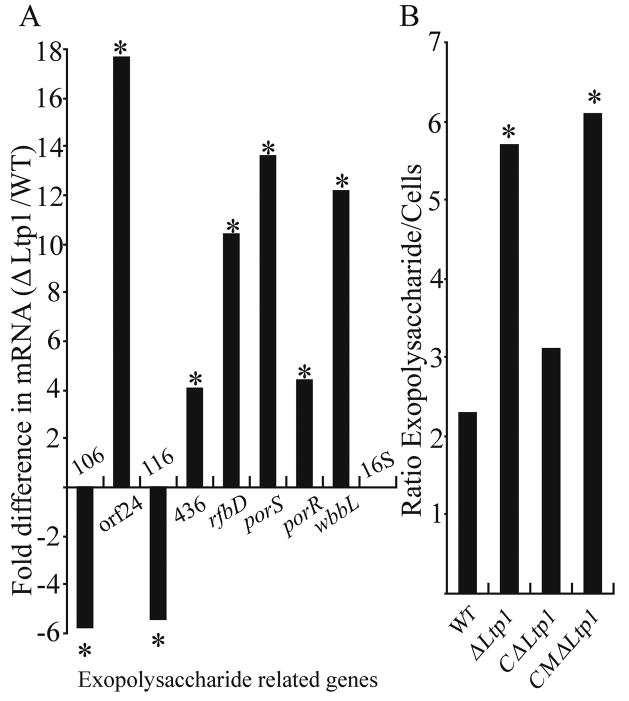
As extracellular polysaccharide is important for the development of biofilm communities, and as LMWTPs are involved in extracellular polysaccharide production in other organisms, we examined the role of Ltp1 in the formation of exopolysaccharide in P. gingivalis biofilms. Several loci are thought to contribute to the production and secretion of distinct polysaccharides in P. gingivalis, and the expression of representative genes of these loci in P. gingivalis biofilms was quantitated by real time RT-PCR (Fig. 4A). The region spanning PG0106-PG0120 is involved in the production of K-antigen capsule, and the gene constituents vary among P. gingivalis strains (Aduse-Opoku et al., 2006). PG0106, a predicted glucosyltransferase, and PG0116, a hypothetical protein, were downregulated in the Ltp1 mutant. Similarly, Davey and Duncan (2006) reported that mutation of PG0106 resulted in enhanced P. gingivalis single species biofilm formation. In contrast, orf24 another putative glucosyltransferase, was upregulated in the Ltp1 mutant. orf24 is absent in the sequenced P. gingivalis strain (W83) but is present in strains 33277 and 381, indicting a specific role in a subset of P. gingivalis strains (Aduse-Opoku et al., 2006). Also upregulated in the Ltp1 mutant were: PG0436, a putative capsular polysaccharide transport protein; rfbD a putative dTDP-4-dehydrorhamnose 3,5-epimerase, involved in the biosynthesis and degradation of surface polysaccharides; porS and porR, involved in anionic polysaccharide biosynthesis and surface retention (Paramonov et al., 2005; Shoji et al., 2002); and wbbL a putative rhamnosyltransferase present in a locus that also contains several glucosyltransferases. The finding that gene expression can be up- or down-regulated indicates that Ltp1 exerts precise and possible multi-level control of gene expression. Collectively, these results led us to predict that exopolysaccharide production in the Ltp1 mutant would be altered. To test this prediction, exopolysaccharide in P. gingivalis biofilms was labeled with concanavalin A- and wheat germ agglutinin- FITC. Exopolysaccharide levels were then determined by confocal microscopy and quantitative image analysis, and normalized to the levels of Syto-17 labeled P. gingivalis cells in the biofilm (Fig 4B). Polysaccharide production was increased over 2.5-fold in ΔLtp1 and in CMΔLtp1, and returned to parental levels in CΔLtp1. Hence, Ltp1 contributes to the regulation of exopolysaccharide production in P. gingivalis through impacting the transcriptional activity of several genes with synthesis and transport functions.
Fig. 4.
