Abstract
The human T-cell leukemia virus type 1 (HTLV-1) Gag polyprotein contains two adjacent proline-rich motifs (sequence PPPYEPTAP) in the C terminus of the matrix domain. Proline-to-alanine mutations were introduced into either or both motifs of HTLV-1 to determine the effect on the release of HTLV-1 virus-like particles from 293T cells. The release of both single mutants was significantly reduced, whereas a double mutation in both motifs abolished the release of the HTLV-1 particles. Two-hybrid and in vitro binding assays showed that the HTLV-1 Gag polyprotein binds both Tsg101 and Nedd4 proteins. The interaction with HTLV-1 Gag required the central WW domain of Nedd4 and the ubiquitin enzyme variant (UEV) domain of Tsg101. We expressed various fragments of Nedd4 and Tsg101 proteins in 293T cells and tested for their ability to interfere with virion release mediated by the HTLV-1 Gag-Pro polyprotein. Fragments consisting of the N-terminal UEV domain of Tsg101 and the central WW and C-terminal Hect domains of Nedd4 protein all caused transdominant inhibition of HTLV-1 particle release. Similarly, inhibition of the proteasome significantly decreased HTLV-1 particle release. Furthermore, the WW domain overexpression caused an early arrest of HTLV-1 particle morphogenesis before the membrane is deformed into the typical half-shell structure. This result suggests that Nedd4 is involved early in budding of HTLV-1.
Gag polyproteins of a number of retroviruses carry domains that promote the efficient exocytosis of the virus particle from the plasma membrane at the final stages of viral budding (12). These proline-rich domains are highly conserved and are located in different locations in Gag polyproteins. Lentivirus human immunodeficiency virus type 1 (HIV-1) Gag polyproteins contain a PTAP motif located in the p6 domain at the C-terminal end of the polyprotein. Deletion of the p6 protein leads to an arrest in HIV-1 virus release (15), and mutations in this region affect both the budding and the processing of p55gag (23). In some isolates of HIV-2 and simian immunodeficiency virus, a second PTAP motif in the matrix domain of the Gag polyprotein may also provide L-domain function. Unlike the other lentiviruses, equine infectious anemia virus (EIAV) contain a unique L domain motif, YXXL, in its C-terminal p9 protein (49). Gag polyproteins of the oncoviruses Rous sarcoma virus (RSV) and murine leukemia virus (MLV) contain an L-domain motif PPPY generally located between the matrix and capsid proteins. The RSV L domain is contained in its small p2 peptide (45), whereas the MLV L domain is found in p12 (68). Mutations or deletions in these L domains result in the retention of virus particles at the plasma membrane, often in groups, indicating that these short motifs play an essential role in budding and release. Similarly, the Mason-Pfizer monkey virus (MPMV) L domain was recently shown to lie within the pp16 protein and to function in its budding (65). Outside the retrovirus family, filoviruses (Ebola virus) and rhabdoviruses (vesicular stomatitis virus) also contain L domains that seem to perform functions similar to those described in retroviruses (17, 18, 33). Interestingly, Ebola virus contains an L domain that is a combination of both PTAP and PPPY motifs (PTAPPEY). This motif, when attached to a truncated HIV-1 Gag, provided L-domain function (1, 58).
The interchangeability of L-domain motifs between various retroviruses (30, 45, 57, 67, 68), along with their ability to function in ectopic locations, suggested that they might serve as docking sites for cellular factors involved in the budding process. The EIAV p9 protein harboring the L-domain motif YXXL was shown to bind the AP-50 subunit of the AP-2 complex (50).The PTAP motif present in the HIV-1 and HIV-2 Gag polyprotein and VP40 in Ebola virus (13, 33, 37, 60) binds the E2 ubiquitin enzyme variant (UEV) Tsg101. This protein lacks the highly conserved cysteine residue (replaced by a tyrosine residue) present in active E2 ubiquitin enzymes (61). Tsg101 was first discovered as a tumor suppressor gene (31) and was more recently described to be an important player in the delivery of cargo proteins to late endosomal compartments (2). Depletion of Tsg101 by RNAi from the cytoplasm of mammalian cells (13) or overexpression of the N-terminal UEV domain of Tsg101 (9, 14) arrested or decreased the release of HIV-1, functionally linking Tsg101 to virus budding and release. E3 ubiquitin ligases of the Nedd4 family were shown to be involved in budding of RSV and MPMV retroviruses (26, 48, 66). Proteasome inhibitors were found to interfere with the release of HIV-1 (55) and RSV (47), suggesting that depletion of free ubiquitin from the cytoplasm of mammalian cells disrupts retroviral budding and release. The role of ubiquitin in retroviral budding and release remains unclear. Interestingly, retroviruses such as EIAV and mouse mammary tumor virus, which use late-domain motifs other than PPPPY and PTAP, were found to be insensitive to proteasome inhibitors (43). Retroviral Gag proteins are ubiquitinated (40, 51, 57), but the ubiquitin ligases performing this function are not yet identified. In addition, a small amount of free ubiquitin is found in various retrovirus particles (40, 51). Recently, monoubiquitination was shown to be a key regulator of the endocytosis pathway, although its role in this process is still not understood (20, 56). Furthermore, Nedd4 and Tsg101 (and their yeast counterparts Rsp5 and Vsp23) were shown to play roles in internalization of plasma membrane receptors and in protein sorting and trafficking; these processes appear to be regulated by monoubiquitination (2, 3, 10, 11, 20-22, 24, 25). Collectively, these observations suggest that these proteins are used to promote efficient release of the viral particles from the plasma membrane.
HTLV-1 is a type C retrovirus that assembles, buds, and releases from the plasma membrane of infected cells (44, 59). Its Gag polyprotein exhibits a typical Gag structure and contains an N-terminal matrix (MA), a central capsid (CA) protein, and a C-terminal nucleocapsid. No protein equivalent to HIV-1 p6 or other short peptide(s) has been described in HTLV-1 Gag. The MA protein is myristylated, and the myristic acid is responsible for targeting and driving the assembly of Gag polyproteins at the plasma membrane of mammalian cells (6), whereas the CA protein harbors regions important for HTLV-1 particle assembly (52). In the C-terminal end of MA, two potential L-domain motifs (PPPYEPTAP) are located (Fig. 1). Their function and their possible physical and functional binding to the ubiquitin ligase Nedd4 and the E2-like protein Tsg101 have not been established. We examined the role of both PPPY and PTAP motifs in the budding and release of the HTLV-1 Gag. We show by substituting the prolines for alanines in both motifs that both PPPY and PTAP motifs function as L domains and that the PPPY motif plays the predominant role in this function. We also examined the binding of E3 ubiquitin ligase Nedd4 and inactive E2 Tsg101 to HTLV-1 Gag by using two-hybrid and in vitro assays and studied their role in the release of HTLV-1 from mammalian cells. Moreover, inhibition of proteasome inhibitors caused a decrease of HTLV-1 particle budding. Our data suggest that HTLV-1 release requires ubiquitin and a combination of proteins used during the budding of both RSV and HIV-1 retroviruses.
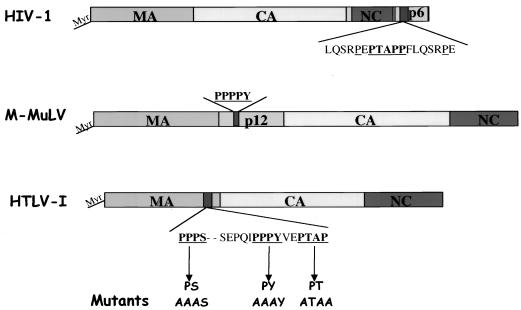
FIG. 1.
Schematic representation of HIV-1, MLV, and HTLV-1 retroviral Gag polyproteins. The proline-rich motifs carrying the L-domain function are underlined. Mutants generated in the HTLV-1 Gag are indicated in boldface.
