Abstract
The positive-stranded RNA genome of Plautia stali intestine virus (PSIV) has an internal ribosome entry site (IRES) in an intergenic region (IGR). The IGR-IRES of PSIV initiates translation of the capsid protein by using CAA, the codon for glutamine. It was previously reported (J. Sasaki and N. Nakashima, J. Virol. 73:1219-1226, 1999) that IGR-IRES extended by several nucleotides into the capsid open reading frame (ORF). Despite the fact that the secondary structure model of the IGR-IRES is highly conserved, we were unable to find structural similarities in the 5′ region of the capsid ORFs in related viruses. Therefore, we reevaluated the role of the capsid ORF in IGR-IRES-mediated translation in PSIV. Mutation of the CAA codon with various triplets did not inhibit IGR-IRES-mediated translation. N-terminal amino acid analyses of mutated products showed that the IGR-IRES could initiate translation by using various elongator tRNAs. By replacement of the capsid ORF with exogenous coding sequences having AUG deleted, translation products were produced in most cases, but capsid-exogenous fusion proteins were produced more efficiently than were the translation products. These data indicate that the 5′ part of the capsid ORF is not an absolute requirement for the IGR-IRES-mediated translation. RNA structure probing analyses showed that the 5′ part of the capsid ORF was a single strand, while that of exogenous reading frames was structured. Exogenous sequences also caused structural distortion in the 3′ part of the IGR-IRES. We hypothesize that the single-stranded capsid ORF helps to form the tertiary structure of the IGR-IRES and facilitates precise positioning of ribosomes.
Internal ribosome entry is an alternative mode of translation initiation that was first found in the RNA genomes of mammalian picornaviruses (15, 23). An RNA tertiary structure that is formed at a region upstream of the coding sequence recruits ribosomes to the mRNA and allows translation to occur independently of the 5′ cap. The RNA element responsible for ribosome entry is called the internal ribosome entry site (IRES) (11, 13, 19, 35).
Plautia stali intestine virus (PSIV) is a member of the family Dicistroviridae (20), formerly known as insect picorna-like or cricket paralysis-like virus (3, 21, 31). The positive-stranded RNA genome of picornaviruses contains one IRES element in the 5′ region. In contrast, the genome of dicistroviruses contains two IRES elements. One IRES is for nonstructural protein precursor translation, and the other is for the capsid protein precursor (37, 39). To distinguish between the two IRES elements, the one found in the intergenic region (IGR) between nonstructural and capsid protein precursor genes in dicistroviruses is called IGR-IRES.
An unusual feature of IGR-IRES-mediated translation is that translation of the capsid open reading frames (ORFs) starts with glutamine (33) or alanine (5, 38) instead of methionine. Usually, the initiation site selection for translation involves base pair formation between an AUG initiation codon and the anticodon triplet of an initiator methionine tRNA (Met-tRNAi). Met-tRNAi is essential for translation initiation, and methionine is the only amino acid that initiates protein synthesis in normal mRNAs (12, 17, 26). In contrast, the formation of a pseudoknot (PK) immediately upstream of the capsid ORF enables methionine-independent initiation of translation in dicistroviruses (5, 33, 38).
Data from mutagenesis studies have predicted that IGR-IRES has four stem-loops, which include three PK structures (16). The predicted structure has been verified by RNA structure probing analysis for both cricket paralysis virus (CrPV) (14) and PSIV (22). These studies show that the structural elements in the IGR-IRES are crucial for the initiation of translation. Because mutations have been previously introduced in the first decoded triplet in IGR-IRES-mediated translation (25, 32, 38), translation initiation involving amino acids other than glutamine or alanine could be possible (25, 27). However, deletion analysis, which was designed to determine the 3′ border of IGR-IRES elements, showed that several nucleotides in the 5′ part of the capsid ORFs were necessary for initiation of translation in PSIV (32) and Rhopalosiphum padi virus (RhPV) (5).
To clarify the role of the 5′ part of the capsid ORF in the IGR-IRES-mediated translation, we made various constructs with both mutations at the first decoded triplet and replacements of the capsid ORF with exogenous sequences. In vitro translation studies indicated that the capsid ORF was not absolutely required but enhanced the IGR-IRES-mediated translation. Data from RNA structure probing analysis suggested that this enhancing effect probably was associated with the single-strandedness of the 5′ part of the capsid ORF.
MATERIALS AND METHODS
Plasmid construction.
A series of constructs with mutations in the first decoded codon of the second cistron in pT7CAT-5375 (32) were generated by PCR-based mutagenesis (16). To determine whether the capsid ORF is required for IGR-IRES-mediated translation, coding sequences for firefly luciferase (Fluc), Renilla luciferase (Rluc), green fluorescent protein (GFP), and enhanced green fluorescent protein (EGFP) having ATG deleted were inserted immediately downstream of PK I of pT7CAT-5375. These four coding sequences having ATG deleted were amplified by PCR from pSP-luc+ and pRL-null (Promega), pGFP (Clontech), and pEGFP (Novagen), with 20-mer forward primers with the fourth nucleotide in the coding sequences at the 5′ end and reverse primers with EcoRI sequences in the 5′ parts for directional cloning. The four amplified fragments were phosphorylated, digested with EcoRI, and then ligated with the EcoRI-digested fragment of pT7CAT-5375, from which the capsid ORF had been deleted by PCR, to construct pT7CAT-IRES-ΔaugFluc, pT7CAT-IRES-ΔaugRluc, pT7CAT-IRES-ΔaugGFP, and pT7CAT-IRES-ΔaugEGFP, respectively. The pT7CAT-IRES-ΔaugEGFPmut was obtained from pT7CAT-IRES-ΔaugEGFP by PCR-based mutagenesis. To conduct translation by ribosome scanning, the coding sequences of Fluc, Rluc, and GFP, either with or without the ATG initiation codon, were amplified by PCR and ligated into pT7Blue (Novagen) that had been digested with HincII and EcoRI to construct pT7Fluc, pT7Rluc, pT7GFP, pT7ΔaugFluc, pT7ΔaugRluc, and pT7ΔaugGFP, respectively. When pT7EGFP was constructed, by methods described above, the translation product was not observed, probably because of an unfavorable interaction between the 5′ untranslated region and the EGFP coding sequence. To facilitate ATG-dependent translation, an omega sequence (obtained from pEU3-NII; Toyobo) was used for the 5′ untranslated region as a translation enhancer (10).