Ltp1 activity modulates exopolysaccharide production in P. gingivalis biofilms. (A) Quantitative Real Time RT-PCR of mRNA levels for genes involved in exopolysaccharide production and secretion. Data are presented as ratio of transcript number ΔLtp1:WT. 16S rRNA was used as a control. * denotes p < 0.01 (t test) compared to WT. (B) Levels of exopolysaccharide production in biofilms of P. gingivalis strains stained with FITC-labeled concanavalin A and wheat germ agglutinin. Bacteria were stained with Syto-17. Fluorescent images were collected by confocal microscopy and quantitated with diame software. Exopolysaccharide levels are expressed ratio of FITC:Syto-17 (exopolysaccharide:cells) fluorescence. * denotes p < 0.01 (t test) compared WT.
luxS production and hemin uptake are controlled by Ltp1
Maturation of dual and single species P. gingivalis biofilms also requires the activity of the AI-2 family of signaling molecules (McNab et al., 2003). We reasoned, therefore, that Ltp1 may also control expression of the LuxS enzyme that is responsible for AI-2 formation. P. gingivalis is one of only a few organisms in which LuxS production and AI-2 activity is regulated at the level of luxS transcription (James et al., 2006). Expression of luxS in biofilms of parental and mutant P. gingivalis cells was monitored by real time RT-PCR. Loss of Ltp1 or disruption of Ltp1 catalytic activity resulted in an approximately 3 fold increase in the levels of luxS mRNA (Fig 5A). Thus, enhanced biofilm formation by the Ltp1 mutant may arise from an increase in AI-2 mediated signaling in addition to increased exopolysaccharide production. However, as AI-2 can control exopolysaccharide production in other organisms (McNab et al., 2003; Waters and Bassler, 2005), a corollary of these data is that the primary effect of Ltp1 may be on LuxS and AI-2 production which subsequently modulate exopolysaccharide synthesis. To address this possibility, differential regulation of polysaccharide related genes was examined in planktonic (depressed Ltp1 expression) and biofilm (elevated Ltp1 expression) cells of parental and LuxS mutant strains. By real time RT-PCR we found that regulation of gene expression appears to be independent of LuxS. rfbD mRNA levels are shown in Fig 5B, and expression of PG0106, orf24, PG0116, PG0436, porS, porR and wbbL exhibited a similar pattern (not shown).
Fig. 5.
Ltp1 controls LuxS dependent signaling pathways in P. gingivalis. (A) Quantitative Real Time RT-PCR of luxS mRNA levels from biofilms of P. gingivalis strains. Data are expressed as ratio of luxS transcript relative to WT. 16S rRNA was used as a control. * denotes p < 0.01 (t test) compared to wild type. (B) Expression of rfbD by WT and ΔLuxS strains in planktonic and biofilm mode. rfbD mRNA transcript number (per microgram of RNA) was determined by quantitative Real Time RT-PCR. 16S rRNA was used as a control. * denotes p < 0.01 (t test) compared to biofilm condition. (C) Uptake of [55Fe]hemin by P. gingivalis strains measured by scintillation spectroscopy. (D) Transcriptional activity across the hmu locus in biofilms of P. gingivalis. hmu gene expression was determined by quantitative Real Time RT-PCR. Data are expressed as ratio of hmu gene transcript WT: ΔLtp1. 16S rRNA was used as a control. Hemin uptake along with mRNA levels for each of the hmu genes was significantly higher in the WT as compared to the Ltp1 mutant, p < 0.01 (t test).
In P. gingivalis LuxS/AI-2 controls pathways involved in uptake of hemin, the primary source of iron for this organism (James et al., 2006). To examine the role of Ltp1 in hemin uptake we assayed parental and Ltp1 mutant cells for uptake of [55Fe]hemin and found that in the absence of Ltp1, hemin uptake was significantly reduced (Fig 5C). As the major hemin uptake system in P. gingivalis is encoded by the hmu locus (Lewis et al., 2006), we next investigated the transcriptional activity of this locus in the Ltp1 mutant. Consistent with the hemin uptake data, all 6 genes in the hmu locus were downregulated in the absence of Ltp1 (Fig 5D). As AI-2 negatively regulates hmuR expression (James et al., 2006), these data are consistent with an indirect role for Ltp1 mediated through LuxS. However, the nature of the Ltp1 dependent control of Hmu requires further investigation.