MATERIALS AND METHODS
Cells and transfection.
Human 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% of fetal bovine serum at 37°C and 5% CO2. Transfections were performed (∼2 × 106 cells in a 10-cm dish) with either calcium or Fugene 6 transfection reagent (Roche) according to the manufacturer's recommendations.
Plasmid construction and mutagenesis.
HTLV-1 Gag-Pro mammalian cell expression plasmid was constructed as described previously (6). Briefly, the fragment containing the entire HTLV-1 gag and pro genes was cut from p5′X13 plasmid (a gift from Therese Astier-Gin, University of Bordeaux, Bordeaux, France) by using AvrII and ApaI sites, and the cDNA fragment was inserted into the plasmid pSV (a gift from Elkharroubi Aboubaker, National Cancer Institute [NCI], Frederick, Md.) between the XbaI and SmaI sites. The plasmid obtained was named pSVgag-pro. Ptax, Prex, and PX, expressing the HTLV-1 X region accessory proteins Tax and Rex, were gifts from David Derse, NCI. The full-length HTLV-1 provirus used in the present study is described in Nicot et al. (38). The provirus was modified by replacing the XhoI-Xho-I fragment in the env gene with green fluorescent protein (GFP).
The human Nedd4 gene was obtained from the Kazuza DNA Research Institute, Chiba, Japan (GenBank accession number KIAA0093). The C2 (amino acids [aa] 1 to 485), WW (aa 631 to 1633), and Hect (aa 1838 to 2820) fragments were generated by PCR and inserted in the mammalian expression plasmid 3X Flag between the NotI and KpnI sites. The plasmid carrying a Hect fragment mutated in the active domain of the Nedd4 ligase (cysteine 894 to tyrosine) was constructed by using a QuickChange site-directed mutagenesis kit (Stratagene). Plasmid pLLI-C, expressing the UEV N-terminal region of Tsg101 (N-Tsg101; a gift from L. Li and S. Cohen, Stanford University, Calif.), is described elsewhere (32).
For in vitro binding assays, the C2 (aa 1 to 485), WW (aa 631 to 1633), and Hect (aa 1838 to 2820) fragments were generated by PCR and cloned in pCRT7CT-TOPO plasmid (Invitrogen). This plasmid allows the direct insertion of PCR products in frame with DNA coding for six histidines (His6). The resulting plasmids pCRCTC2, pCRCTWW, and pCRCTHect express three human Nedd4 fragments, C-terminally His6-tagged ∼21-, ∼38-, and ∼42-kDa proteins, respectively. Plasmid ptybHTLVMA was constructed by inserting the entire HTLV-1 MA gene between NdeI and SapI sites in the PTYBI plasmid (IMPACT T7 System; New England Biolabs). The resulting plasmid contains the MA gene cloned in frame with DNA encoding the intein and the chitin-binding domain genes (IMPACT T7 System). This expresses an ∼75-kDa protein that contains the N terminus of intein and chitin-binding domain proteins fused to MA protein and allows the purification and elution of a native 19-kDa HTLV-1 MA protein. For expression of proteins fused to glutathione S-transferase (GST) protein, we used pGEX-2T-PL that is a modified pGEX-2T plasmid (Pharmacia Amersham) with a multisite polylinker. The N-terminal region of Tsg101 (aa 1 to 270) containing the UEV domain coding sequence was inserted at the 3′ end of the GST gene at the BamHI and SalI sites. The resulting plasmid pGSTN-Tsg101expresses a GST-N-Tsg101 fusion protein of ca. 45 kDa.
For two-hybrid assays, Tsg101 and Nedd4 genes were cloned into the pGADNOT plasmid (34) between NotI and SalI sites, and the HTLV-1 Gag gene was cloned into pSH2-1 plasmid (16) between the BamHI and SalI sites. Two plasmids containing MLV and HIV-1 Gag, pSH2-1MLVGag and pMA424, were described previously (34).
Two-hybrid assay.
Protein-protein interactions in yeast were performed with pGADNOT and pSH2-1 plasmids in CTY10-5d and GGY::171 strains (generously provided by R. Sternglanz, SUNY, Stony Brook, N.Y.). The pSH2-1 plasmid encodes an N-terminal LexA DNA domain (LexADB) and pGADNOT encodes an N-terminal Gal4 activation domain (Gal4AD). The assay was performed as described previously (34).
Protein purification and in vitro binding assays.
His6-tagged proteins were expressed in BL21(DE3)/pLysS OneShot Escherichia coli (Invitrogen). Bacteria were collected (50 ml) and resuspended in cold lysis buffer (50 mM Tris [pH 8], 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride). Lysozyme was added to 1 mg/ml, and the suspension was incubated on ice for 30 min. The suspension was sonicated and ultracentifuged at 100,000 × g for 30 min. NaCl was adjusted to 500 mM, and 50 μl of Ni-agarose beads (Qiagen) was added, followed by incubation for 1 h at 4°C. The Ni-agarose beads were spun down and washed twice with 500 mM NaCl-50 mM Tris (pH 8) and then washed three times with buffer containing 50 mM NaCl, 50 mM Tris (pH 8), and 10 mM imidazole. Bound proteins were then eluted with buffer containing 50 mM NaCl, 50 mM Tris (pH 8), and 200 mM imidazole. HTLV-1 MA was expressed by using the IMPACT T7 System and PTYBI plasmid. Purification of the protein was performed as described previously (7), and then the purified HTLV-1 MA was dialyzed against 50 mM NaCl-50 mM Tris (pH 8) buffer overnight at 4°C. Interaction between His-C2, His-WW, and Hect proteins and HTLV-1 MA was evaluated in 50 mM NaCl-50 mM Tris (pH 8), and beads were extensively washed with the same buffer. The complex of proteins bound to the beads was eluted with 200 mM imidazole buffer.
GST fusion protein N-Tsg101-GST was expressed in BL21 E. coli. One gram of cell pellet was resuspended in cold phosphate-buffered saline containing the protease inhibitor cocktail Complete Mini (Roche). Lysozyme was added (0.5 mg/ml), and the suspension was incubated for 1 h on ice. After sonication, the lysates were centrifuged at 13,000 rpm for 30 min. Cleared lysates were incubated with 100 μl of glutathione-Sepharose beads (Amersham Pharmacia) for 1 h at 4°C. Beads were washed three times with 200 mM NaCl-50 mM Tris (pH 8)-5 mM EDTA-5 mM dithiothreitol and twice with 200 mM NaCl-50 mM Tris (pH 8)-5 mM EDTA. A binding assay between N-Tsg101-GST and HTLV-1 MA was performed in 200 mM NaCl-50 mM Tris (pH 8)-5 mM EDTA-1 mM dithiothreitol buffer for 2 h at 4°C. Beads were washed three times with same buffer and twice with 200 mM NaCl-50 mM Tris (pH 8)-5 mM EDTA buffer. Bound protein complexes were eluted with 10 mM reduced glutathione.
Immunoprecipitation and Western blotting.
Cells were lysed on ice in 300 to 500 μl of radioimmunoprecipitation assay buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS]), sonicated, and centrifuged for 20 min at 4°C at 13,000 rpm. HTLV-1 Gag proteins were immunoprecipitated with rabbit anti-HTLV-1 capsid antibody (Advanced Biosciences Laboratories) and collected on protein A-Sepharose beads. Proteins were analyzed on SDS-12.5% polyacrylamide gels and then transferred to nitrocellulose membranes (Millipore). Gag was subsequently detected by Western blotting with mouse anti-capsid or goat anti-capsid antibodies. In other Western blots, the membranes were probed with anti-GST mouse monoclonal (Covance), anti-His mouse monoclonal (Qiagen), mouse anti-Na+/K+ ATPase subunit (Biomol), mouse anti-Flag M2 (Sigma), or mouse anti-Myc (Santa Cruz) antibodies. The HTLV-1 MA was detected by using a rabbit polyclonal antibody generously provided by Stephen Orozslan (NCI).