To isolate translation products, we constructed plasmids that produce glutathione S-transferase (GST) fusion proteins from the second cistrons of pT7CAT-5375-cgu and pT7CAT-5375-aaa. The GST coding sequence that corresponded to nucleotides 261 to 1047 of pGEX-6P-3 (Amersham Biosciences) was obtained by PCR. The amplified fragments were phosphorylated, treated with EcoRI, and ligated into large fragments of pT7CAT-5375-cgu and pT7CAT-5375-aaa that had been digested with XhoI, blunted with Klenow enzyme, and then digested with EcoRI, to construct pT7CAT-cguCP-GST and pT7CAT-aaaCP-GST. To examine methionine aminopeptidases in wheat germ extracts, the second cistrons of the two plasmids were amplified with ATG-containing forward primers and cloned into pEU3-NII, generating pEU-cguCP-GST and pEU-aaaCP-GST.
To quantify the translation products mediated by the IGR-IRES, we constructed monocistronic plasmids that had EGFP and Fluc coding sequences with ATG initiation codons immediately downstream of PK I. The ATG triplets were introduced into pT7CAT-IRES-ΔaugEGFP and pT7CAT-IRES-ΔaugFluc by PCR-based mutagenesis. To delete the chloramphenicol acetyltransferase (CAT) coding sequence from the ATG-containing plasmids, the plasmids were digested with HindIII and EcoRV, blunt ended, and ligated to construct pT7-IRES6192-EGFP and pT7-IRES6192-Fluc. To construct plasmids containing the capsid coding sequence at the 5′ end of the downstream coding sequence, a SalI-EcoRI-digested EGFP fragment obtained from pEGFP was ligated into the SalI and EcoRI sites of pT7Blue to construct pT7-EGFP2. The PSIV sequences from nucleotides 5668 to 6240 and 5668 to 6297 were amplified by PCR with forward primers that contained HindIII sequences in their 5′ parts and reverse primers containing SalI sequences in their 5′ parts. The amplified PCR products were digested with HindIII and SalI and then ligated into the HindIII and SalI sites of pT7-EGFP2 to construct pT7-IRES6240-EGFP and pT7-IRES6297-EGFP. To replace the coding sequence of EGFP in the two plasmids with that of Fluc, these plasmids were digested with NcoI and EcoRI. The resultant 3.4-kb fragments from each plasmid were ligated with the Fluc coding sequence, which was obtained by NcoI and EcoRI digestion from pSP-luc+ to construct pT7-IRES6240-Fluc and pT7-IRES6297-Fluc.
For structure probing assays, we constructed monocistronic plasmids because dicistronic transcripts did not give clear results. The wild-type PSIV sequence between nucleotides 5950 and 6279 was amplified from pT7CAT-5375 with a forward primer containing HindIII sequence in the 5′ part and a reverse 18-mer primer. To obtain sequences for IRES-ΔaugEGFP, IRES-ΔaugFluc, and IRES6195-LUC, 18-mer reverse primers were designed at 87 nucleotides downstream of the 5′ ends of the coding regions and then amplified from pT7CAT-IRES-ΔaugEGFP, pT7CAT-IRES-ΔaugFluc, and pCAT-IRES6195-LUC. The forward primers contained the HindIII sequences described above. The amplified fragments were ligated with pT7Blue, which had been treated with HindIII and HincII, to yield pT7-IRES-CP, pT7-IRES-ΔaugEGFP, pT7-IRES-ΔaugFluc, and pT7-IRES6195-LUC.
RNA synthesis and in vitro translation.
Plasmids were linearized with restriction enzymes before in vitro transcription. Both capped and uncapped RNAs were synthesized with a Ribomax large-scale RNA production system (Promega) in the presence or absence of a cap analog, 7mGpppG (Promega). Capped RNAs were used for translation mediated by a scanning mechanism in monocistronic RNAs. Usually, 180 pmol of RNAs/ml was translated in wheat germ extracts in the presence of 0.4 nmol of biotinylated lysyl-tRNA (Roche)/ml. To detect small amounts of translation products, 430 pmol of RNAs/ml and 1.6 nmol of biotinylated lysyl-tRNA/ml were used for the experiments depicted in Fig. 4. The biotinylated translation products were separated and detected as described previously (16).
FIG. 4.
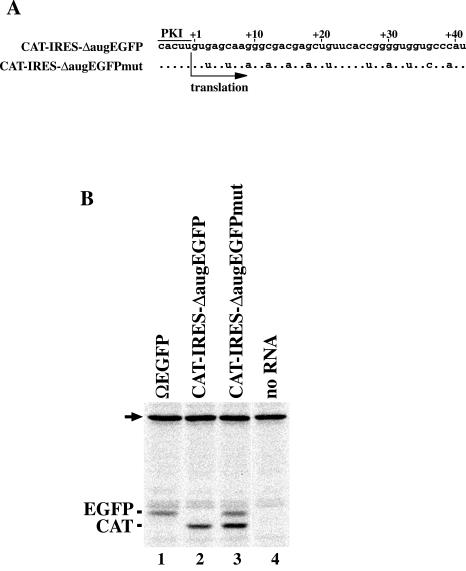
Recovery of translation efficiency by decreasing GC content in the 5′ region of EGFP coding sequence having AUG deleted. (A) Nucleotide sequences of 5′ regions of second cistrons in the transcripts from pT7CAT-IRES-ΔaugEGFP and pT7CAT-IRES-ΔaugEGFPmut. The numbers on the sequences represent nucleotide positions of the second cistrons. Dots represent nucleotides identical to the ones shown above. (B) Comparison of electrophoretic mobilities of translation products. Positions of the products are shown at left of the panel. The arrow indicates endogenous biotinylated protein in the wheat germ extract.
