Abstract
Although plasma human immunodeficiency virus type 1 (HIV-1) RNA concentration is a major determinant of the rate of HIV-1 disease progression, the reasons for variability in plasma virus loads among infected individuals are not fully understood. We conducted investigations with 15 HIV-1-infected individuals who were not receiving antiretroviral therapy to evaluate the hypothesis that HIV-1 replication rate in vitro is a significant determinant of plasma virus load. Virus could not be isolated from one subject. Two subjects were excluded because they had features previously associated with distinct plasma virus loads and altered rates of disease progression; one harbored a syncytium-inducing virus and the second was heterozygous for a 32-bp deletion from the CCR5 gene. HIV-1 replication rates were determined by culturing autologous virus isolates in phytohemagglutinin-treated peripheral blood mononuclear cells (PBMC) and determining the rate of p24 antigen production during the logarithmic phase of viral replication. The contribution of HIV-1 reverse transcriptase (RT) and protease (PR) alleles to replication capacity was assessed using recombinant viruses in a single-cycle infection assay. HIV-1 replication rates ranged from 0.15 to 0.76 log10 pg/ml/day and were reproducible within the same donor PBMC (coefficient of variation ± 4%). RT-PR replication capacity ranged from 14 to 95% of that of control virus and was linearly related to replication rate (r2 = 0.53; P = 0.007). Plasma HIV-1 RNA concentration was linearly related to replication rate (r2 = 0.71; P < 0.001) and RT-PR replication capacity (r2 = 0.44; P = 0.019). These data suggest that different RT-PR alleles are important determinants of HIV-1 replication rates and that HIV-1 replication rate explains much of the variability in plasma virus load in chronic HIV-1 infection.
In the absence of antiretroviral therapy, most human immunodeficiency virus type 1 (HIV-1)-infected individuals progress to AIDS and death. The median time between seroconversion and the development of AIDS is approximately 10 years (16). Rates of disease progression, however, are highly variable, ranging from rapid progression to AIDS within 1 year to long-term asymptomatic survival for over 15 years. The plasma HIV-1 RNA concentration in untreated HIV-1-infected individuals is a significant surrogate marker of the rate of disease progression (32). Although both host factors and viral factors are important determinants of the rate of HIV-1 disease progression, the reasons for variability in plasma HIV-1 RNA concentration and disease course among individuals with chronic HIV-1 infection are not fully understood (23, 24).
Multiple host factors have been associated with altered plasma virus loads and rates of disease progression (reviewed in references 23 and 24). A 32-bp deletion from the chemokine receptor CCR5 gene (CCR5Δ32), which prevents normal expression of this HIV-1 coreceptor, has been associated with diminished susceptibility to HIV-1 infection, lower plasma HIV-1 RNA concentrations, and delayed disease progression (13, 15). Levels of CCR5 expression and CD4+ T cells differ greatly even among individuals who are homozygous wild type at this locus, and in one study the density of CCR5 molecules on CD4+ T cells correlated with the plasma HIV-1 RNA concentration (37). Host CD4+ cell susceptibility independent of levels of CCR5 expression has been found to be a critical determinant of plasma virus concentration and disease progression in macaques infected with the same strain of simian immunodeficiency virus (19), suggesting that other factors besides CCR5 expression affect host cell susceptibility. Levels of expression of RANTES, a β-chemokine that competitively inhibits HIV-1 entry through the CCR5 coreceptor, have been associated with altered rates of disease progression (34a). Virus-specific CD8+ T cells, which demonstrate potent antiretroviral activity in vitro (43), have been implicated in disease progression as well. Long-term nonprogressors have vigorous HIV-1-specific CD8+ T-cell responses compared to other HIV-1-infected individuals (21, 38), suggesting that these responses are critical determinants of disease progression. Removal of CD8+ cells from simian immunodeficiency virus-infected macaques augments plasma viremia (28, 39), further bolstering the hypothesis that CD8+ lymphocytes are critical to virus control in vivo.
Several lines of evidence support an association between viral phenotype and rate of HIV-1 disease progression. Early in the HIV-1 epidemic it was recognized that infection with viruses that induced syncytia on transformed T-cell lines (SI), and that were subsequently found to use the CXCR4 coreceptor for entry, was associated with higher plasma HIV-1 RNA concentrations and more-rapid disease progression than infection with viruses that did not induce syncytia on these cell lines (NSI) and that used the CCR5 coreceptor (12, 42). Long-term nonprogressors who harbor HIV-1 with mutations in nef have been described, and the viruses infecting those individuals have been characterized as less fit than wild-type viruses from individuals with progressive disease (5). An assay that measures the contribution of reverse transcriptase (RT) and protease (PR) to virus replication has been used to show that drug-resistant HIV-1 isolates have impaired replicative capacity (14). Diminished fitness of these isolates has been hypothesized to explain the clinical benefit of antiretroviral therapy in the setting of persistent virus replication. Further evidence for a link between virus replication rate and disease progression is suggested by the results of a study that showed that HIV-1 harbored by three long-term survivors had significantly less replicative fitness in growth competition experiments compared with HIV-1 harbored by three individuals with progressive disease (36); viral replicative fitness in these assays was significantly correlated with subjects' plasma HIV-1 RNA concentrations.