Proteases are Client Proteins of Ltp1
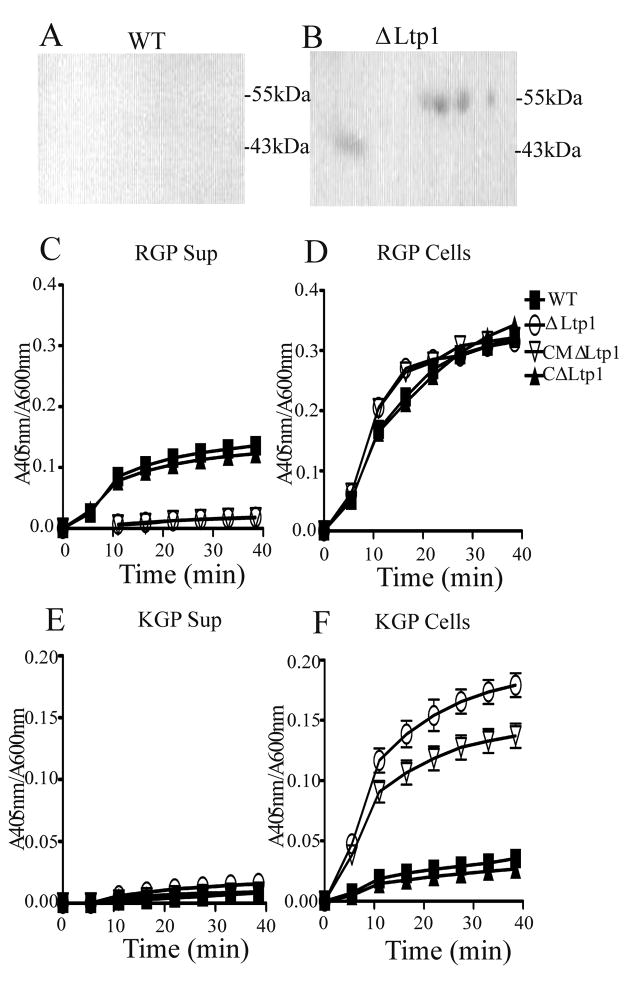
To begin to identify the set of client proteins for Ltp1, a proteomic-mass spectrometry approach was adopted. 2D gel electrophoresis followed by blotting with phosphotyrosine specific antibodies, was performed on parent and Ltp1 mutant cells. As shown in Fig. 6A&B spots of 43 and 55 kDa remained phosphorylated in the Ltp1 mutant. Amino acid sequencing by tandem mass spectrometry revealed that the 43 kDa spot contained peptides derived from the lysine specific protease Kgp, and the arginine specific protease RgpB, while the 55 kDa spots contained peptides derived from the arginine specific protease RgpA. Hence the major proteases of P. gingivalis, that contribute to nutrition and virulence through degradation of numerous host proteins (O’Brien-Simpson et al., 2003; Potempa et al., 2003), would appear to be substrates for the Ltp1 enzyme. To examine the potential role of phosphorylation of the proteases, Kgp and RgpA/B activity was measured in whole cells and culture supernatant of parent and mutant cells (Fig. 6C-F). Loss of Ltp1 activity resulted in a significant decrease in Rgp activity in culture supernatants, but not in whole cells, indicating that Ltp1 phosphatase activity is required for efficient secretion of the Rgp enzymes. In contrast, in the absence of Ltp1 the cell-associated KGP activity of P. gingivalis was increased, indicating that dephosphorylation diminishes the proteolytic activity of Kgp. Neither the rgp or kgp genes were regulated at the transcriptional level (not shown), and although this does not completely exclude the possibility that changes in the amount of proteases occurred, it does increase the likelihood that the phosphorylation status of the proteases is responsible for the observed phenotypes. However an indirect effect mediated through other pathways controlled by Ltp1 can not be excluded and indeed is likely occur concomitantly as LuxS can regulate Kgp (James et al., 2006) expression, and PorR is involved in protease secretion (Shoji et al., 2002).
Fig. 6.
Ltp1 regulates proteolytic activity. P. gingivalis WT (A) or ΔLtp1 (B) cell lysates were immunoprecipitated with phosphotyrosine antibodies, separated by 2D electrophoresis, blotted and probed with phosphotyrosine antibodies. MS/MS identified peptides derived from the lysine specific protease Kgp, and the arginine specific protease RgpB in the 43 kDa spot; and the arginine specific protease RgpA in the 55 kDa spot. Proteolytic activity was determined in culture supernatants (C, E) and whole cells (D, F) of P. gingivalis WT and ΔLtp1 strains. Release of p-nitroanilide cleaved from synthetic peptide substrates was measured by A405 and protease activity normalize to cell number (A600).