N-terminal sequence analysis.
RNAs were translated in a Proteios wheat germ extract kit (Toyobo) that was prepared from highly purified wheat germ to prevent inactivation of ribosome by tritin (18). A 50-μl reaction mixture containing 50 μg of dicistronic RNA or 25 μg of monocistronic RNA was incubated at 25°C for 20 h. GST fusion proteins were purified with a bulk GST purification module (Amersham Biosciences). N-terminal sequences were determined as described previously (31).
Quantification of reporter proteins.
To reevaluate the IGR-IRES activity of the transcripts from pCAT-IRES6195-LUC (32), translation was performed using a rabbit reticulocyte lysate system (Promega) in a 12.5-μl reaction mixture containing 11, 45, and 180 pmol of RNAs/ml. After incubation at 30°C for 1 h, 5 μl of the reaction mixture was diluted to 25 μl with distilled water and mixed with an equal volume of Steady Glo luciferase assay reagent (Promega). The luminescence intensity was measured using a Gene Light 55 luminometer (Microtech Nition). To examine the effect of the 5′ region of the capsid ORF on translation efficiency, 4 μg of RNAs was translated in a 25-μl reaction mixture by using the Proteios kit. After incubation at 25°C for 2 h, 1 μl of the reaction mixture was diluted to 25 μl and the luminescence intensity was measured. Fluorescence emission of EGFP was measured at 535 nm with an ABI Prism 7700 sequence detector (Applied Biosystems). The quantity of reporter proteins in the reaction mixtures was calculated by the use of standard curves that were constructed by adding known amounts of recombinant EGFP (Clontech) and firefly luciferase (Toyo Inki Inc.) to wheat germ extracts. All experiments were independently repeated three times.
Structure probing analysis of RNA.
Preliminary experiments for RNase T1 cleavage assay were done using the method of Brown et al. (1). Conditions were subsequently modified for the IGR-IRES of PSIV. RNAs were dissolved in buffer A (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 50 mM KCl), heated at 70°C for 5 min, and then slowly cooled to room temperature. The RNAs (2.8 pmol) were digested with 5 U of RNase T1 (Ambion) in 40 μl of buffer A at 37°C for 6 min. After phenol extraction and isopropanol precipitation, digested RNAs were dissolved in 10 μl of diethyl pyrocarbonate-treated water. A half volume of the dissolved RNAs was mixed with 2 pmol of 33P-labeled primers, heated at 90°C for 2 min, and then cooled quickly on ice. For primer extension, 5 U of ReverTra Ace (Toyobo) was added and the mixtures were incubated at 51°C for 15 min. Modifications with dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimidemetho-p-toluene sulfonate (CMCT) were made as described previously (22). Primers corresponding to nucleotides 6279 to 6262 in the PSIV sequence, 90 to 74 in the EGFP coding sequence, and 90 to 73 in the firefly luciferase coding sequence were used for extension reactions.
RESULTS
The wild-type initiator triplet can be replaced by various triplet codons.
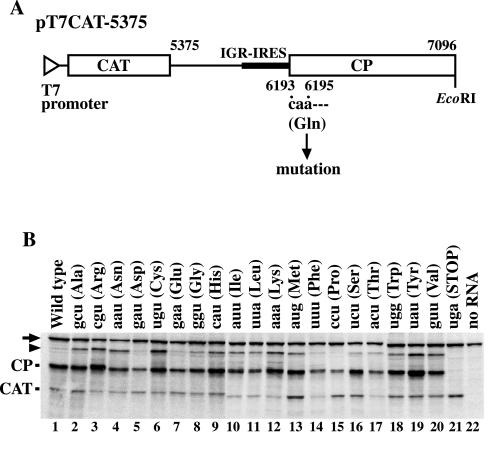
To examine the effect of changing the first decoded triplet (CAA, nucleotides 6193 to 6195) on IGR-IRES-mediated translation, we introduced mutations into pT7CAT-5375 (Fig. 1A). This plasmid contains a T7 promoter sequence followed by a CAT gene and nucleotides 5375 to 7096 of PSIV and produces CAT protein from the first cistron and the truncated capsid protein from the second cistron (32). Products from the second cistron were translated from the mutated transcripts, with the single exception of a transcript with the UGA stop codon at nucleotides 6193 to 6195 (Fig. 1B). Mutations in the CAA triplet resulted in quantitative variations in products from the second cistron, but a number of mutant RNAs gave levels of products comparable to those of the wild type (Fig. 1B, lanes 2, 3, 6, 9, 12, 13, 16, 18, 19, and 20). These results indicate that the wild-type CAA triplet codon is not essential for IGR-IRES-mediated translation.
FIG. 1.
Effect of the first decoded triplet on IGR-IRES-mediated translation in PSIV. (A) Schematic diagram of the dicistronic plasmid pT7CAT-5375. The open triangle indicates a T7 promoter, and open boxes indicate ORFs for CAT and capsid protein (CP). The bold line indicates IGR-IRES. The numbers indicate nucleotide positions corresponding to the PSIV genome. The first codon of the capsid ORF, nucleotides 6193 to 6195, was mutated. (B) Translation products of the mutated constructs. The mutated first triplets are indicated on the lanes. Deduced amino acids encoded by the triplets are shown in parentheses. Positions of the products are indicated at left of the panel. The arrow indicates endogenous biotinylated protein in the wheat germ extract. The arrowhead indicates aggregates of the capsid protein and its template RNA. These extra bands disappear when the reaction mixtures are treated with puromycin or RNases after incubation for translation. Also, introduction of a stop codon in the 3′ part of the second cistron prevents the appearance of these bands.