Electron microscopy.
Cultured cells were scraped with a disposable scraper, centrifuged at 150 × g for 5 min, and fixed in cacodylate-buffered (0.1 M, pH 7.4) glutaraldehyde (2% [vol/vol]). The cell pellet was rinsed in cacodylate buffer, postfixed in osmium tetroxide (1% [vol/vol]) in the same buffer, and then dehydrated in graded ethanol and propylene oxide. The pellet was infiltrated overnight in equal mixtures of propylene oxide and epoxy resin. The pellet was embedded in pure resin and cured at 55°C. Thin sections were stained in uranyl acetate and lead citrate and stabilized by carbon evaporation. The electron microscopic images were obtained with a Hitachi H7000 electron microscope equipped with Gatan digital camera.
Proteasome inhibitor assay.
293T cells were transfected with a HTLV-1-expressing construct as described above. One day after transfection, the culture medium was replaced with DMEM alone (control) or DMEM containing 1 μg of Z-Leu-Leu-Leu-CHO (MG-132) proteasome inhibitor (BostonBiochem, Cambridge, Mass.)/ml, followed by further incubation for another 24 h. Cells were harvested and lysed as desribed above; the expression of Gag was analyzed by immunoprecipitation with a rabbit anti-CA antibody, followed by Western blotting with a goat anti-CA antibody. Virions were pelleted through a 20% sucrose cushion and analyzed by Western blot with a goat anti-CA antibody.
RESULTS
Both adjacent motifs in the short sequence PPPYEPTAP in MA are L domains of HTLV-1.
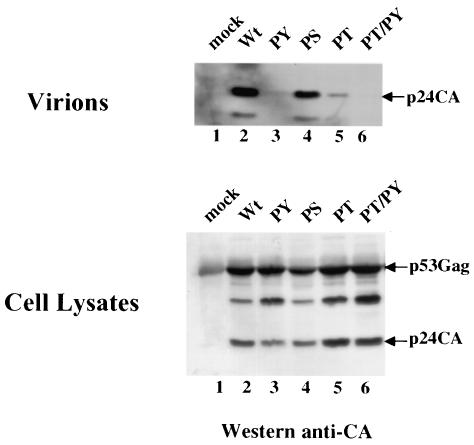
The HTLV-1 Gag polyprotein p53gag contains a region in the C-terminal end of the p19 MA protein composed of three short proline-rich motifs (Fig. 1). To determine the role of these sequences in the release of HTLV-1 Gag, we expressed the gag-pro genes under the control of an simian virus 40 promoter (pSVgag-pro) and tested the expression, processing, and release of HTLV-1 Gag- and Gag-Pro-related proteins. We substituted the prolines in each of the PPPPS, PPPY, or PTAP motifs with alanine residues. 293T cells were transfected with wild-type pSVgag-pro, pSVPY (mutant PPPY to AAAY), pSVPT (mutant PTAP to ATAA), pSVPS (mutant PPPS to AAAS), or PSVPY-PT (mutant PPPYEPTAP to AAAYEATAA), and the expression of intracellular and release of p53gag was analyzed. p53gag polyproteins were immunoprecipitated by using a rabbit anti-CA polyclonal antibody, and the virion particles were pelleted through a 20% sucrose cushion. Both were analyzed by Western blotting with a mouse anti-CA monoclonal antibody. The wild-type Gag-Pro was efficiently processed and released into the culture medium (Fig. 2, lane 2) yielding a 24-kDa CA protein. In contrast, release of Gag-Pro-PY and Gag-Pro-PT proteins was significantly reduced, with no (lane 3) or very little CA (lane4) detected in the culture medium. Mutation in the PPPY motif resulted in a reproducibly greater reduction of Gag release (lane 3). The double mutation of both motifs in the Gag-Pro-PYPT polyprotein resulted in total loss of Gag release, with no p24 CA protein detected in the culture medium (lane 6). Mutation of the PPPS motif had little or no effect on Gag release (lane 4). None of the mutations in either PPPS or PPPYEPTAP motifs affected the intracellular accumulation or processing of Gag (Fig. 2, bottom panel, compare lane 2 to lanes 3, 4, 5, and 6). Together, the data indicate that both motifs in the PPPYEPTAP sequence functions as L domains in HTLV-1 Gag release. The PPPY motif seems to play a dominant role in this process.
FIG. 2.
Late-domain function in HTLV-1 Gag. Wild-type HTLV-1 Gag (lane 1), single-mutant PY, PS, and PT (lanes 2, 4, and 5), and double mutant PYPT (lane 6) were expressed in 293T cells. Gag and CA expressed in the cells were immunoprecipitated from the cell lysates by using a rabbit anti-CA antibody, and the proteins were detected by Western blotting with a goat anti-CA antibody. Proteins released as virions were pelleted through a 20% sucrose cushion, and Gag proteins were detected by using a goat anti-CA antibody.
Both Tsg101 and Nedd4 bind HTLV-1 Gag polyprotein.
The results presented above show that the PPPY and PTAP motifs in HTLV-1 Gag are required for the release of viral particles. Based on findings for other retroviruses, these motifs could be sites of binding for both Tsg101 and Nedd4 proteins. To test this, the entire Tsg101 (31) and Nedd4 (GenBank accession number KI0093) proteins were fused to the C-terminal end of the Gal4 activation domain (Gal4AD), and full-length HTLV-1 Gag was cloned as a fusion with the C-terminal end of LexA in pSH2-1. The yeast two-hybrid strain CTY-5D was cotransformed with plasmids encoding Gal4AD Tsg101 or Gal4ADNedd4 and LexA-HTLV-1 Gag, and the strength of the interaction between the fusion proteins was monitored by β-galactosidase staining of yeast colonies. Mutations of PPPY in pSH2-1 HTLV-1 Gag polyprotein abolished the interaction with Nedd4, and binding to Tsg101 was not affected (Table 1). In contrast, substitutions of prolines for alanines in the PTAP motif abolished the interaction with Tsg101, and the binding to Nedd4 remained unaffected. Replacement of prolines in both PPPY and PTAP motifs by alanines abolished the interaction of the mutated protein with Nedd4 and Tsg 101. The positive controls confirmed the activities of the constructs: the HIV-1 Gag bound Tsg101 as previously described (13, 35, 61) but not with Nedd4, and MLV Gag interacted with Nedd4 protein but not with Tsg101. These data show that the L domain of HTLV-1 Gag, the PPPYEPTAP sequence, bound both Tsg101 and Nedd4 cellular proteins and that the binding of HTLV-1 Gag to Nedd4 via PPPY is independent of its binding to Tsg101 via PTAP.
TABLE 1.
Two-hybrid interaction between HTLV-1 Gag, Tsg101, and Nedd4a
| Protein | Interaction with:
|
||
|---|---|---|---|
| GadNedd4 | GadTsg101 | V-GadNot | |
| PSH-2HTLV-I Gag | +++ | ++++ | −/+ |
| PSH-2HTLV-I PPPY-Gag | −/+ | ++++ | −/+ |
| PSH-2HTLV-I PTAP-Gag | ++++ | −/+ | −/+ |
| PSH-2HIV-1 Gag | − | ++++ | − |
| PSH-2MLV Gag | +++ | − | − |
| V-PSH-2 | − | − | − |
The plus (+) symbol indicates detection of interaction as shown by the β-galactosidase assay (blue colonies), and the minus (−) symbol indicates its absence (white colonies). The larger the number of + symbols, the stronger the interaction between the two proteins tested. −/+, background.
UEV domain of Tsg101 binds HTLV-1 MA in vitro.