In the present study, we hypothesized that if the HIV-1 replication rate is a significant determinant of disease progression, a simple measure of the virus replication rate in vitro would be linearly related to plasma virus concentration. In a cohort of chronically HIV-1-infected individuals who were not receiving antiretroviral therapy, we evaluated the relationship between plasma HIV-1 RNA concentration and the in vitro replication rate of low-passage-number HIV-1 isolates as well as the replication capacity of recombinant viruses that contained autologous RT and PR. To avoid potential confounding effects from other factors known to be associated with HIV-1 disease progression, our analysis was restricted to subjects who were homozygous for wild-type CCR5 and who were infected with NSI viruses. The observation that expression of the chemokine receptors CXCR4 and CCR5 is differentially affected in peripheral blood mononuclear cells (PBMC) under culture conditions similar to those used in our in vitro replication assay provided an additional rationale for the exclusion of individuals infected with SI viruses (6).
(These data were presented in part at the 9th Conference on Retroviruses and Opportunistic Infections, Seattle, Wash., abstr. 346, 27 February 2002.)
MATERIALS AND METHODS
Human subjects. The subjects of this study were HIV-1-infected individuals recruited for a study of lymphoid tissue immune responses who were not receiving antiretroviral therapy. Individuals who were known or suspected to have become infected within the preceding 6 months were excluded. HIV-1-seronegative subjects were laboratory workers who were tested every 6 months for HIV-1 antibodies. Informed consent was obtained from all participants in this study in accordance with the Colorado Multiple Institutional Review Board.
Plasma virus measurements. The HIV-1 Monitor Assay (Roche Diagnostics, Indianapolis, Ind.) was used according to the manufacturer's instructions to measure HIV-1 RNA levels in plasma.
CCR5Δ32 genotyping. DNA was isolated from subjects' cells, amplified by PCR, and analyzed by electrophoresis on ethidium-stained agarose gels as previously described (33). Homozygous wild-type DNA and heterozygous DNA were run in parallel as controls.
SI phenotype. SI phenotypes were determined on MT2 cells as previously described (26).
HIV-1 isolation and titration. HIV-1 was isolated from PBMC or inguinal lymph node tissue by cocultivation with PBMC from multiple HIV-1-seronegative donors according to standard techniques (25). Viral stock titers were determined using a single HIV-1 seronegative donor's PBMCs that were stimulated with phytohemagglutinin (PHA) and interleukin-2 (IL-2) (PHA lymphoblasts) for 3 days as previously described (27). This donor's PBMCs were never used for initial virus isolation. The 50% tissue culture infective dose was calculated by the Spearman-Karber method (J. Hubert, Bioassay, p. 65-66; Spearman-Karber method, 2nd ed., Hunt Publishing, Dubuque, Iowa, 1984).
HIV-1 replication assays. Replication of HIV-1 isolates was assayed by culturing 2,000 50% tissue culture infective doses of each isolate with 2 × 106 3-day-old PHA lymphoblasts in 2 ml of growth medium consisting of RPMI 1640 (Gibco BRL, Gaithersburg, Md.) supplemented with 20% heat-inactivated fetal bovine serum (Sigma, St. Louis, Mo.), penicillin (Sigma) (100 U/ml), streptomycin (Sigma) (0.1 mg/ml), IL-2 (Roche Diagnostics) (10 U/ml), and l-glutamine (Gibco BRL) (200 nM). PHA lymphoblasts were obtained from the same seronegative donor who provided PBMCs for virus titration. Assays were performed using four different batches of PHA lymphoblasts obtained from this donor on four different days. Cultures were incubated with virus for 2 h, washed two times with phosphate-buffered saline, and incubated in the medium described above for 10 days at 37°C, 95% humidity, and 5% CO2. A 0.2-ml aliquot of culture supernatant was harvested on days 0, 1, 2, 3, 4, 6, 8, and 10 and assayed for p24 antigen by enzyme-linked immunosorbent assay (Beckman Coulter, Miami, Fla.). Cultures were fed with 10% medium changes on the days that the supernatant was harvested.
Drug susceptibility testing. GeneSeq HIV and PhenoSense HIV assays (ViroLogic, Inc., South San Francisco, Calif.) were used as genotypic and phenotypic assays, respectively, for the determination of virus isolate antiretroviral drug resistance.