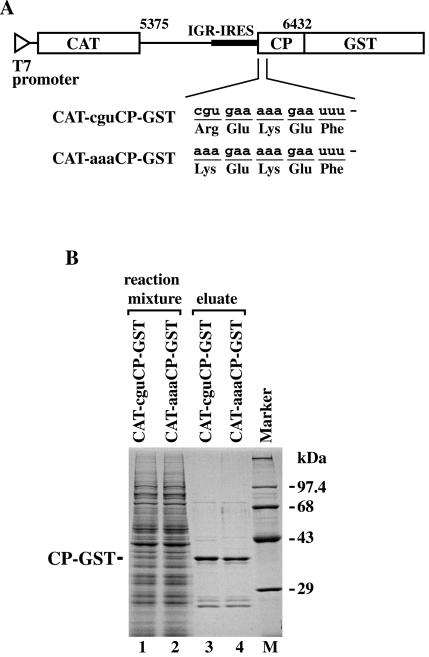
To confirm that incorporation of the first amino acid depends on the mutated triplets in the second cistron, we determined N-terminal amino acid sequences of translation products mediated by the IGR-IRES. To isolate products from the second cistrons, we constructed two plasmids that produce C-terminal GST fusion proteins, pT7CAT-cguCP-GST and pT7CAT-aaaCP-GST (Fig. 2A). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of each affinity-purified eluate showed that a major protein band was stained with Coomassie brilliant blue (Fig. 2B). The N-terminal amino acid sequences of the products from CAT-cguCP-GST and CAT-aaaCP-GST were Arg-Glu-Lys-Glu-Phe and Lys-Glu-Lys-Glu-Phe, respectively. To examine the role of methionine aminopeptidases in the wheat germ extract translation system, the N-terminal sequences of the products of AUG-dependent translation from pEU-cguCP-GST and pEU-aaaCP-GST were also determined. The sequences were Met-Arg-Glu-Lys-Glu-Phe and Met-Lys-Glu-Lys-Glu-Phe, respectively, indicating that methionine aminopeptidases were not active in the extract when the penultimate residues were lysine or arginine. Because the N-terminal sequence of the wild-type PSIV 33-kDa capsid protein was Gln-Glu-Lys-Glu-Phe (31), the mutations introduced in the first CAA triplet caused mutations in the N-terminal amino acid. These results indicate that the IGR-IRES can initiate translation by using elongator tRNAs for many different triplet codons.
FIG. 2.
Isolation of the IGR-IRES-mediated translation products for N-terminal amino acid sequence analysis. (A) Plasmids for GST fusion protein expression. The open triangle indicates a T7 promoter, and open boxes indicate ORFs for CAT and GST-fused capsid protein (CP). A bold line indicates IGR-IRES. The numbers indicate nucleotide positions corresponding to the PSIV genome. Nucleotide sequences of transcripts immediately downstream of PK I and the expected N-terminal amino acid sequences are shown below. (B) Separation of GST fusion proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis after isolation with glutathione-Sepharose beads. Protein bands were stained with Coomassie brilliant blue. Positions of the products are shown at left of the panel.
The capsid ORF is not an absolute requirement for IGR-IRES-mediated translation.
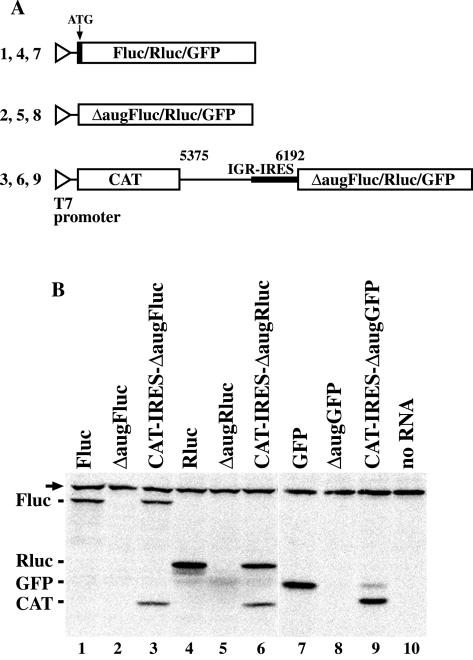
Deletion studies have shown that several nucleotides in the 5′ region of the PSIV capsid coding sequence were necessary for IGR-IRES-mediated translation (32). However, mutations in the first triplet of the coding region did not impair IGR-IRES-mediated translation (Fig. 1B). To examine the effect of the downstream coding sequence on IGR-IRES-mediated translation, we replaced the capsid ORF in pT7CAT-5375 with the Fluc, Rluc, and GFP genes having ATG deleted (Fig. 3A). The electrophoretic mobilities of polypeptides from the IGR-IRES-mediated translation mechanism were compared with those from the scanning mechanism. Translation from CAT-IRES-ΔaugFluc produced two proteins, CAT and a protein with an electrophoretic mobility similar to that of Fluc (Fig. 3B, lanes 1 and 3). However, there were no translation products from ΔaugFluc with the authentic AUG initiation codon deleted (Fig. 3B, lane 2). Similar results were obtained using Rluc (Fig. 3B, lanes 4 to 6) and GFP (Fig. 3B, lanes 7 to 9). These results suggest that the IGR-IRES of PSIV can translate exogenous sequences having AUG deleted without the capsid ORF.
FIG. 3.
IGR-IRES-mediated translation in exogenous reading frames without the capsid ORF. (A) Schematic diagrams of plasmids for translation mediated by a scanning mechanism, either with or without authentic AUG triplets, and for translation mediated by IGR-IRES. Open triangles indicate T7 promoters. Open boxes indicate coding sequences to be translated. The bold line indicates IGR-IRES. The numbers above the line indicate nucleotide positions corresponding to the PSIV genome. Numbers on the left of constructs correspond to lane numbers in panel B. (B) Comparison of electrophoretic mobilities of translation products. Positions of the products are indicated at left of the panel. The arrow indicates endogenous biotinylated protein in the wheat germ extract.