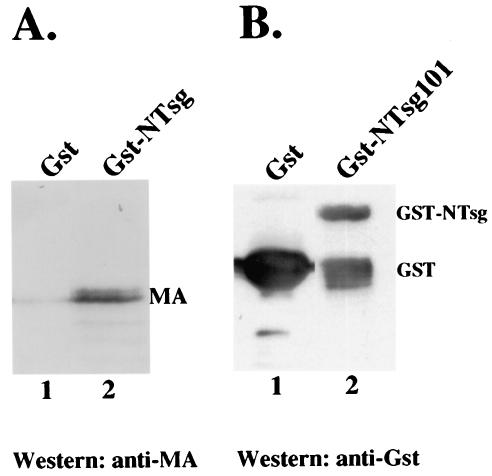
To test for the interaction between Tsg101 and MA of HTLV-1 Gag in vitro, we expressed an N-terminal fragment of Tsg101 protein (aa 1 to 270) as a GST fusion protein in E. coli. HTLV-1 MA protein was expressed in E. coli and purified as described previously (7). This method comprises a two-step purification system that allowed the production of large amounts of untagged HTLV-1 MA. GST-NTsg101 (∼45 kDa, Fig. 3B) or GST alone was immobilized on glutathione beads and incubated with untagged HTLV-1 MA for 2 h at 4°C. The unbound proteins were removed, the beads were washed extensively, and the bound proteins were eluted with buffer containing 10 mM reduced glutathione. Proteins in the eluted fractions were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with a rabbit polyclonal anti-HTLV-1 MA antibody. HTLV-1 MA bound strongly to GST-NTsg101 (Fig. 3A, lane 2) and not to GST protein (lane 1). Thus, the N-terminal region of Tsg101 containing the UEV domain is sufficient to bind HTLV-1 MA in vitro.
FIG. 3.
UEV domain in Tsg101 binds HTLV-1 MA. The N-terminal region in Tsg101 (aa 1 to 270) was expressed in fusion with GST protein (Gst), purified, and detected with a mouse anti-GST antibody as an ∼45-kDa GST-N-Tsg101 protein (B, lane 2). When GST-N-Tsg101 immobilized on glutathione beads was incubated with untagged HTLV-1 MA, the HTLV-1 MA protein was captured by the GST-N-Tsg101 (A, lane 2) as detected by using a rabbit anti-MA antibody. However, no MA bound beads carrying GST alone (A, lane 1).
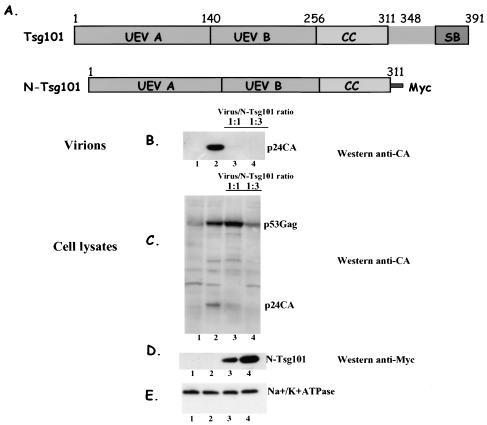
Overexpression of UEV domain of Tsg101 inhibits HTLV-1 Gag release.
To test for the role of the N-terminal UEV domain of Tsg101 in the budding and release of HTLV-1 particles, we expressed various ratios of this region of Tsg101 (the N-Tsg region is expressed as a fusion with Myc tag to yield a protein of ∼32 kDa [Fig. 4E, lanes 3 and 4]) to HTLV-1 Gag-Pro and evaluated its effect on virus particle production (Fig. 4B, lanes 3 and 4). At an N-Tsg101/Gag-Pro DNA ratio of 1:1, viral particle release was reduced 90% (Fig. 4B, compare lanes 2 and 3); at DNA ratio of 1:3, particle release was no longer detected (lane 4). Overexpression of N-Tsg101 also affected processing of HTLV-1 Gag and an intracellular accumulation of uncleaved p53Gag was observed (Fig. 4C, compare lanes 2 and 3). This defect seems to occur in late stages of assembly, since no effect on Gag expression level was detected and only processing appears to be altered. Expression of other intracellular proteins such as the membrane protein Na+/K+ ATPase remained unaffected as shown by the detection of a constant amount of its alpha-3 subunit (∼110 kDa) (Fig. 4E, lanes 1, 2, 3, and 4). Collectively, the data suggest that N-Tsg101 specifically disrupts HTLV-1 Gag release by physically interfering with Tsg101 binding to the PTAP motif in HTLV-1 Gag and thus impairing a Tsg101 function in late stages of budding and exocytosis of viral particles.
FIG. 4.
Overexpression of UEV domain in Tsg101 inhibits HTLV-1 budding. Schematic representation (A) of Tsg101 domains and the N-Tsg101 construct that expresses the UEV and the coiled-coil domains of TSG101 with a C-terminal Myc tag (B), 293T cells transfected with the HTLV-1 construct alone (B and C, lanes 2), or with the HTLV-1 construct and increasing amounts of N-Tsg101 DNA (B and C, lanes 3 and 4). Gag proteins released as virions were pelleted through a 20% sucrose cushion, and Gag was detected by Western blotting with a goat anti-CA antibody (B, lanes 2 to 4). Intracellular expression and processing of Gag were analyzed by immunoprecipitation with a rabbit anti-CA antibody, followed by Western blotting with a goat anti-CA antibody (C, lanes 1 to 4). The N-Tsg101 DN fragment was detected by Western blot with a mouse anti-myc antibody (D, lanes 1 to 4). The Na+/K+ ATPase protein was detected by using a mouse anti-Na+/K+ ATPase antibody (E, lanes 1 to 4).
WW domain in Nedd4 E3 ligase binds HTLV-1 MA p19 protein.
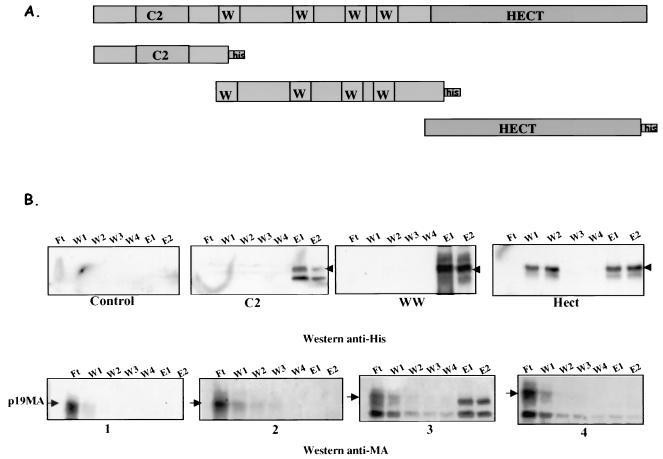
The WW domains of Nedd4 family members are 40-aa sequences that contain two highly conserved tryptophans and an invariant proline (5); they are typically involved in protein-protein interactions. These domains bind proline-rich sequences, including the PPPY motif, in their target proteins. We hypothesized that the PPPY motif in HTLV-1 Gag recruits Nedd4 protein through its WW domain to the site of budding. To test this, we generated DNA constructs expressing polyhistidine-tagged versions of the separate C2, WW, and Hect domains (Fig. 5A) in E. coli (18, 42, and 38 kDa, respectively, Fig. 5B, lanes E1 and E2 in subpanels C2, WW, and Hect). Lysates of bacteria expressing the Nedd4 domains were prepared, the His-tagged proteins were recovered on Ni-agarose beads, and the immobilized proteins were mixed with purified untagged HTLV-1 MA p19 protein. The binding reaction was allowed to proceed at 4°C for 2 h; the beads were separated from the unbound proteins in the supernatants (flowthrough [Ft] fraction) and washed four times (W1, W2, W3, and W4 fractions), and the bound proteins were eluted from the beads (E1 and E2 fractions) (Fig. 5). The fractions were then tested for the presence of MA by Western blot with a rabbit anti-MA antibody. When only Ni-agarose beads were added to MA p19 protein, MA was found in the unbound flowthrough fraction (lane Ft in subpanel 1). Similar results were obtained when MA p19 was added to beads containing C2 and Hect domains (lanes Ft in subpanels 2 and 4). In contrast, substantial amounts of MA were bound and released in the “eluted proteins” when the WW domain was used in the binding assay (Fig. 5B, lanes E1 and E2 in subpanel 3). These data show a direct and specific interaction between the central WW domain of Nedd4 E3 ligase and the MA protein of HTLV-1.