Discussion
P. gingivalis has the potential to be an aggressive pathogen in periodontal disease. The organism colonizes the biofilms that accumulate on oral non-shedding surfaces and can damage periodontal tissues through the production of arginine and lysine specific proteases. Paradoxically, P. gingivalis is also present in the oral cavity in the absence of tissue destruction, implying that mechanisms to restrain pathogenic potential are operational in the organism. The results of this study indicate that the LMWTP Ltp1 occupies a central position in regulating several aspects of P. gingivalis virulence (summarized in Fig. 7). At the phenotypic level, Ltp1 activity constrains accumulation of both monospecies P. gingivalis biofilms and heterotypic P. gingivalis-S. gordonii communities. Biofilm development in general proceeds through a series of ordered developmental steps (Stanley and Lazazzera, 2004). In the case of P. gingivalis-S. gordonii consortia, the first step is a multivalent coadhesive interaction mediated by two distinct adhesin receptor pairs. The P. gingivalis long fimbriae (FimA) bind to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) present on the streptococcal surface (Maeda et al., 2004). In addition, the P. gingivalis short fimbriae (Mfa) engage the streptococcal SspA/B (antigen I/II) adhesins (Park et al., 2005) through an approximately 27 aa binding epitope of SspA/B termed BAR (Daep et al., 2006; Demuth et al., 2001). Mfa expression is negatively regulated by the Clp proteolytic chaperone system components ClpXP (Capestany et al., 2008). Following co-adhesion, LuxS dependent signaling is required for further development of the heterotypic biofilm communities (McNab et al., 2003). With regard to monospecies P. gingivalis biofilms, initial attachment depends on an internalin family protein, InlJ (Capestany et al., 2006), and development requires expression of the short fimbriae and the universal stress protein UspA (Chen et al., 2006; Lin et al., 2006). Conversely, loss of several gene products results in enhanced biofilm growth of P. gingivalis. Inhibitors of homotypic biofilm accumulation include ClpXP along with ClpC, and GalE (UDP-galactose 4-epimerase)(Capestany et al., 2008; Nakao et al., 2006). Limitation of biofilm development appears to be important for P. gingivalis, and such mechanisms in general are thought to arise in order to optimize exposure to oxygen (either maximal or minimal), or to facilitate influx of nutrients and efflux of waste (Rainey and Rainey, 2003).
Fig. 7.
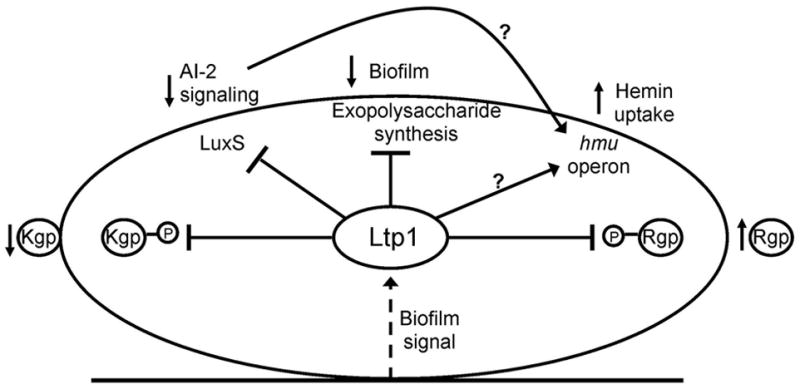
Schematic representation of the functionality of Ltp1. Contact with biotic or abiotic surfaces transduces a signal (dotted arrow) that upregulates Ltp1 expression. Ltp1 downregulates (flat arrow) expression of exopolysaccharide and LuxS, and upregulates (arrow) expression of the hmu locus. Ltp1 also dephosphorylates Rgp and Kgp. Phenotypic outcomes of Ltp1 activity include restricted biofilm development, reduced AI-2 signaling, increased uptake of hemin (as a consequence of either lower AI-2 levels or direct stimulation of hmu transcription), decreased cell associated Kgp activity and increased Rgp activity in culture supernatants.
Control of P. gingivalis biofilm development by Ltp1 was associated with a decrease in exopolysaccharide production. P. gingivalis possesses multiple loci with a potential role in exopolysaccharide production based on the presence of genes encoding products with synthesis and export functions (Aduse-Opoku et al., 2006). Ltp1 activity regulated expression of representative genes from all four of these loci:PG0106-0120 (involved in K-antigen production); PG1135–1142 (involved in anionic polysaccharide production); PG0435–0437; and PG1560–14565. Given the multi-locus involvement in the production of exopolysaccharides, the existence of a broadly active regulator such as Ltp1 can be predicted to facilitate temporal and spatial co-ordination of the synthesis and export of polymers. Exopolysaccharide production is an integral feature of many biofilm communities where it provides a matrix in which bacterial cells are embedded, and can protect from external threats (Xavier and Foster, 2007). However, as polysaccharide production is energetically costly, some organisms terminate polymer secretion at high cell density (Nadell et al., 2008). Of additional relevance to P. gingivalis, exopolysaccharide can physically propel individual cells into a more oxygenated environment (Xavier and Foster, 2007), hence the benefit to the anaerobic P. gingivalis of exopolysaccharide control mechanisms when in a community structure.