The absence of translation products from the previously constructed second cistrons in pCAT-IRES6192-LUC and pCAT-IRES6195-LUC led us to the conclusion that the IGR-IRES of PSIV extends slightly into the capsid ORF (32), as has also been concluded for RhPV (5). The prior reports, as well as our present results, point to the discrepancy regarding the role of the capsid ORF in IGR-IRES-mediated translation. To address this issue, we reevaluated the translation efficiencies of transcripts from pCAT-IRES6195-LUC, used in a previous experiment (32), by measuring luciferase activity. In vitro translation of transcripts in rabbit reticulocyte lysates showed that increases in luciferase activity were related to increases in quantities of RNAs (Table 1). When the RNA transcripts from pCAT-IRES6195-LUC were translated using a concentration of 180 pmol/ml, equivalent to conditions described for Fig. 3B, the luminescence intensity of the translation mixture was 32,191 compared to 184,753 for pT7CAT-IRES-ΔaugFluc. To examine nonspecific translation initiation in our in vitro system, we used the construct pCAT-IRES6195-LUC/fs, which has a frameshift mutation made by deleting the 13th adenine from the second cistron of pCAT-IRES6195-LUC. Since the background levels of luminescence intensity detected in transcripts from pCAT-IRES6195-LUC/fs were 831, the intensity observed in the pCAT-IRES6195-LUC construct was not the result of nonspecific initiation. When the activity of 11 pmol of RNA/ml was measured under conditions equivalent to those previously reported (32), the luminescence intensity of pCAT-IRES6195-LUC decreased to 258, indicating translation activity of a background level. These results show that translation of transcripts from pCAT-IRES6195-LUC, although very ineffective, was not abolished. The introduction of a BamHI sequence (GGATCC) during the construction of pCAT-IRES6195-LUC between the PSIV and a luciferase coding sequence, originally done to define the 3′ border of the IRES (32), was associated with decreased translation efficiency of this construct. Therefore, the capsid ORF of PSIV is not an absolute requirement for the in vitro IGR-IRES-mediated translation.
TABLE 1.
Influence of insertion of a BamHI sequence on IGR-IRES activity
| Construct | Luminescence intensity (RLUa) at RNA concn (pmol/ml):
|
||
|---|---|---|---|
| 11 | 45 | 180 | |
| pCAT-IRES6195-LUC | 258 ± 14 | 5,073 ± 378 | 32,191 ± 2,123 |
| pT7CAT-IRES-ΔaugFluc | 2,260 ± 334 | 34,524 ± 4,031 | 184,753 ± 7,835 |
| pCAT-IRES6195-LUC/fs | 102 ± 14 | 238 ± 14 | 831 ± 44 |
RLU, relative light units.
The wild-type capsid ORF has an enhancing effect on IGR-IRES-mediated translation.
Reevaluation of translation activity in pCAT-IRES6195-LUC showed that the 5′ part of the coding sequence affected IGR-IRES-mediated translation, but the quantitative relationship between the capsid ORF and the yield of translation products is unknown. To quantify the effect of the capsid ORF on IGR-IRES-mediated translation, we constructed monocistronic plasmids with EGFP and Fluc reading frames at 0, 48, and 105 nucleotides downstream of the 5′ end of the capsid ORF. Quantification of the translation products is shown in Table 2. No detectable protein was detected from the transcripts of pT7-IRES6192-EGFP, which had no capsid ORF. However, when the 48- and 105-nucleotide capsid ORFs were inserted, the amount of EGFP increased to 13.95 and 19.04 μg/ml, respectively (Table 2). Because it was possible to detect 0.20 μg of EGFP/ml under our experimental conditions, insertion of the 48-nucleotide sequence indicated stimulation of the amount of products by about 70-fold. With the luciferase gene, transcripts from pT7-IRES6192-Fluc produced 0.40 μg of luciferase/ml, while those from pT7-IRES6240-Fluc and T7-IRES6297-Fluc produced 3.79 and 5.03 μg/ml, respectively (Table 2), indicating that insertion of the 48-nucleotide sequence stimulated the amount of products by about ninefold. These results indicate that the wild-type capsid ORF has an enhancing effect on the translation.
TABLE 2.
Enhancing effect of 5′ region of capsid ORF on IGR-IRES-mediated translation
| Construct | Capsid ORF (no. of bases) | EGFP
|
FLUC
|
||
|---|---|---|---|---|---|
| Fluorescence intensityc | Amt of protein (μg/ml) | Luminescence intensity (RLUa) | Amt of protein (μg/ml) | ||
| pT7-IRES6192-EGFP/Fluc | 0 | NDb | <0.20 | 33,416 ± 772 | 0.40 ± 0.01 |
| pT7-IRES6240-EGFP/Fluc | 48 | 1,743 ± 91 | 13.95 ± 0.73 | 314,510 ± 6,120 | 3.79 ± 0.08 |
| pT7-IRES6297-EGFP/Fluc | 105 | 2,447 ± 39 | 19.04 ± 0.30 | 417,647 ± 6,473 | 5.03 ± 0.08 |
RLU, relative light units.
ND, not detectable.
Arbitrary units.
The IGR-IRES prefers lower GC contents at the 5′ part of the coding sequence.