RT and PR replication capacity. The contribution of RT and PR to replication capacity was assessed using a modified version of the PhenoSense assay (ViroLogic, Inc.). A retroviral vector capable of measuring replication capacity was constructed using the NL4-3 infectious molecular clone of HIV-1. The vector contains a luciferase expression cassette replacing the HIV-1 envelope gene. RT and PR sequences were amplified from autologous virus stocks and inserted into the vector using defined restriction enzyme sites. Recombinant viruses were generated by introducing retroviral vector DNA into 293 cells by transfection along with an expression vector that produces the envelope protein of murine leukemia virus. At 2 days after transfection, virus was harvested and used to inoculate new cell cultures, which were incubated for an additional 2 to 3 days. Input virus at infection was normalized by the efficiency of transfection, as measured by the luciferase expressed by the vector DNA in the transfected cells. After normalization, the amount of luciferase activity detected in the infected cells was used as a direct measure of RT and PR replication capacity. The RT and PR replication capacity of recombinant viruses derived from patient isolates was expressed as the percent replication of a recombinant reference virus derived from NL4-3. The replication capacity of the NL4-3 reference virus (100%) closely approximates the mean replication capacity of recombinant viruses derived from more than 1,000 wild-type isolates (T. Wrin and C. J. Petropoulos, unpublished data).
RESULTS
Study subjects.
HIV-1 was isolated from 14 of 15 subjects from whom virus isolation was attempted. One subject was excluded from further analysis because he was a CCR5Δ32 heterozygote, and a second subject was excluded because he harbored a SI virus. The clinical and demographic characteristics of the remaining 12 individuals who are the subjects of the following analyses are presented in Table 1. All subjects were male, three-quarters of the group were Caucasian, and the median age was 36 years. The median CD4+ T-cell count was 403 cells/mm3 (range, 107 to 726 cells/mm3), and the median plasma HIV-1 RNA concentration was 26,145 copies/ml (range, 538 to 166,875 copies/ml). Eight subjects were antiretroviral therapy naïve, whereas four had received single- or dual-nucleoside analog RT inhibitor therapy for periods of time ranging from 2 to 12 months. No subject received antiretroviral therapy within the 3 months preceding virus isolation. None of the virus isolates contained well-recognized PR or RT inhibitor resistance mutations or exhibited drug susceptibility outside of the natural variation of wild-type viruses.
TABLE 1.
Demographic and clinical characteristics of study subjects
| Subject | Age (yr) | Race | Prior therapya | HIV-1 riskb | CD4+ T cell count (cells/mm3) | HIV-1 RNA in plasma (copies/ml) |
|---|---|---|---|---|---|---|
| 1 | 37 | Caucasian | Yes | M | 169 | 74,335 |
| 3 | 43 | Black | No | M | 464 | 149,049 |
| 4 | 33 | Hispanic | Yes | M, I | 163 | 18,742 |
| 5 | 24 | Caucasian | Yes | M, I | 282 | 35,331 |
| 10 | 32 | Caucasian | No | M, I | 342 | 29,144 |
| 11 | 44 | Caucasian | No | M | 591 | 166,875 |
| 13 | 38 | Caucasian | No | M, I | 107 | 46,390 |
| 17 | 40 | Caucasian | No | M | 571 | 1,673 |
| 20 | 50 | Caucasian | No | I | 215 | 4,380 |
| 23 | 32 | Caucasian | No | M | 726 | 10,022 |
| 25 | 35 | Caucasian | No | M | 553 | 23,145 |
| 26 | 35 | Hispanic | Yes | M | 473 | 538 |
Subjects with a prior history of antiretroviral therapy, which in all instances was limited to nucleoside analog reverse transcriptase inhibitor treatment.
M, men having sex with men; I, intravenous drug use.
HIV-1 replication assays.
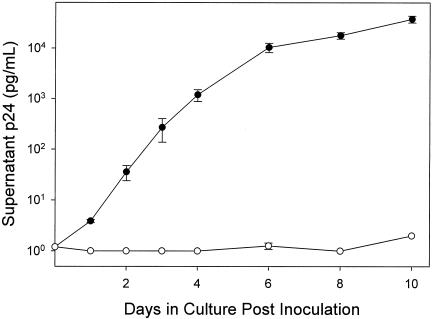
HIV-1 replication assays from all subjects demonstrated a triphasic pattern of p24 antigen production, as has previously been observed (11, 31). For all isolates, there was an initial lag phase between days 0 and 1 after inoculation, a logarithmic phase between day 1 and day 4 or 6, and a plateau phase between day 4 or 6 and day 10. To investigate the cause of the plateau phase, PHA lymphoblasts were inoculated with HIV-1 on days 3 and 7. Inoculation of PHA lymphoblasts on day 3 produced the expected triphasic kinetics of supernatant p24 antigen accumulation (Fig. 1). In contrast, no significant p24 antigen production was observed in PHA lymphoblasts inoculated on day 7 (Fig. 1). Similar results were obtained in six experiments evaluating growth of three different HIV-1 isolates in lymphoblasts derived from two different donors (data not shown). These findings suggested that the plateau in p24 antigen production observed in HIV-1 replication assays could have resulted from loss of ability of PHA lymphoblasts to become newly infected after day 4 of the assay. Alternatively, the plateau could be explained by infection of all susceptible cells in the culture prior to day 4. In either case, the continued accumulation of p24 antigen between days 4 and 6 likely resulted from virus production in cells infected prior to day 4.