FIG. 5.
HTLV-1 MA binds WW domain in Nedd4. (A) C2, WW, and Hect domains of human Nedd4 were cloned in fusion with the His6 tag, expressed in E. coli, and purified on Ni-agarose beads. (B) Detection of fusion proteins and binding MA in fractions. When untagged HTLV-1 MA (19 kDa) was mixed with beads, no protein was detected by Western blotting with the anti-His mouse antibody (control panel). This antibody recognized C2 (∼18 kDa), WW (∼42 kDa), and Hect (∼38 kDa) that were purified on beads (subpanels C2, WW, and Hect). HTLV-1 MA was only complexed with WW fragment (subpanel 2), whereas no MA was found bound to Ni-agarose beads, to C2, or Hect fragment (subpanels 1, 3, and 4) when membranes were probed with rabbit anti-MA antibody. The positions of MA, C2, WW, and Hect proteins are indicated by arrows.
Overexpression of Nedd4 central domain WW interferes with HTLV-1 release.
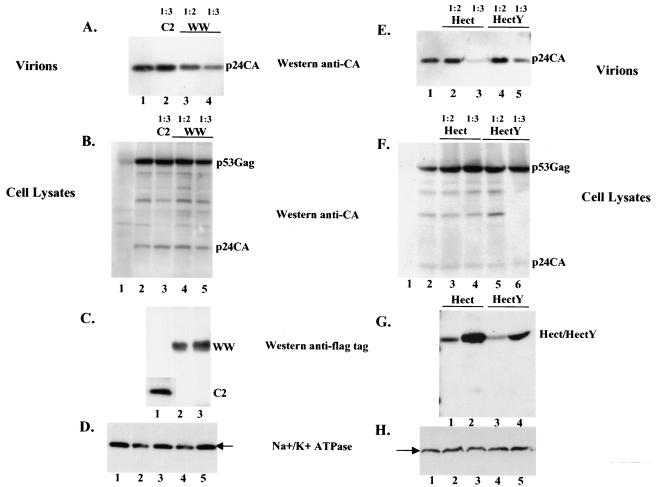
To test whether the WW domain of human Nedd4 can act as a dominant-negative inhibitor of release of HTLV-1, we expressed HTLV-1 Gag-Pro polyprotein in 293T cells, along with one of the three separate domain of Nedd4 E3 ligase: C2, WW, Hect, and Hect Y(a Hect fragment in which the active cysteine in position 894 was substituted with a tyrosine) fused to the Flag epitope tag (Fig. 6C, lanes 1, 2, and 3, and G, lanes 1, 2, 3, and 4) and monitored the effect on the release of HTLV-1. The yield of particles was assessed by Western blotting with a mouse monoclonal anti-CA antibody. High levels of C2 fragment did not affect the release of HTLV-1 Gag (Fig. 6A), whereas WW, Hect, and Hect Y expression reproducibly interfered with this process (Fig. 6A, lanes 3 and 4, and E, lanes 2, 3, 4, and 5). The coexpression of Gag-Pro and C2, WW, Hect, or HectY had no impact on the intracellular expression or processing of Gag-Pro polyprotein (Fig. 6B and F, lanes 2, 3, 4, and 5). Examination of the expression of other cellular proteins such as Na+/K+ ATPase, which is a plasma membrane protein, showed that the cells were not grossly affected by the overexpression of the various domains of E3 Nedd4 ligase (Fig. 6D and H). Taken together, these data demonstrate that the overexpression of the central WW and Hect domains specifically interferes with the release of HTLV-1 Gag. The transdominant effect exerted by WW domain on HTLV-1 Gag exocytosis and the fact that Nedd4 protein binds to HTLV-1 Gag through its PPPY L domain motif suggests that Nedd4 is normally recruited by HTLV-1 Gag to positively regulate the functions of budding and release of HTLV-1.
FIG. 6.
Overexpression of WW and Hect fragments interferes with HTLV-1 budding. 293T cells were transfected with HTLV-1 Gag-Pro alone (A and E, lanes 1, and B and F, lanes 2), with HTLV-1 Gag-Pro and C2 (A, lane 2, and B, lane 3), WW (A, lanes 3 and 4, and B, lanes 4 and 5), Hect (E, lanes 2 and 3, and F, lanes 3 and 4), or Hect Y (E, lanes 4 and 5, and F, lanes 5 and 6). Gag proteins released as virions were pelleted through a sucrose cushion and analyzed by Western blotting with a mouse anti-CA antibody (A and E); intracellular expression and processing were analyzed by immunoprecipitation with a rabbit anti-CA antibody, followed by Western blotting with a mouse anti-CA antibody (B and F). The HTLV-1 construct/dominant-negative DNA ratio is indicated above the panels. The dominant-negative fragments were detected with a mouse anti-Flag antibody (C and G). The Na+/K+ ATPase protein was detected by using a mouse anti-Na+/K+ ATPase antibody (D and H). The positions of HTLV-1 p53Gag and p24CA and the dominant-negative fragments C2, WW, Hect, and HectY are indicated.
HTLV-1 budding is sensitive to proteasome inhibitors.
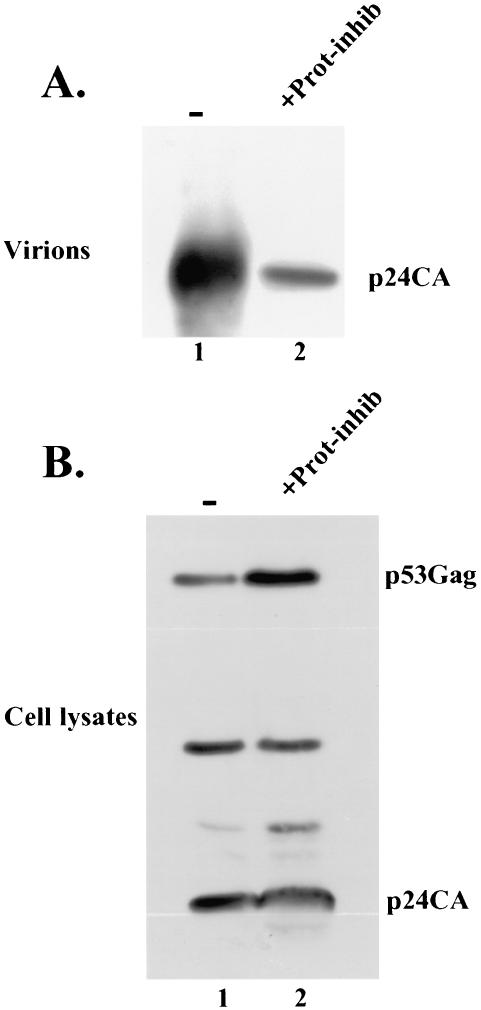
A functional proteasome was shown to be required for an efficient release of most tested retroviruses (43). To determine whether this is also the case for HTLV-1, 293T cells expressing HTLV-1-expressing plasmid were treated with proteasome inhibitor, and its effect on viral release was examined. When left untreated, 293 cells released HTLV-1, and intracellular expression and processing of Gag occurred normally (Fig. 7A and B, lanes 1). However, when cells were treated with proteasome inhibitor, a significant reduction of HTLV-1 release was observed (Fig. 7A, compare lanes 1 and 2), and larger amounts of Gag accumulated inside the cytoplasm, whereas the processing remained unaffected (Fig. 7B, compare lanes 1 and 2). These results indicate that the function of proteasome is required to an efficient release of HTLV-1.
FIG. 7.