The structure of IGR-IRES has multiple base pair interactions involving four stem-loops and three PKs (16). Structure probing analysis of IGR-IRES elements of CrPV (14) and PSIV (22) showed that the predicted model fundamentally represents a functional structure. While the predicted structures of the IGR elements in dicistroviruses are highly conserved (16) and 5′ parts of the capsid ORFs seemed to be necessary for the translation in both PSIV (32) and RhPV (5), we were unable to find any structural similarities in the 5′ regions of dicistroviruses by using the MFOLD program. However, comparison of nucleotide sequences in the 5′ parts of capsid ORFs in dicistroviruses suggested that these viruses have AU-rich sequences in these regions. The average GC content in the first 48 nucleotides in the capsid ORFs of 10 dicistroviruses is 29.3%, while the EGFP coding sequence has a GC content in the 5′-terminal 48 nucleotides of 66.7%. These observations could mean that the GC-rich sequence in the 5′ region in a coding sequence is incompatible with IGR-IRES-mediated translation.
To test the importance of GC content, we constructed pT7CAT-IRES-ΔaugEGFPmut, which has a wild-type GFP coding sequence in the first 41 nucleotides in the second cistron of pT7CAT-IRES-ΔaugEGFP, in order to decrease GC content in this region (Fig. 4A). Translation from CAT-IRES-ΔaugEGFP produced only the CAT from the first cistron (Fig. 4, lane 2), while that from CAT-IRES-ΔaugEGFPmut produced both CAT and a protein whose electrophoretic mobility was indistinguishable from that of EGFP (Fig. 4, lane 3). These results suggested that a low GC content of the 5′ part of the coding sequence was preferred for IGR-IRES-mediated translation.
Exogenous coding sequences distort RNA structures around the initiation site.
Because structural elements in the IGR-IRES have multiple base pair interactions, it is likely that inefficiently translated RNAs would be structurally distorted. To test this hypothesis, we compared structures of RNAs that have exogenous reading frames immediately downstream of PK I.
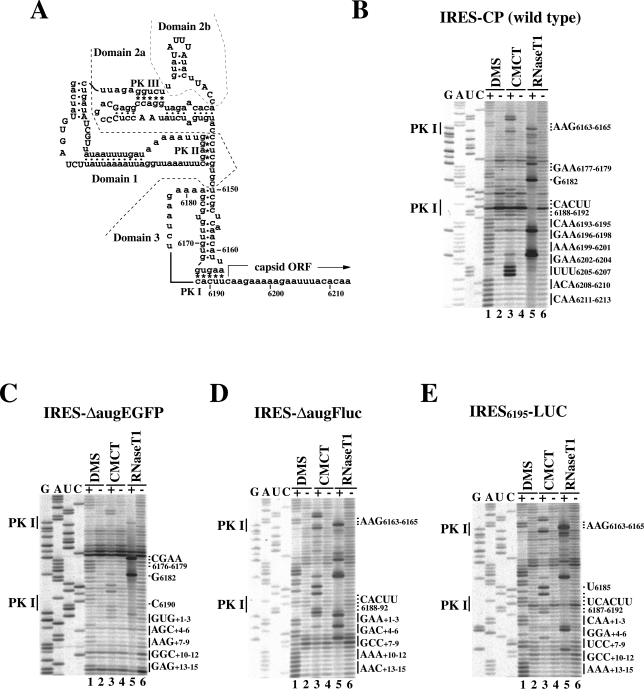
RNase T1 specifically cuts the 3′ part of the G residue that is located in a single-stranded region. After partial digestion of IRES-CP with RNase T1, G6202 and G6196, which are located in the 5′ part of the capsid ORF, were most efficiently cleaved (Fig. 5B, lane 5). In contrast, no obvious cleavage of IRES-ΔaugEGFP was detected in the coding region despite the presence of guanines at +1, +3, +5, +9, +10, +11, +13, and +15 (Fig. 5C, lane 5). These data indicate that the EGFP coding region is inaccessible to the enzyme. RNA modified with DMS prevents primer extension at unpaired adenine and cytosine and, when modified with CMCT, prevents extension at unpaired uracil and guanine. DMS- and CMCT-modified IRES-CP arrested primer extension at the first 21 nucleotides in the ORF (Fig. 5B, lanes 1 and 3). In contrast, IRES-ΔaugEGFP arrested extension at a few nucleotides, such as +4A, +7A, and +8A, and ambiguous arrests were observed at +2U and +14A (Fig. 5C, lanes 1 and 3). These results indicate that the coding region of the wild-type construct is a single strand, but the coding region of the ΔaugEGFP, which was not translated by the IGR-IRES (Fig. 4, lane 2), is structured.
FIG. 5.
RNA structure probing analysis around initiation sites. (A) Secondary structure model of the IGR-IRES and the 5′ region of the capsid ORF in PSIV. Asterisks and dots indicate base pair interactions for pseudoknots (PK I, PK II, and PK III) and for helical segments, respectively. Uppercase letters represent highly conserved sequences in dicistroviruses. Nucleotide positions of the PSIV genome are indicated. (B to E) Primer extension profiles in transcripts from pT7-IRES-CP (B), pT7-IRES-ΔaugEGFP (C), pT7-IRES-ΔaugFluc (D), and pT7-IRES6195-LUC (E). Nucleotides for PK I are shown at the left of panels. Nucleotides either significantly modified or cleaved in the IRES sequence are indicated at the right of panels. Codons downstream of PK I are marked at right of panels. The nucleotide immediately downstream of PK I was counted as +1 in exogenous constructs.
G6182 is located in a single-stranded region in domain 3 of the IGR-IRES in PSIV, while G6177 is in a helical segment (Fig. 5A). IRES-CP was cleaved more efficiently at G6182 than at G6177 (Fig. 5B, lane 5), while IRES-ΔaugEGFP was cleaved efficiently at both G6182 and G6177 (Fig. 5C, lane 5). Also, A6178-A6179 were unmodified in the wild type (Fig. 5B, lane 1) but modified in the ΔaugEGFP construct (Fig. 5C, lane 1). A nonspecific termination, which is probably caused by the PK I base pair formation at C6190 in the wild type (Fig. 5B), was not observed, and another nonspecific termination appeared at C6176 in the ΔaugEGFP construct (Fig. 5C). These observations indicate that the coding sequence of ΔaugEGFP distorts the structure of domain 3.