FIG. 1.
Growth curves for an HIV-1 isolate in PHA lymphoblasts inoculated on day 3 (closed circles) and day 7 (open circles). Closed circles show the typical triphasic growth curve for an HIV-1 isolate after inoculation of lymphoblasts on day 3, with a lag phase (days 0 to 1), exponential phase (days 1 to 6) and plateau phase (days 6 to 10). Open circles show growth of the same HIV-1 isolate on PHA lymphoblasts inoculated on day 7. All data points are the mean ± range of two parallel cultures.
HIV-1 replication rates.
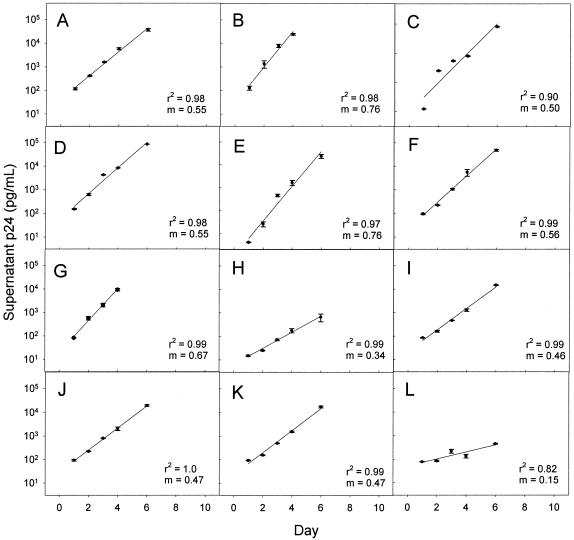
The replication rate of each HIV-1 isolate was determined by linear regression analysis of log10-transformed culture supernatant p24 antigen concentration (Fig. 2). To avoid influences of lag and plateau phases of HIV-1 replication on measurement of HIV-1 replication rate, all replication rates were determined from the slope of the exponential phase of p24 accumulation. In 10 subjects the exponential phase of p24 accumulation occurred between days 1 and 6, and in 2 subjects the exponential phase occurred between days 1 and 4. All infections were performed in duplicate. For each isolate there was a strong linear relationship between mean log10-transformed supernatant p24 antigen concentration and time (r2 range, 0.82 to 1.0). There was a fivefold range of HIV-1 replication rates (0.15 to 0.76 log10 pg/ml/day; median, 0.53 log10 pg/ml/day). Replication rate determinations were highly reproducible. The coefficient of variation for replicate determinations with PBMC from a single donor was ± 4% (intradonor variability). To evaluate interdonor variability, the replication rates of three different viruses were measured using PHA lymphoblasts from three different donors known to be homozygous wild type at the CCR5 locus (Table 2). The coefficient of variation for interdonor variability was ± 9%.
FIG. 2.
Measurement of HIV-1 replication rate. PHA lymphoblasts from a single donor were infected with NSI HIV-1 isolates from 12 different subjects on day 0. Data during the phase of exponential increase of supernatant p24 were fitted by linear regression. The coefficient of determination (r2) and slope (m) for each regression are shown. The slope (m) is the viral replication rate. All data points are the mean ± range of two parallel cultures. (A) Subject 1. (B) Subject 3. (C) Subject 4. (D) Subject 5. (E) Subject 10. (F) Subject 11. (G) Subject 13. (H) Subject 17. (I) Subject 20. (J) Subject 23. (K) Subject 25. (L) Subject 26.
TABLE 2.
Measurement of HIV-1 replication rates in PBMC from different donors
PR and RT replication capacity.
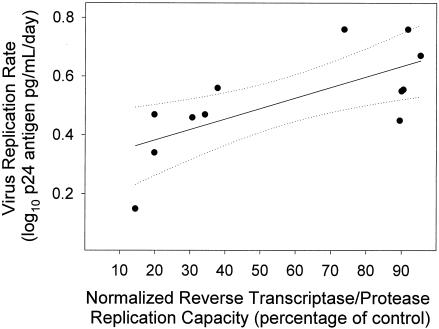
PR and RT replication capacity of recombinant viruses derived from patient isolates relative to the NL4-3-derived reference virus (100%) ranged from 14 to 95% (median, 56%). PR and RT replication capacity was linearly related to the HIV-1 replication rate measured in PHA lymphoblasts (r2 = 0.53; P = 0.007) (Fig. 3).