HTLV-1 budding is sensitive to proteasome inhibitors. 293T cells were transfected with HTLV-1 plasmid DNA. At 24 h, control cells were left “untreated” (A and B, lanes 1 marked with a “−” symbol), and “treated” cells were exposed to proteosome inihibitor Z-Leu-Leu-Leu-CHO (MG-132) at a final concentration of 1 μl/ml for 24 h (A and B, lanes 2, +Prot-inhib), and incubation was prolonged for another 24 h. Gag proteins expressed inside the cells were immunoprecipitated by using a rabbit anti-CA antibody and detected by Western blotting with a goat anti-CA antibody (B). Gag proteins released as virions were pelleted through a 20% sucrose cushion and detected by Western blotting with a goat anti-CA antibody (A). Positions of HTLV-1 Gag and CA are indicated.
Overexpression of WW and N-Tsg101 domains causes relocalization and early arrest of HTLV-1 budding.
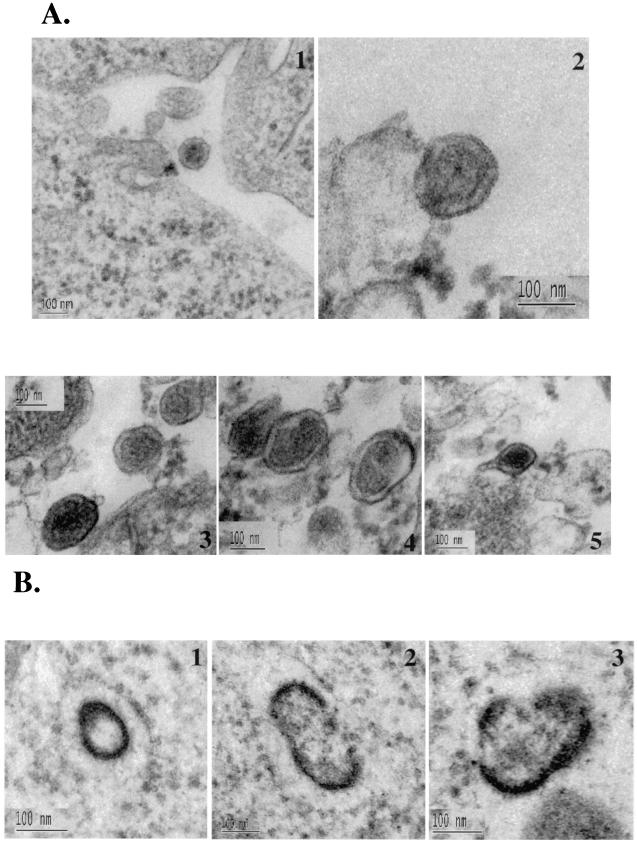
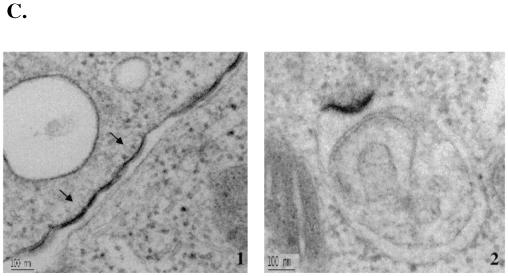
Overexpression of the Nedd4 WW and N-Tsg101 domains exerted a dominant-negative effect on the release of HTLV-1 Gag, suggesting that the WW and UEV fragments can interfere with the budding of HTLV-1 from mammalian cells. To examine the location of Gag in these cells, 293T cells were transfected with HTLV-1 and Flag-WW constructs or HTLV-1 and N-Tsg101 at a DNA ratio of 1:3, fixed, and sectioned; viral particles released in culture media were centrifuged through a sucrose cushion and fixed, and both cells and pellets were examined by transmission electron microscopy (TEM). In cells expressing HTLV-1 alone, viral particles were seen released from the plasma membrane (Fig. 8A1). In the viral pellets, numerous viral particles were observed (Fig. 8A2 to 5). These particles display a size and morphology typical for mature extracellular HTLV-1 virions; they are 120 to 150 nm in size and contain a central spherical electron-dense core surrounded by an electron-lucent layer and by membrane. In addition, viral particles, with a short stick-like structure protruding from one side of the particle, were also observed (Fig. 8A5). Similar structures have been previously described for wild-type RSV particles (8). Spherical electron-dense structures reminiscent of immature particles were also found inside the cytoplasm of cells expressing HTLV-1 proteins (Fig. 8B1). Furthermore, other, more irregular structures containing either two (Fig. 8B2) or multiple (Fig. 8B3) budding structures were found in the cytoplasm. These images suggest that HTLV-1 particles typically assemble at the plasma membrane, but virus-like structures can form intracellularly as well.
FIG. 8.
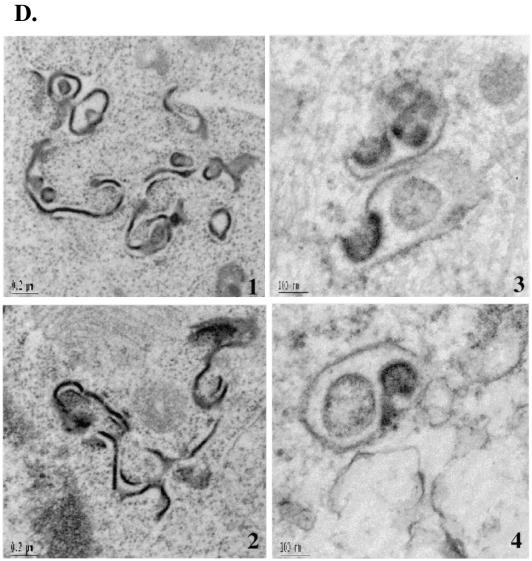
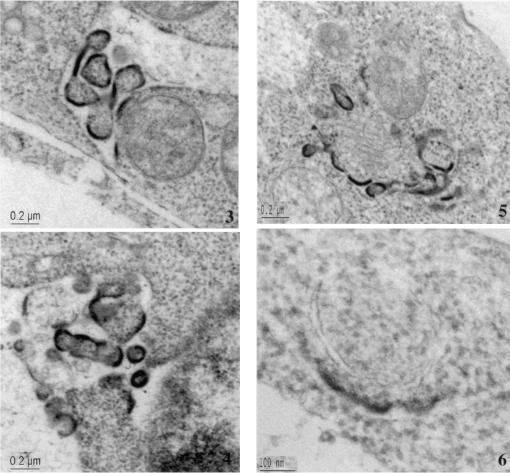
WW and N-Tsg101 DN fragments cause HTLV-1 Gag relocalization and arrest in an early step of budding. 293T cells transfected with HTLV-1 construct alone (A1 to 5 and B1 to 3), with HTLV-1 and WW-Flag plasmids (C1 to 6), or with HTLV-1 and N-Tsg101 plasmids (D1 to 4) were fixed, sectioned, and examined by TEM. (A1 to 5) Electron micrograph A1 shows an HTLV-1 viral particle released at the plasma membrane; images A2 to A5 show HTLV-1 released particles. (B1 to 3) Intracellular structures reminiscent of immature particles. (C and D) Electron micrographs show Gag features found when HTLV-1 is expressed in presence of the WW fragment (C1 to 6) or the N-Tsg101 fragment (D1 to 4).
When HTLV-1 Gag was expressed in the presence of the WW fragment of Nedd4, cells show that Gag accumulated in patches of thick electron-dense material condensed underneath the plasma membrane; the structures were relatively flat or only slightly bulged out of the cell surface (Fig. 8C1). These arrested early-assembly features were not found in untransfected or cells expressing HTLV-1 alone. Moreover, examination of the cytoplasm of these cells revealed the presence of electron-dense assembly structures initiating budding toward the inside of cellular vesicles (Fig. 8C2). Larger electron-dense structures, mostly in groups, were found in large intracellular vacuoles (Fig. 8C3) and in the perinuclear region as well (Fig. 8C4). These arrested structures were often found inside the cell, albeit close to the cell surface (Fig. 8C4 and 5), suggesting that an intracytoplasmic Gag assembly and budding may have been initiated on cytoplasmic internal membranes.