We also carried out structure probing assays against IRES-ΔaugFluc (Fig. 5D). This construct was able to be translated (Fig. 3 and Table 1), but its efficiency was probably low because insertion of the capsid sequence enhanced luciferase activity (Table 2). Obvious structural distortions were not observed, but moderate structural changes that paralleled the translation activity were detected in comparison to the wild type. In the RNase T1 cleavage assay, differences of band intensities between the 3′ part of the IGR-IRES and the coding sequence were normalized (Fig. 5D, lane 5). The termination at C6190 in the wild type disappeared; however, nonspecific terminations were observed at +7G, +8C, and +9C (Fig. 5D). Except for these few nucleotides, the coding sequence appeared to be a single strand.
Profiles of the primer extension of IRES6195-LUC that showed ineffective translation (Table 1) were similar to those of IRES-ΔaugFluc, but additional significant changes were detected. Specifically, RNase T1 cleavage in IRES6195-LUC was decreased in its coding region but was intensified at G6165 (Fig. 5E, lane 5), indicating that the coding region was less accessible for the enzyme than was the loop region in the 3′ part of the IGR-IRES. C6188 and A6189, which comprise PK I, were not modified in either the wild type (Fig. 5B, lane 1) or IRES-ΔaugFluc (Fig. 5D, lane 1) but were modified in IRES6195-LUC, suggesting that PK I formation was somewhat impaired in this construct. In the wild type, A6163-A6164 and U6191-U6192 were weakly reactive with DMS and CMCT. Because these four nucleotides were located at the edge of PK I, the base pair interactions were thought to be unstable in our experimental conditions (22). In the case of IRES6195LUC, A6163-A6164 were modified effectively (Fig. 5E, lane 1) in comparison to the wild type. Therefore, the band intensities at U6191-U6192, which should be base paired with A6163-A6164 for PK I, should also be increased. However, when intensities were compared with U6185 and U6187, the modification at U6191-U6192 was decreased (Fig. 5E, lane 3). Because this result was reproducible, it is possible that a part of U6191-U6192 may fail to interact with A6163-A6164 because of structural distortion in this construct. This construct contains a BamHI sequence at the position +4 to +9. Although +4G and +5G were weakly cleaved with RNase T1, modification of these two G residues with CMCT was not detected (Fig. 5D, lane 3). In addition, modifications at +6A and +7U were also decreased compared with upstream regions in this construct. These observations suggest that a part of the BamHI sequence, GGAU, is somewhat structured and may have caused ineffective translation mediated by the IGR-IRES.
DISCUSSION
Previous deletion analysis suggested that the 5′ regions in the capsid ORFs were required for IGR-IRES-mediated translation (5, 32). The data presented here, however, show that the IGR-IRES of PSIV can translate exogenous reading frames having AUG deleted without the capsid ORF (Fig. 3). When we previously examined the 3′ boundary of the IGR-IRES of PSIV, a BamHI sequence (GGATCC) was generated (32). In the case of RhPV, a restriction sequence of AatII (GACGTC) was also used in constructing the plasmids (5). In this study, plasmids were constructed by direct ligation with phosphorylated PCR products and most of the constructs produced IGR-IRES-mediated translation products. Comparison of the nucleotide sequences indicated that these viruses preferentially utilize an AU-rich sequence in this region. The previous deletion analyses, however, increased the GC content in this region by insertion of recognition sequences for restriction enzymes. As a result, translation efficiency of transcripts from pCAT-IRES6195-LUC decreased to a background level (Table 1) and RNA structure around the initiation site was distorted (Fig. 5E), indicating that translation efficiency mediated by IGR-IRES is very sensitive to sequences at the 5′ part in coding regions.
Unlike IGR-IRES elements in dicistroviruses, the IRES element of hepatitis C virus (HCV) requires base pairing between an AUG initiation codon and Met-tRNAi (28), but IRES from either virus can bind the 40S ribosome at the initiation site on its RNA (24, 38). A study of the HCV IRES showed that a short length of the 5′ part of the HCV coding sequence was necessary for activity (29). However, another study showed that the HCV coding sequence was not essential for activity of the HCV IRES based on data showing that a luciferase coding sequence was efficiently translated without the HCV coding sequence (36). This controversy was resolved by experiments showing that the unstructured viral coding region makes translation mediated by the IRES more efficient (30). More recently, chemical probing analysis has shown a strong correlation between the pestivirus IRES activity and absence of secondary structure near the initiation codon (9). The structure of IGR-IRES is of primary importance for initiation (14, 16, 22, 33). Therefore, if the GC-rich sequences at the 5′ region of coding sequences are structured and interact with the IGR-IRES sequence, conditions would not be favorable for the IGR-IRES-mediated translation initiation.
Recent papers have dissected in detail the initiation process mediated by IGR-IRES. Chemical footprint analysis reveals that the 40S ribosomal subunit interacts with domains 1 and 2 but not with domain 3 of the PSIV IRES (22). However, domain 3 competed with the anticodon stem-loop of tRNAPhe when salt-washed 80S ribosomes were preincubated with poly(U), indicating that domain 3 is located in the ribosomal decoding site (22). Analysis of factorless assembly of the 80S ribosome on the IGR-IRES element of CrPV shows that ribosomes first enter near PK III and then reposition themselves at PK I (14). Toeprint analyses in the CrPV IRES indicate that pseudotranslocation allows the first aminoacylated elongator tRNA to enter the ribosomal P site and is an essential event in the IGR-IRES-mediated initiation of translation (14, 25, 38). Finally, elongation in the absence of any eukaryotic initiation factors (eIFs) has been shown elsewhere for CrPV (25). Data presented here show that the 5′ region of the capsid ORF is not structured (Fig. 5B). Because the translation efficiency of exogenous reading frames was improved by either the insertion of the wild-type ORF (Table 2) or mutations increasing A and U nucleotides (Fig. 4), it has been assumed that single-strandedness in the 5′ region of the downstream coding sequence is important for efficient translation. The 80S ribosome covers 14 nucleotides of the mRNA downstream of the P site (4). When the 5′ region of a coding sequence is structured, the precise targeting of ribosomes on the initiation site, which involves the repositioning of the ribosomal P site to PK I and pseudotranslocation, would be inhibited.