FIG. 3.
HIV-1 replication rate in PHA lymphoblasts was linearly related to HIV-1 RT and PR replication capacity. HIV-1 replication rate is the slope (m) of the regressions in Fig. 2. RT and PR replication capacity were determined in a single cycle-based assay using recombinant virus that contained the RT and PR genes of each HIV-1 isolate. The replication capacity is the percentage of virus replication relative to the reference virus strain, NL4-3. Solid line indicates fit of data by linear regression (r2 = 0.53; P = 0.007). Dashed lines indicate the 95% confidence interval for the regression.
Relationship between in vitro measurements of HIV-1 replication and plasma HIV-1 RNA concentration.
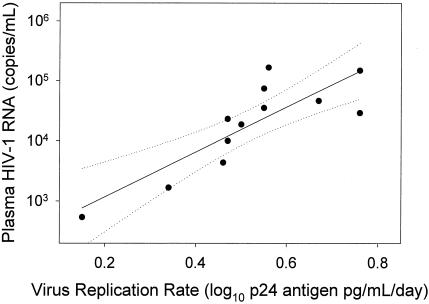
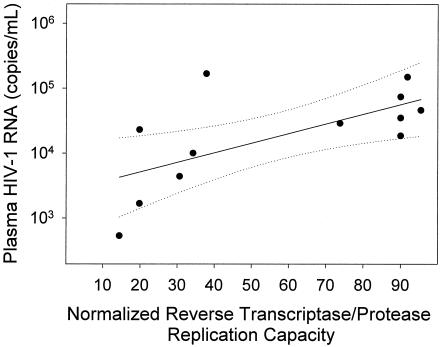
There was a strong linear relationship (r2 = 0.71; P < 0.001) between the HIV-1 replication rate and plasma HIV-1 RNA concentration (Fig. 4). This relationship spanned a range of plasma HIV-1 RNA levels of 3 orders of magnitude. The relationship between the growth rate and plasma HIV-1 RNA concentration was statistically significant even when the highest (r2 = 0.66; P = 0.003) or the lowest (r2 = 0.52; P = 0.013) values were omitted from the calculation. However, when data from the subjects with the two lowest plasma HIV-1 RNA concentration results (subjects 17 and 26) were removed from the analysis, the linear relationship did not reach statistical significance (r2 = 0.33; P = 0.08). Thus, the strong overall relationship between the replication rate and plasma HIV-1 RNA concentration was not overly influenced by data points at the extreme high and low ends of the spectrum of plasma HIV-1 RNA concentration but was dependent on analysis of data from subjects with a broad range of plasma virus load. RT and PR replication capacity levels were linearly related (r2 = 0.44; P = 0.019) with plasma HIV-1 RNA concentration as well (Fig. 5), although the relationship was not as strong as that for the HIV-1 replication rate and plasma HIV-1 RNA concentration.
FIG. 4.
HIV-1 replication rate in PHA lymphoblasts was linearly related to plasma HIV-1 RNA concentration. Plasma HIV-1 RNA values are from Table 1 and virus replication rate is the slope of the regressions in Fig. 2. Solid line indicates fit of data by linear regression (r2 = 0.71; P < 0.001). Dashed lines indicate the 95% confidence interval for the regression.
FIG. 5.
HIV-1 RT and PR replication capacity was linearly related to plasma HIV-1 RNA concentration. RT and PR replication capacity was determined in a single cycle-based assay using recombinant virus as described in the legend to Fig. 3. Solid line indicates fit of data by linear regression (r2 = 0.44; P = 0.019). Dashed lines indicate the 95% confidence interval for the regression.
DISCUSSION
In chronic HIV-1 infection, the plasma HIV-1 RNA concentration represents a quasi-steady-state equilibrium between virus production and virus clearance. Over the past decade, much attention has been focused on the latter part of this equation, namely, on virus clearance. It has become widely accepted, on the basis of compelling data that CD8+ T cells mediate potent antiretroviral activity both in vitro (8, 10, 43, 45) and in vivo (28, 39) and that differences in HIV-1-specific CD8+ T-cell responses account for most differences in plasma virus concentration among HIV-1-infected individuals. Nevertheless, increasing evidence suggests that this paradigm might not be correct. With few exceptions (9, 34), the results of most studies of chronically infected individuals have shown that the magnitude and breadth of HIV-1-specific CD8+ cell responses are not inversely correlated with the plasma HIV-1 RNA concentration (1, 2, 20, 29), contrary to what would be expected if these immune responses were the primary determinant of the plasma HIV-1 RNA concentration. Indeed, in some instances HIV-1-specific CD8+ T-cell responses have correlated directly with the plasma HIV-1 RNA concentration (4, 17). Studies within our own laboratory that included some of the same subjects in the present study failed to reveal significant correlations between the plasma HIV-1 RNA concentration and HIV-1-specific CD8+ cell response in either PBMCs or lymph node cells (R. Schlichtemeier, J. E. Forster, S. MaWhinney, A. M. Mian, J. M. Folkvord, A. H. Harken, and E. Connick, unpublished data) and provided the impetus for the present investigation of HIV-1 replication rate.