P. gingivalis mutants that are deficient in AI-2 production are unable to develop into mature biofilm communities (McNab et al., 2003). AI-2 dependent signaling is required for recruitment of P. gingivalis cells from the fluid phase and incorporation into the sessile communities. Consistent with this, Ltp1 activity in P. gingivalis biofilm cells negatively regulates expression of LuxS, an enzyme responsible for AI-2 production. Thus, Ltp1 exerts negative regulation of biofilm development through limitation of cell-cell communication, in addition to exopolysaccharide production. The broader LuxS regulon includes the Hmu hemin uptake system that is suppressed at the transcriptional level by AI-2 (James et al., 2006). Hence, as Ltp1 levels increase in biofilms, the resulting reduction in AI-2 alleviates suppression of transcriptional activity across the hmu locus and enhances hemin uptake. In this manner biofilms would be ensured an adequate supply of hemin. Elevated hemin can increase the virulence of P. gingivalis in various animal models (Kesavalu et al., 2003; Marsh et al., 1994; McKee et al., 1988); however, Ltp1 may balance this effect and mitigate virulence by concordantly restricting increases in biofilm biovolume.
The major gingipains of P. gingivalis, RgpA, RgpB and Kgp were identified as potential client proteins of Ltp1. These proteases provide peptide substrates for the asaccharyolytic P. gingivalis, and can degrade the structural components of the periodontium along with immune effector molecules. In addition, proteolytic activity is necessary for processing of P. gingivalis proteins such as fimbriae (Genco et al., 1999; Lamont and Jenkinson, 1998; O’Brien-Simpson et al., 2003), and the gingipains are considered major virulence determinants of the organism. Proteases can be posttranslationally processed for retention on the cell surface or secretion into the extracellular milieu. The arginine-specific Rgp enzymes are glycosylated and the carbohydrate domain is thought to contain phosphorylated branched mannans (Paramonov et al., 2005). The phospho-mannans are not present on secreted Rgps and hence these domains may play a role in outer membrane attachment (Veith et al., 2002). In the absence of Ltp1, Rgp activity in the supernatant was reduced, which could indicate that dephosphorylation by Ltp1 is involved in disrupting the phosphorylated branched mannan dependent attachment. In contrast, loss of Ltp1 did not affect secretion of the lysine specific protease Kgp that has not been shown to possess the phosphorylated carbohydrate epitope. Cell associated Kgp activity, however, was higher in the absence of Ltp1 activity. Consequently, in comparison to their planktonic counterparts, P. gingivalis cells in a biofilm environment will secrete high levels of Rgps possibly to ensure the nutritional requirements of the biofilm microcolony are achieved. Kgp activity, which was predominantly cell associated, will be reduced in biofilms which may prevent organisms damaging neighboring cells in the closely packed communities.
As the existence of tyrosine phosphorylation in bacteria was confirmed little more than a decade ago (Grangeasse et al., 2007), it is not surprising that the functional characterization of this posttranslational modification system is in its infancy. While numerous autophosphorylating tyrosine kinases and phosphotyrosine phosphatases have now been identified in bacteria, most of these display catalytic mechanisms that are distinct from eukaryotic enzymes. Only a limited number of eukaryotic-like LMWTP have been biochemically characterized and their cellular roles are poorly understood. In P. gingivalis the overall outcome of Ltp1 activity amounts to a virulence rheostat that facilitates persistence of P. gingivalis in the absence of overt disease. These results extend the repertoire of physiological processes controlled by LMWTP dephosphorylation in bacteria.
Experimental Procedures
Bacteria and culture conditions
P. gingivalis strains ATCC 33277, Δltp1 (Simionato et al., 2006), CΔltp1, CMΔltp1, and ΔluxS (McNab et al., 2003) were cultured in Trypticase Soy Broth (TSB), supplemented hemin and menadione, anaerobically at 37°C. When necessary, erythromycin at 10 μg ml−1, tetracycline at 1 μg ml−1, or gentamicin at 100 μg ml−1 were incorporated into the medium. For RNA extraction, monospecies biofilms were generated in tissue culture flasks by inoculation with P. gingivalis at A600 of 1.0. After 24 h incubation, the supernatant was removed and the remaining surface-associated cells were used as biofilm cells. S. gordonii DL1 was cultured anaerobically at 37°C in TSB. Escherichia coli strains were grown aerobically at 37°C in LB media containing when necessary kanamycin (50 μg ml−1) and chloramphenicol (30 μg ml−1).