The IGR-IRES activity of RhPV has been detected in Spodoptera frugiperda Sf9 cells that express T7 RNA polymerase (6). We examined the IGR-IRES activity of PSIV in Drosophila melanogaster S2 cells, which were transfected with a capped transcript containing nucleotides 5668 to 6240 of PSIV followed by Fluc coding sequence, the 3′ untranslated region of PSIV, and poly(A) tail. Luciferase activity of the transfected cells was only 3%, in comparison to that of cells transfected with RNAs, which were designed to be translated by a canonical ribosome-scanning mechanism (data not shown). In mammalian and yeast cell lines, the IGR-IRES activity of CrPV has been stimulated by eIF2α phosphorylation (8, 34). In CrPV-infected cells, eIF2α is heavily phosphorylated when the capsid protein synthesis is at a maximum level (2). These findings suggest that IGR-IRES-mediated translation would be activated after shutdown of host protein synthesis by viral infection. The inefficient activity of the IGR-IRES in cells did not allow us to detect translation of an IRES-ΔaugFluc construct in Drosophila S2 cells. The question of whether the IGR-IRES facilitates translation of exogenous reading frames in cells will be answered after the viral factors that control shutdown of host protein synthesis are identified.
It is known that mutated initiator tRNA molecules with altered anticodon triplets function in both eukaryotic and prokaryotic translation systems (7). When we examined IGR-IRES-mediated translation in a prokaryote system with an S30 extract of Escherichia coli, no translation product was detected (data not shown). Because ribosomes in prokaryotes and eukaryotes are structurally different, the IGR-IRES of PSIV would not be recognized by the prokaryotic ribosomes. However, the results of our study suggest that the IGR-IRES elements of dicistroviruses direct initiation, starting with elongator tRNAs and using the normal translation apparatus in eukaryotic cell-free systems.
Acknowledgments
This work was supported by grants from the pioneer program from MAFF, the COE program from Special Coordination Funds for Promoting Science and Technology, and PROBRAIN, Japan, to N.N.
REFERENCES
- 1.Brown, E. A., S. P. Day, R. W. Jansen, and S. M. Lemon. 1991. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J. Virol. 65:5828-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christian, P., E. Castens, L. Domier, K. Johnson, N. Nakashima, P. Scotti, and F. van der Wilk. 2000. Cricket paralysis-like viruses, p. 678-683. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 4.Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. [DOI] [PubMed] [Google Scholar]
- 5.Domier, L. L., N. K. McCoppin, and C. J. D'Arcy. 2000. Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF 2. Virology 268:264-271. [DOI] [PubMed] [Google Scholar]
- 6.Domier, L. L., and N. K. McCoppin. 2003. In vivo activity of Rhopalosiphum padi virus internal ribosome entry sites. J. Gen. Virol. 84:415-419. [DOI] [PubMed] [Google Scholar]
- 7.Drabkin, H. J., and U. L. RajBhandary. 1998. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 18:5140-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez, J., I. Yaman, P. Sarnow, M. D. Snider, and M. Hatzoglou. 2002. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 277:19198-19205. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, S. P., I. K. Ali, A. Kaminski, P. Digard, and R. J. Jackson. 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8:1558-1571. [PMC free article] [PubMed] [Google Scholar]
- 10.Gallie, D. R., D. E. Sleat, J. W. Watts, P. C. Turner, and T. M. A. Wilson. 1987. The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15:8693-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 12.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 14.Jan, E., and P. Sarnow. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324:889-902. [DOI] [PubMed] [Google Scholar]
- 15.Jang, S. K., H. G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanamori, Y., and N. Nakashima. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7:266-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 18.Madin, K., T. Sawasaki, T. Ogasawara, and Y. Endo. 2000. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 97:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Salas, E., R. Ramos, E. Lafuente, and S. López de Quinto. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82:973-984. [DOI] [PubMed] [Google Scholar]
- 20.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima, N., J. Sasaki, K. Tsuda, C. Yasunaga, and H. Noda. 1998. Properties of a new picorna-like virus of the brown-winged green bug, Plautia stali. J. Invertebr. Pathol. 71:151-158. [DOI] [PubMed] [Google Scholar]
- 22.Nishiyama, T., H. Yamamoto, N. Shibuya, Y. Hatakeyama, A. Hachimori, T. Uchiumi, and N. Nakashima. 2003. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 24.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, T. V., and C. U. T. Hellen. 2003. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RajBhandary, U. L., and C. M. Chow. 1995. Initiator tRNAs and initiation of protein synthesis, p. 511-528. In D. Söll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. American Society for Microbiology, Washington, D.C.
- 27.RajBhandary, U. L. 2000. More surprises in translation: initiation without the initiator tRNA. Proc. Natl. Acad. Sci. USA 97:1325-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds, J. E., A. Kaminski, A. R. Carroll, B. E. Clarke, D. J. Rowlands, and R. J. Jackson. 1996. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2:867-878. [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. M. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki, J., N. Nakashima, H. Saito, and H. Noda. 1998. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology 244:50-58. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki, J., and N. Nakashima. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki, J., and N. Nakashima. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97:1512-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, S. R., K. D. Gulyas, and P. Sarnow. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. USA 98:12972-12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, J. E., M. J. Powell, S. E. Hoover, and P. Sarnow. 2000. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20:4990-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, J. E., T. V. Pestova, C. U. T. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 39.Woolaway, K. E., K. Lazaridis, G. J. Belsham, M. J. Carter, and L. O. Roberts. 2001. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J. Virol. 75:10244-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]