We evaluated the relationship between HIV-1 replication rate in vitro and plasma HIV-1 RNA concentration in a cohort of individuals who were not receiving antiretroviral therapy. Subjects with SI virus or CCR5Δ32 heterozygosity, both features known to be associated with distinct rates of disease progression and viral loads, were excluded from the analysis. It is notable that most individuals are initially infected with NSI viruses (46) and that the majority of HIV-1-infected individuals are homozygous wild type at the CCR5 locus (33, 41). Thus, only a minority of individuals (14%) were excluded from our analyses as a result of these restrictions. We found a strong linear relationship between HIV-1 replication rate and plasma HIV-1 RNA concentration that spanned a fourfold range of viral replication rate and a 3 log10 range of plasma HIV-1 RNA concentration. These data suggest that HIV-1 replication rate is a major determinant of plasma virus concentration, and this finding is likely widely applicable to many HIV-1-infected individuals.
The notion that virus replication rate is a significant determinant of plasma virus load and disease progression is not new but was proposed early in the HIV-1 epidemic, when it was observed that virus was more readily isolated from individuals with highly symptomatic HIV-1 infection and replicated more quickly in vitro than virus from less symptomatic or asymptomatic individuals (3). The switch from usage of the CCR5 coreceptor (NSI virus) to usage of the CXCR4 coreceptor (SI virus) for virus entry was frequently found to be associated with increased cytopathicity and viral load (12). Subsequent studies demonstrated that HIV-1 NSI virus often increased in cytopathicity and replication rate during the course of progressive HIV-1 infection even without changing coreceptor usage (11, 30). Studies of long-term nonprogressors, which used techniques similar, but not identical, to those used in our laboratory, found evidence of impaired HIV-1 replicative capacity in approximately half of the individuals who were long-term nonprogressors (5, 7). More recently, Quiñones-Mateu et al. (36) used a dual infection competition assay to show that virus from three long-term nonprogressors was significantly less fit than that from three progressors; a significant correlation between plasma virus concentration in vivo and virus fitness in vitro was observed. Our study is unique in that it utilized virus isolates from a cohort of HIV-1-infected individuals and did not preselect for individuals with rapid or slow disease progression. Furthermore, the present study demonstrates that during a short period of in vitro culture, substantial differences in virus replication rate can be detected with a simple viral growth assay.
There are several potential limitations to the replication rate assay that we employed in this study. First, the assay was performed with PBMC from a single donor, as measurements of cells from different donors had greater variability than cells from a single donor. Indeed, studies using macaque PBMCs have demonstrated that CD4+ cells from different individuals may differ greatly in their ability to support retrovirus replication, independent of CCR5 expression, and CD4+ cell susceptibility to infection is a predictor of the virus set point (19). Thus, measurements of replication rate among different donors are not likely to be directly comparable. A second potential limitation of the replication rate assay is that it involves treatment of PBMC with PHA and IL-2. These culture conditions have been shown to upregulate CXCR4 expression rapidly on CD4+ cells, whereas CCR5 expression increases more slowly (6). This differential effect on coreceptor expression could alter the replication rate of viruses that use the different coreceptors with respect to each other. It remains to be determined whether replication rates of SI and NSI viruses are comparable in this system. Another potential limitation of the assay's reliance upon PHA lymphoblasts is that they are highly activated cells that are not necessarily physiologically similar to the cells in lymphoid tissues that are infected in vivo and produce the majority of plasma virus. The impact of viral proteins, such as Tat and Nef, which have been shown to play a critical role in activating CD4+ T-cells and enhancing HIV-1 replication (44), would likely be minimized in our assay. Thus, it is conceivable that this assay underestimates the contribution of virus replication rate to plasma HIV-1 RNA concentration. More physiologically relevant models, such as lymphoid tissue histoculture systems (18), may be necessary to fully discern the relationship between virus replication rate and plasma virus concentration.