Construction of complemented strains
DNA sequence containing the PG1641 ORF along with 1,000 bp upstream of the PG1641 initiation codon was amplified from P. gingivalis chromosomal DNA using primers Ltp1-F and Ltp1-R (Table S1), and cloned into shuttle vector plasmid pT-COW (Gardner et al., 1996). The resulting plasmid pT-CΔltp1, was introduced into the Δltp1 mutant by conjugation (Simionato et al., 2006) creating strain CΔltp1. A mutation in the predicted catalytic site of Ltp1 was created by using primers MLtp1-F and MLtp1-R (Table S1) to alter the cysteine 10 to serine. The mutated gene was cloned into pT-COW and the resultant plasmid pT-CMΔltp1 was conjugated into the Δltp1 mutant creating strain CMΔltp1. Transconjugants were selected with erythromycin and tetracycline and the presence of plasmids was confirmed by PCR and Southern hybridization. Plasmids were sequenced to confirm the construct.
Expression of recombinant protein
The ltp1 coding region was amplified using primers rLtp1-F and rLtp1-R (Table S1), confirmed by sequencing, cloned into pET30b and transformed into E. coli strain TunerDE3 pLysS (Novagen). His-tag protein was purified with a BioLogic DuoFlow chromatography system loaded with a Nickel-NTI column. Purity was greater than 97%, as determined by SDS-PAGE Coomassie staining, and the identity of the band confirmed by LC MS/MS sequencing.
Phosphatase enzyme assays
Phosphatase released from phosphopeptides was measured with a molybdate:malachite green assay. Phosphatase assays were performed with 1μg protein at RT, for 15 min in phosphate-free water, with Molybdate Dye/Additive mixture (Promega). Peptide substrates were: tyrosine phosphopeptide DADEpYLIPQQG (Promega); serine phosphopeptide RRApSVA (Upstate,) and threonine phosphopeptide RRApTVA (Promega). Phosphate release was detected by measuring the absorbance of a molybdate:malachite green:phosphate complex according the manufacturer’s directions. Km and Kcat values were calculated from substrate concentration curves using non-linear regression of at least 3 replicates for each concentration point (GraphPad Prism). Orthovanadate, a tyrosine phosphatase inhibitor was tested at 1 mM and sodium fluoride, a serine phosphatase inhibitor, was tested at 50 mM. Results were compared with a Student’s unpaired two-tailed t test.
P. gingivalis biofilms
Homotypic biofilm formation by P. gingivalis was quantified by a microtiter plate assay (O’Toole and Kolter, 1998), as adapted for P. gingivalis (Capestany et al., 2008). Parental and mutant strains in early log phase (2 ×108 cells) were incubated in microtiter plate wells at 37°C anaerobically for 24 h. The resulting biofilms were washed, stained with 1% crystal violet, and destained with 95% ethanol. Absorbance at 595 nm was determined with a Benchmark microplate reader. Heterotypic P. gingivalis-S. gordonii communities were generated and analyzed as described previously (Kuboniwa et al., 2006). S. gordonii cells were labeled with hexidium iodide (15 μg ml−1), then cultured for 16 h with rocking in a CultureWell coverglass system (Grace Biolabs). Fluorescein-labeled P. gingivalis cells (2 × 106 in pre-reduced PBS) were reacted with the S. gordonii biofilm for 24 h anaerobically at 37°C with rocking. The resultant communities were examined on a Yokogawa spinning disc confocal scanning laser microscope system with a 60x 1.4 N.A. objective. Images were digitally reconstructed (2D image; x-z section, y-z section and x-y section, 3D image; x-y-z section), and quantitation of P. gingivalis-specific fluorescence was determined with Imaris software (Bitplane). Quantitation of S. gordonii-specific fluorescence ensured equivalent levels of the streptococcal substratum were present in each experiment. Biofilm assays were repeated independently three times with each strain in triplicate and analyzed with a Student’s unpaired two-tailed t test.