When HTLV-1 was expressed in presence of the N-Tsg101 fragment of Tsg101, HTLV-1 Gag seemed to be retained in the cytoplasm to form electron-dense “worm-like” structures (Fig. 8D1 and 2). These structures were also found concentrated in groups and were shaped as long extended winding sheets occasionally bent into half-shell structures (Fig. 8D1 and 2). Assembly structures were often observed budding into vacuoles (Fig. 8D3) or attached to membranes with a long stalk (Fig. 8D4).
These results suggest that overexpression of the WW domain of Nedd4 and the UEV domain of Tsg101 interfered with proper budding of HTLV-1 by causing Gag to be relocalized to regions of the cell other than the plasma membrane and by preventing the membrane bending necessary for budding and release of viral particles.
DISCUSSION
PPPYEPTAP sequence functions as a late domain in HTLV-1.
The results presented here show that the HTLV-1 Gag polyprotein contains two adjacent proline-rich motifs, PPPY and PTAP, located between the MA and CA proteins, which are important for L-domain function. Replacement of prolines with alanines in either motif significantly decreased exocytosis of HTLV-1 particles, indicating that both motifs are required to achieve efficient release. Mutations of prolines in both motifs simultaneously completely abolished particle production. The accumulation and processing of mutant Gag polyproteins remained unaffected. The nearby PPPS motif, which is also conserved in the p12 protein in MLV Gag polyprotein (68), was apparently unimportant for the release of HTLV-1 particles. The results suggest that the entire PPPYEPTAP sequence functions as the late domain of HTLV-1. In this regard, HTLV-1 Gag presents a unique case among retroviruses in that it needs both PPPY and PTAP late-domain motifs to be intact to efficiently release from the cell. Most retroviruses have one obvious L domain: a PPPY motif in RSV, MPMV, and MLV Gag polyproteins (64, 65, 67, 68), a PTAP motif as in HIV-1 and HIV-2 (15, 23, 37), or a YXXL motif in EIAV (49). Similarly, only one PPPY motif is conserved in viruses closely related to HTLV-1, such as HTLV-2 and BLV; no PTAP motif was found in either virus. In HIV-2 and simian immunodeficiency virus, a second PTAP is also located in the C-terminal end of the MA protein in Gag; however, the contribution of this second PTAP to budding has not been clearly determined. The two overlapping L-domain motifs in PTAPPEP sequence in Ebola virus VP40 function independently as L domains and remain individually functional when moved to other locations of the protein (33). However, when the PTAPP sequence was used in the context of minimal HIV-1 Gag (1, 58), this motif alone was not sufficient for the exocytosis of viral particle, and the addition of the PPPY motif was required to restore both ubiquitination and release of HIV-1 Gag (58). The requirement of both PPPY and PTAP for proper and efficient release of HTLV-1 particles suggests that, in the context of HTLV-1 Gag, both motifs are necessary to achieve exocytosis. Recently, the PPPY motif located in the C-terminal region of MA protein in bovine leukemia virus Gag was shown to cause viral particles to remain tethered to the plasma membrane (63). Similarly, the PPPY motif was shown to be necessary for budding and release of HTLV-1 (28). Interestingly, the PPPY motif in HTLV-1 was required in an early stage of budding before the formation of the typical type C membrane curvature, thus distinguishing the HTLV-1 budding process from other retroviruses such as RSV and MPMV that use PPPY as the L domain to separate particles from the membrane.
Tsg101 and HTLV-1 release.
In the present study, we present evidence of binding between the PTAP motif of HTLV-1 MA and Tsg101. This binding appears to be essential for the budding and release of HTLV-1 since interfering with it by overexpression of an N-terminal UEV fragment of Tsg101 (N-Tsg101) completely inhibited the release of HTLV-1. This indicates an important role of Tsg101 in the budding and release of HTLV-1 particles. This is consistent with the role attributed to Tsg101 in the budding of retroviruses such as HIV-1 (9, 13), as well as other viruses (33, 60). Tsg101 also binds PTAP motif in VP40 Ebola virus (33, 60); both proteins colocalize at the plasma membrane (35), where Tsg101 directed the budding of Ebola virus particles to occur in membrane raft microdomains (33). Tsg101 is a protein known to function in vacuolar protein sorting machinery (2) by binding to monoubiquitinated proteins, thus driving their entry into the multivesicular body sorting machinery (24). Tsg101 (and its yeast counterpart Vps23) is believed to execute this function as part of a multiprotein complex (3) that is also called ESCRT-I (2). Interestingly though, Tsg101 and Vps23 are not interchangeable in rescuing yeast mutants incapable of internalizing plasma membrane defective proteins (4; F. Bouamr and C. Carter, unpublished data). Recently, a component of ESCRT-I complex, Vps28, was shown to be involved in HIV-1 budding through binding to the C-terminal region of Tsg101 (36), suggesting that HIV-1 may be using the mammalian ESCRT-I complex to efficiently achieve budding. This may also be the case for HTLV-1, since it requires Tsg101 to release particles.
The TEM data presented here showed viral particles budding inside cytoplasmic vacuoles, suggesting that N-Tsg101 often blocked HTLV-1 Gag budding at an earlier stage and before Gag reached the plasma membrane. This suggests that, in the case of HTLV-1, the block of budding may take place earlier in the process than with HIV-1, which seems to be blocked at a later stage in budding at the plasma membrane (9, 13). Thus, in order to bud efficiently, HTLV-1 requires functions of Tsg101 that naturally involve the protein in both the cell membrane trafficking and fission.
Nedd4 ubiquitin ligase involvement in HTLV-1 release.
We determined that the WW domain in human Nedd4 ubiquitin ligase physically binds the PPPY motif in the MA protein of HTLV-1 Gag. Overexpression of either of two dominant-negative fragments of Nedd4 comprising WW and Hect domains caused inhibition of HTLV-1. Furthermore, the WW domain caused an early arrest of HTLV-1 budding before the typical half-shell curvature was formed. Early budding structures were also observed in the cytoplasm before they reached the plasma membrane, suggesting that Gag was either targeted to internal membrane or retained at these locations and prematurely initiated assembly. These observations suggest that Nedd4 ubiquitin ligase is involved in Gag trafficking and plasma membrane curvature formation during budding and release of HTLV-1.
Nedd4 is an active E3 ubiquitin ligase that transfers ubiquitin molecule(s) to a substrate as a consequence of a sequential cascade of events initiated by an activating E1 enzyme and a conjugating E2 enzyme. The WW domains in Nedd4 bind PPPY motif in HTLV-1, as has been reported for RSV and MPMV retroviruses (26, 48, 66) and Ebola virus (17, 18, 33, 60). Consequently, a subpopulation of retroviral Gag (∼2%) and VP40 is believed to become monoubiquitinated (17, 18, 47, 57, 58). This step is crucial in late stages of retroviral budding, since preventing Nedd4 ubiquitin ligases from functioning by using WW dominant-negative fragments (26, 46, 65) or by using proteasome inhibitors that reduce the amount of free ubiquitin in the cytoplasm (43, 47, 55) and monoubiquitination (43) inhibits viral budding and release. This defect can be partially corrected in the case of RSV by fusing ubiquitin to Gag polyprotein carrying a functional L-domain motif (47). This implies that the recruitment of members of the ubiquitin E3 ligase family, which are highly conserved in mammals and yeast and thus could possibly be functionally interchangeable, is a crucial step in the late stages of retroviral budding. Similarly, HTLV-1 seems to recruit Nedd4 ubiquitin ligase through its PPPY motif to achieve efficient exocytosis from the cell. Similarly, the WW domain of Yap protein caused a block in RSV budding at the plasma membrane (48). The Hect domain of Nedd4 also exerted a strong inhibition on HTLV-1 release, although no physical interaction between MA and the Hect domain was detected. This suggests that the binding of dominant-negative fragments of Nedd4 to Gag is not required to cause inhibition of HTLV-1 release. It is likely that WW and Hect domains affect HTLV-1 release by competing with full-length Nedd4 for essential partners.