A number of HIV-1 gene products may contribute to overall virus replication rate. The RT and PR replication capacity assay measures solely the contribution of these two viral proteins to replication rate. Nevertheless, and somewhat surprisingly, we observed a significant linear relationship between RT and PR replication capacity and HIV-1 replication rate, suggesting that different RT-PR alleles contribute significantly to virus replication rate in vitro. Several studies have documented diminished replication capacity or diminished fitness of HIV-1 isolates with drug resistance mutations in these proteins (35). Virus from subjects with partial virus suppression in the context of antiretroviral therapy has been shown to have impaired RT and PR replication capacity compared to the wild-type virus of these same individuals in the absence of antiretroviral therapy (14). The magnitude of difference in replication capacity in this earlier study was found to correlate with the magnitude of increase in plasma HIV-1 RNA concentration when antiretroviral therapy was discontinued. Drug resistance, however, is unlikely to explain the differences in virus replication rate among subjects in the present study, since the majority of subjects were antiretroviral therapy naïve and none harbored significant drug resistance mutations. The present study demonstrates that RT and PR contribute significantly to virus replication rate and plasma virus concentration in antiretroviral naïve or minimally treated individuals. It is conceivable that the RT and PR replication capacity assay may be even more strongly related to plasma HIV-1 RNA concentration and virus fitness in antiretroviral treatment-experienced populations of HIV-1-infected individuals than in antiretroviral treatment-naïve populations.
We observed a significant linear relationship between HIV-1 replication rate in vitro and plasma HIV-1 level in vivo. These data suggest that differences in HIV-1 replication rates among HIV-1 isolates are a major determinant of disease progression. It is likely that differences among individuals in HIV-1 replication rate have obscured other relevant determinants of virus replication in vivo. For example, although HIV-1-specific CD8+ cell responses have often not correlated with plasma HIV-1 RNA concentration, it is conceivable that a relationship might emerge after controlling for virus replication rate. Other host cellular factors have also been implicated in control of virus replication (22, 40), and their role in vivo may be more easily discerned after controlling for virus replication rate as well. A better understanding of the contribution of virus replication rate to plasma HIV-1 RNA concentration is critical to unraveling other determinants of disease progression in HIV-1 infection.
Acknowledgments
This work was supported by Public Health Service grants R29AI-42499 and AI41536 from the National Institutes of Health. GeneSeq HIV, PhenoSense HIV, and replication capacity assays were performed at ViroLogic, South San Francisco, Calif., which also supported those studies. Development of the replication capacity assay was supported in part by a Small Business Innovative Research grant (1 R43 AI50321) from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
We express our gratitude to the subjects who participated in this study. We also thank Matthew Purner and Monique Givens for technical assistance; Shirley Nelson for performing the assays for plasma HIV-1 RNA; Arshia Mian for assistance in preparation of the manuscript; and Michael Grodesky, Robert Schooley, Susan Valone, Bev Putnam, Larry Sharkey, Cheryl McDonald, Ruth Berggren, Wheaton Williams, M. Graham Ray, Julie Subiadur, Ron Schimmel, Eileen Dunne, Steven Johnson, and Alex Kallen for assistance in recruiting subjects to this study.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193:169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asjo, B., L. Morfeldt-Manson, J. Albert, G. Biberfeld, A. Karlsson, K. Lidman, and E. M. Fenyo. 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet ii:660-662. [PubMed] [Google Scholar]
- 4.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. [DOI] [PubMed] [Google Scholar]
- 6.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. L.-Y., F. Francois, A. Mosoian, and M. E. Klotman. 2003. CAF-mediated human immunodeficiency virus (HIV) type 1 transcriptional inhibition is distinct from α-defensin-1 HIV inhibition. J. Virol. 77:6777-6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chouquet, C., B. Autran, E. Gomard, J. M. Bouley, V. Calvez, C. Katlama, D. Costagliola, and Y. Riviere. 2002. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS 16:2399-2407. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., H. Mohri, Y. Cao, and D. D. Ho. 1993. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 67:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, and S. J. O'Brien. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 14.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 15.de Roda Husman, A. M., M. Koot, M. Cornelissen, I. P. Keet, M. Brouwer, S. M. Broersen, M. Bakker, M. T. Roos, M. Prins, F. de Wolf, R. A. Coutinho, F. Miedema, J. Goudsmit, and H. Schuitemaker. 1997. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann. Intern. Med. 127:882-890. [DOI] [PubMed] [Google Scholar]