Quantitative real time RT-PCR
Primers (Table S1) were designed by using Beacon Designer V7 software. 16S rRNA was included as a control. Predicted product sizes were in the 100- to 200-bp range. Bacterial cells were lysed using Trizol (Invitrogen), and RNA was extracted with phenol-chloroform and precipitated with isopropanol. RNA preparations were washed with 70% ethanol, dissolved in RNase-free H2O, and treated with RNase-free DNase I (QIAGEN). Treated RNA samples were further purified with a second on-column DNase I treatment using the RNeasy minikit (QIAGEN). The iScript cDNA synthesis kit (Bio-Rad) was used to generate cDNA from RNA (1 μg) templates. Specific DNA standards for the genes under investigation were synthesized from chromosomal DNA using standard PCR. Real time RT-PCR was performed on a Bio-Rad iCycler using SYBR Green Supermix (Bio-Rad). Results were analyzed with the iCycler iQ Optical System software version 3.0a. The melt curve profiles were examined to verify a single peak for each sample, and transcript copy number was calculated as described (Yin et al., 2001). RNA extracts were prepared in duplicate from independent experiments and cDNA samples were loaded in triplicate.
Exopolysaccharide measurement
Syto-17-labelled P. gingivalis strains were cultured in individual wells of CultureWell coverglass multi-chambered slides. The biofilms were washed and labeled Concanavalin A-FITC and Wheat germ agglutinin-FITC (100 μg ml−1) for 30 min at room temperature. After washing, images were collected by confocal microscopy and analyzed with daime software.
[55Fe]Hemin uptake assay
Bacterial cells were depleted of hemin by serial passage in Mycoplasma medium. Cells from logarithmically growing cultures were harvested and suspended in Mycoplasma medium to an OD of 660nm. 96-well filtration plates (Millipore, Bedford MA) were prewetted with Mycoplasma broth and 100 μl (5 × 107 cells) of bacterial cells were added to designated wells. [55Fe]hemin [specific activity 0.26 mCi mg−1 (9.6 MBq mg−1)] was then added at various time intervals to wells and the plate was incubated at 37oC, anaerobically. Cells treated with the uncoupler CCCP served as a control for energy-independent uptake, and these background levels were subtracted from the final value. Following vacuum filtration to remove the liquid the cells were washed three times with 200 μl of TBS supplemented with 0.1% Tween-20. Plates were air -dried and filters were using the MultiScreen assay system (Millipore). Radioactivity was detected by scintillation counting with “wide open” window setting, using Beckman LS6500 scintillation counter.
2D electrophoresis and protein identification
P. gingivalis cells were lysed in 10 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl, 0.1%SDS, 1% Triton X-100, 1 mM PMSF, 1 mM EDTA, 1 mM orthovanadate. Tyrosine phosphorylated proteins were enriched by immunoprecipitation with phosphotyrosine monoclonal antibody conjugated agarose (ExAlpha) and elution with 10 mM Tris-HCl pH6.8 containing10 mM O-phospho-L-tyrosine. Proteins were separated by 2D electrophoresis, transferred to nitrocellulose membranes and probed with HRP-conjugated tyrosine phosphate antibodies (Amersham). Reactions were developed by ECL. Spots of interest were excised from duplicate gels, digested with trypsin and sequenced by LC MS/MS on an ABI 4700 MALDI-TOF/TOF mass spectrometer.
Proteinase assay
P. gingivalis cells were pelleted and after 24 h the supernatant removed and filtered. The pelleted cells were washed and resuspended in 10 mM HEPES/NaOH pH 7.4. Kgp and Rgp activities were determined by hydrolysis of the synthetic substrates, N-p-tosyl-Gly-Pro-Lys-p-nitroanilide (GPKpNA) and N-α-benzoyl-L-Arg-p-nitroanilide (BApNA), respectively. Release of the cleaved product, p-nitroanilide, was measured by A405. Proteinase activities of whole cells were normalized to A600 values.
Acknowledgments
Supported by NIDCR DE12505, DE11111 (RJL), DE14605 (DRD) and DE18039 (JPL).