Our TEM results showed that HTLV-1 Gag accumulated at inappropriate regions of the cell, implying that Nedd4 activity could be involved in proper trafficking of Gag to the site of budding. Mutations in the yeast orthologue Rsp5 that disrupted its function caused mislocalization of the protein in yeast (62). It is possible that, in mammalian cells, Nedd4 cellular localization, perhaps as a part of a complex, is important for HTLV-1 Gag proper localization to the site of budding. The TEM data also showed that the arrested HTLV-1 structures had less curvature than necessary to form a viral particle. This result shows a new role for Nedd4 and involves the ubiquitin E3 ligase in the early step of membrane bending that is required for retroviral budding. Perhaps this reflects Nedd4's normal role during endocytosis in inducing membrane invagination (53). However, the role of Nedd4 in this process remains unclear, and Nedd4 may carry out multiple discrete functions. The N-terminal domain of Nedd4, C2, which binds to plasma membranes, causing proteins to direct their redistribution to internal membranes (19), could be playing an as-yet-unappreciated function of the protein that is pertinent to its role in membrane curvature. In addition, Nedd4 localizes to the highly fluid lipid raft domains (27), which are believed to favor assembly and release of HIV-1 (39). Thus, it is possible that in early stages of budding, HTLV-1 requires multiple functions of the ubiquitin E3 ligase Nedd4.
Free ubiquitin itself is found in retroviral particles (40, 41, 51) although the significance of its encapsidation in retroviral particle is not understood. Furthermore, although a small proportion of retroviral Gags is ubiquitinated (∼2%), mutations of lysines in the HIV-1 p6 Gag domain did not prevent budding of the virus (40). This suggests that Gag is not the direct substrate or the only substrate of ubiquitin ligase, even though Gag directly binds an active E3 ligase of Nedd4 family (26, 66; the present study). In contrast, mutations of all lysines—specifically, the lysines in positions 47, 48, and 51 in the L domain of HTLV-1 MA—did affect both processing and release of infectious particles, suggesting that transfer of ubiquitin to the protein might be required (29). The direct ubiquitination of HTLV-1 MA is yet to be experimentally demonstrated. In vitro binding between HTLV-1 MA and the N-Tsg101 or WW domain did not require ubiquitination of the protein. However, a role of ubiquitination in facilitating this interaction in vivo is not excluded because the fusion of ubiquitin to HIV-1 p6 improved its binding to Tsg101 (13). Interestingly, it was suggested that a ubiquitin-like motif found in EIAV p9 protein rendered the virus insensitive to proteosome inhibitors (42, 46). In fact, the use of proteasome inhibitors, which cause reduction of free ubiquitin and monoubiquitination, interfered with budding of numerous retroviruses (43, 55). It was hypothesized that the inhibition of retroviral budding is due to accumulation of polyubiquitinated proteins, possibly polyubiquitinated defective Gag proteins normally destined to degradation (54). These findings suggest that the physical presence of ubiquitin by itself or its conjugation to a yet-unknown protein physically near to assembling Gag could play a key role in budding.
In summary, HTLV-1 recruits both the ubiquitin E2-like enzyme Tsg101 and the active ubiquitin E3 enzyme Nedd4 via its PTAP and PPPY L-domain motifs and requires the function of the proteasome to efficiently exit from the cell. Tsg101 is believed to bind ubiquitin, and Nedd4 transfers ubiquitin to many targets. These observations suggest that ubiquitin machinery members play a pivotal role in the budding and release of HTLV-1. The distinctive requirements of HTLV-1 as a model should help define the role of the different cellular partners involved in the complex process of retroviral budding.
Acknowledgments
We thank D. Derse for providing the Tax, Rex, and Px plasmids; A. Elkharroubi for pSV plasmid and for helpful suggestions; L. Li and S. Cohen for providing Tsg101-expressing plasmid; S. Oroszlan for providing the anti-HTLV-1 MA antibodies; the Kazuza Institute for providing Nedd4 plasmid; and C. Carter for support and helpful discussion.
This work was supported in part by NCI contract N01-CO-12400. F.B. is an associate and S.P.G. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, S., and P. A. Lazo. 2003. Human TSG101 does not replace Saccharomyces cerevisiae VPS23 role in the quality control of plasma membrane proteins. FEMS Microbiol. Lett. 221:151-154. [DOI] [PubMed] [Google Scholar]
- 5.Bork, P., and M. Sudol. 1994. The WW domain: a signalling site in dystrophin? Trends Biochem. Sci. 19:531-533. [DOI] [PubMed] [Google Scholar]
- 6.Bouamr, F., L. Garnier, F. Rayne, A. Verna, N. Rebeyrotte, M. Cerutti, and R. Z. Mamoun. 2000. Differential budding efficiencies of human T-cell leukemia virus type I (HTLV-1) Gag and Gag-Pro polyproteins from insect and mammalian cells. Virology 278:597-609. [DOI] [PubMed] [Google Scholar]
- 7.Cornilescu, C. C., F. Bouamr, X. Yao, C. Carter, and N. Tjandra. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306:783-797. [DOI] [PubMed] [Google Scholar]
- 8.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, R., and L. Hicke. 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanes, S. D., and R. Brent. 1989. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell 57:1275-1283. [DOI] [PubMed] [Google Scholar]
- 17.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey, K. F., and S. Kumar. 1999. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9:166-169. [DOI] [PubMed] [Google Scholar]
- 20.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 21.Hicke, L. 1997. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 11:1215-1226. [DOI] [PubMed] [Google Scholar]
- 22.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 23.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 26.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafont, F., and K. Simons. 2001. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE-triggered cell signaling. Proc. Natl. Acad. Sci. USA 98:3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Blanc, I., A. R. Rosenberg, and M. C. Dokhelar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luban, J., K. B. Alin, K. L. Bossolt, T. Humaran, and S. P. Goff. 1992. Genetic assay for multimerization of retroviral Gag polyproteins. J. Virol. 66:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase e2, interacts specifically with human immunodeficiency virus type 2 Gag polyprotein and results in increased levels of ubiquitinated Gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicot, C., T. Astier-Gin, E. Edouard, E. Legrand, D. Moynet, A. Vital, D. Londos-Gagliardi, J. P. Moreau, and B. Guillemain. 1993. Establishment of HTLV-1-infected cell lines from French, Guianese, and West Indian patients and isolation of a proviral clone producing viral particles. Virus Res. 30:317-334. [DOI] [PubMed] [Google Scholar]
- 39.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 41.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, E., and C. S. Goldsmith. 1988. Ultrastructure of human retroviruses. J. Electron Microsc. Tech. 8:3-15. [DOI] [PubMed] [Google Scholar]
- 45.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patnaik, A., and J. W. Wills. 2002. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76:2789-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 52.Rayne, F., F. Bouamr, J. Lalanne, and R. Z. Mamoun. 2001. The NH2-terminal domain of the human T-cell leukemia virus type 1 capsid protein is involved in particle formation. J. Virol. 75:5277-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 54.Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 55.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shih, S. C., K. E. Sloper-Mould, and L. Hicke. 2000. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 19:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taffarel, M., and M. J. Andrada-Serpa. 1998. Electron microscopic observation of a Brazilian HTLV-1 isolated. J. Submicrosc. Cytol. Pathol. 30:393-397. [PubMed] [Google Scholar]
- 60.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 61.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, G., J. M. McCaffery, B. Wendland, S. Dupre, R. Haguenauer-Tsapis, and J. M. Huibregtse. 2001. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell. Biol. 21:3564-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]