- 16.Easterbrook, P. J. 1999. Long-term non-progression in HIV infection: definitions and epidemiological issues. J. Infect. 38:71-73. [DOI] [PubMed] [Google Scholar]
- 17.Ferbas, J., E. S. Daar, K. Grovit-Ferbas, W. J. Lech, R. Detels, J. V. Giorgi, and A. H. Kaplan. 1996. Rapid evolution of human immunodeficiency virus strains with increased replicative capacity during the seronegative window of primary infection. J. Virol. 70:7285-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glushakova, S., J. C. Grivel, K. Suryanarayana, P. Meylan, J. D. Lifson, R. Desrosiers, and L. Margolis. 1999. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J. Virol. 73:3968-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, S., C. R. Brown, H. Dehghani, J. D. Lifson, and V. M. Hirsch. 2000. Intrinsic susceptibility of rhesus macaque peripheral CD4+ T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J. Virol. 74:9388-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, C. M., J. Lawrence, J. M. Schapiro, J. D. Altman, M. A. Winters, M. Crompton, M. Loi, S. K. Kundu, M. M. Davis, and T. C. Merigan. 1999. Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART). J. Immunol. 162:1780-1788. [PubMed] [Google Scholar]
- 21.Harrer, T., E. Harrer, S. A. Kalams, T. Elbeik, S. I. Staprans, M. B. Feinberg, Y. Cao, D. D. Ho, T. Yilma, A. M. Caliendo, R. P. Johnson, S. P. Buchbinder, and B. D. Walker. 1996. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res. Hum. Retrovir. 12:585-592. [DOI] [PubMed] [Google Scholar]
- 22.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan, C. M., and S. M. Hammer. 2001. Host determinants in HIV infection and disease. Part 1: cellular and humoral immune responses. Ann. Intern. Med. 134:761-776. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, C. M., and S. M. Hammer. 2001. Host determinants in HIV infection and disease. Part 2: genetic factors and implications for antiretroviral therapeutics. Ann. Intern. Med. 134:978-996. [DOI] [PubMed] [Google Scholar]
- 25.Hollinger, F. B., J. W. Bremer, L. E. Myers, J. W. Gold, L. McQuay, and the NIH/NIAID/DAIDS/ACTG Virology Laboratories. 1992. Standardization of sensitive human immunodeficiency virus coculture procedures and establishment of a multicenter quality assurance program for the AIDS Clinical Trials Group. J. Clin. Microbiol. 30:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Japour, A. J., S. A. Fiscus, J. M. Arduino, D. L. Mayers, P. S. Reichelderfer, and D. R. Kuritzkes. 1994. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J. Clin. Microbiol. 32:2291-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Japour, A. J., D. L. Mayers, V. A. Johnson, D. R. Kuritzkes, L. A. Beckett, J.-M. Arduino, J. Lane, R. J. Black, P. S. Reichelderfer, R. T. D'Aquila, C. S. Crumpacker, the RV-43 Study Group, and the AIDS Clinical Trials Group Virology Committee Resistance Working Group. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalams, S. A., S. P. Buchbinder, E. S. Rosenberg, J. M. Billingsley, D. S. Colbert, N. G. Jones, A. K. Shea, A. K. Trocha, and B. D. Walker. 1999. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J. Virol. 73:6715-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 31.Lu, W., and J. M. Andrieu. 1992. Similar replication capacities of primary human immunodeficiency virus type 1 isolates derived from a wide range of clinical sources. J. Virol. 66:334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellors, J. W., L. A. Kingsley, C. R. Rinaldo, Jr., J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 33.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. W. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338-340. [DOI] [PubMed] [Google Scholar]
- 34.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 34a.Paxton, W. A., A. U. Neumann, S. Kang, et al. 2001. RANTES production from CD4+ lymphocytes correlates with host genotype and rates of human immunodeficiency virus type 1 disease progression. J. Infect Dis. 183:1678-1681. [DOI] [PubMed] [Google Scholar]
- 35.Quinones-Mateu, M. E., and E. J. Arts. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resistance Updates 5:224-233. [DOI] [PubMed] [Google Scholar]
- 36.Quiñones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynes, J., P. Portales, M. Segondy, V. Baillat, P. Andre, B. Reant, O. Avinens, G. Couderc, M. Benkirane, J. Clot, J. F. Eliaou, and P. Corbeau. 2000. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 181:927-932. [DOI] [PubMed] [Google Scholar]
- 38.Rinaldo, C., X. L. Huang, Z. F. Fan, M. Ding, L. Beltz, A. Logar, D. Panicali, G. Mazzara, J. Liebmann, M. Cottrill, et al. 1995. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J. Virol. 69:5838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 40.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 41.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, and S. J. O'Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 42.Tersmette, M., J. M. Lange, R. E. de Goede, F. de Wolf, J. K. Eeftink-Schattenkerk, P. T. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedema. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed] [Google Scholar]
- 43.Walker, B. D., S. Chakrabarti, B. Moss, T. J. Paradis, T. Flynn, A. G. Durno, R. S. Blumberg, J. C. Kaplan, M. S. Hirsch, and R. T. Schooley. 1987. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328:345-348. [DOI] [PubMed] [Google Scholar]
- 44.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]