References
- Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, Boutaga K, Laine ML, Van Winkelhoff AJ, Curtis MA. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun. 2006;74:449–460. doi: 10.1128/IAI.74.1.449-460.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albandar JM. Epidemiology and risk factors of periodontal diseases. Dent Clin North Am. 2005;49:517–532. v–vi. doi: 10.1016/j.cden.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Black DS, Marie-Cardine A, Schraven B, Bliska JB. The Yersinia tyrosine phosphatase YopH targets a novel adhesion-regulated signalling complex in macrophages. Cell Microbiol. 2000;2:401–414. doi: 10.1046/j.1462-5822.2000.00061.x. [DOI] [PubMed] [Google Scholar]
- Capestany CA, Kuboniwa M, Jung IY, Park Y, Tribble GD, Lamont RJ. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun. 2006;74:3002–3005. doi: 10.1128/IAI.74.5.3002-3005.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190:1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Palmer RJ, Kuramitsu HK. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect Immun. 2002;70:4708–4715. doi: 10.1128/IAI.70.8.4708-4715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Honma K, Sharma A, Kuramitsu HK. A universal stress protein of Porphyromonas gingivalis is involved in stress responses and biofilm formation. FEMS Microbiol Lett. 2006;264:15–21. doi: 10.1111/j.1574-6968.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- Cozzone AJ, Grangeasse C, Doublet P, Duclos B. Protein phosphorylation on tyrosine in bacteria. Arch Microbiol. 2004;181:171–181. doi: 10.1007/s00203-003-0640-6. [DOI] [PubMed] [Google Scholar]
- Cozzone AJ. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J Mol Microbiol Biotechnol. 2005;9:198–213. doi: 10.1159/000089648. [DOI] [PubMed] [Google Scholar]
- Daep CA, James DM, Lamont RJ, Demuth DR. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect Immun. 2006;74:5756–5762. doi: 10.1128/IAI.00813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol. 2006;188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun. 2001;69:5736–5741. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Zhou D. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A. 2000;97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl Environ Microbiol. 1996;62:196–202. doi: 10.1128/aem.62.1.196-202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genco CA, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, Demuth DR, Lamont RJ. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Holt SC, Ebersole JL. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol Immunol. 2003;18:226–233. doi: 10.1034/j.1399-302x.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Tribble GD, James CE, Kilic AO, Tao L, Herzberg MC, Shizukuishi S, Lamont RJ. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JP, Plata K, Yu F, Rosato A, Anaya C. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152:3367–3382. doi: 10.1099/mic.0.29011-0. [DOI] [PubMed] [Google Scholar]
- Lin SL, Le TX, Cowen DS. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell Microbiol. 2003;5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Wu J, Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect Immun. 2006;74:6011–6015. doi: 10.1128/IAI.00797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, Shizukuishi S. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 2004;6:1163–1170. doi: 10.1016/j.micinf.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Marsh PD, McDermid AS, McKee AS, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis W50. Microbiology. 1994;140:861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- McKee AS, McDermid AS, Wait R, Baskerville A, Marsh PD. Isolation of colonial variants of Bacteroides gingivalis W50 with a reduced virulence. J Med Microbiol. 1988;27:59–64. doi: 10.1099/00222615-27-1-59. [DOI] [PubMed] [Google Scholar]
- McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona JK, Paton JC, Miller DC, Morona R. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol. 2000;35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- Morona JK, Morona R, Miller DC, Paton JC. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J Bacteriol. 2002;184:577–583. doi: 10.1128/JB.184.2.577-583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona JK, Morona R, Paton JC. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci U S A. 2006;103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murli S, Watson RO, Galan JE. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cell Microbiol. 2001;3:795–810. doi: 10.1046/j.1462-5822.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- Musumeci L, Bongiorni C, Tautz L, Edwards RA, Osterman A, Perego M, Mustelin T, Bottini N. Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis. J Bacteriol. 2005;187:4945–4956. doi: 10.1128/JB.187.14.4945-4956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R, Senpuku H, Watanabe H. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun. 2006;74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003;4:409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- Preneta R, Jarraud S, Vincent C, Doublet P, Duclos B, Etienne J, Cozzone AJ. Isolation and characterization of a protein-tyrosine kinase and a phosphotyrosine-protein phosphatase from Klebsiella pneumoniae. Comp Biochem Physiol B Biochem Mol Biol. 2002;131:103–112. doi: 10.1016/s1096-4959(01)00490-0. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. Nature. 2003;425:72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- Simionato MR, Tucker CM, Kuboniwa M, Lamont G, Demuth DR, Tribble GD, Lamont RJ. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun. 2006;74:6419–6428. doi: 10.1128/IAI.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- Tierrez A, Garcia-del Portillo F. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell Microbiol. 2005;7:901–909. doi: 10.1111/j.1462-5822.2005.00540.x. [DOI] [PubMed] [Google Scholar]
- Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci U S A. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Talbo GH, Slakeski N, Dashper SG, Moore C, Paolini RA, Reynolds EC. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem J. 2002;363:105–115. doi: 10.1042/0264-6021:3630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci U S A. 2007;104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- Yin JL, Shackel NA, Zekry A, McGuinness PH, Richards C, Putten KV, McCaughan GW, Eris JM, Bishop GA. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol Cell Biol. 2001;79:213–221. doi: 10.1046/j.1440-1711.2001.01002.x. [DOI] [PubMed] [Google Scholar]